Abstract
Chromosomal translocation involving the BCL6 gene affects not only immunoglobulin (Ig) genes but also a number of non-Ig genes as partners. The molecular anatomy of the BCL6 gene rearrangements in 39 cases with diffuse large B-cell lymphoma (DLBCL) by long-distance polymerase chain reaction–based assays was determined. The results showed that Iggenes were affected in 21 cases; non-Ig genes, 15 cases; a deletion of more than a 1-kb segment, 2 cases; and a point mutation, 1 case. Comparative studies between the 21 cases withIg gene partners and the 17 cases with non-Iggene partners, including 2 cases with the deletion, showed that the overall survival of the latter group of patients was significantly inferior to that of the former (P = .0440), and the estimated 2-year overall survival rates were 58.3% vs 17.6% (P = .005). Non-Ig/BCL6 fusion is a poor prognostic indicator of DLBCL, and DLBCL with BCL6translocation could be subclassified according to the individual partner locus and/or gene.
Introduction
Diffuse large B-cell lymphoma (DLBCL) constitutes a heterogeneous spectrum of non-Hodgkin's lymphoma (NHL) with diverse molecular genetic abnormalities.1 The 3 most common genetic lesions associated with chromosomal translocations are rearrangements of cMYC, BCL2, and BCL6proto-oncogenes resulting from t(8;14)(q24;q32) (translocation of chromosome 8, long arm, region 2, band 4, to chromosome 14, long arm, region 3, band 2); t(14;18)(q32;q21); and 3q27 abnormalities, respectively.2 Many studies have focused upon whether these molecular lesions are associated with particular clinical features and whether they can predict the response and outcome for the treatment.2-5 The BCL6 gene was first identified at the breakpoints on 3q27 involved in t(3;14)(q27;q32) and t(3;22)(q27;q11),6-8 and rearrangements of the gene have been observed in up to 35.5% of patients with DLBCL.4,5,9An earlier study showed that the BCL6 rearrangement more frequently occurs in extranodal DLBCL than in node-based disease and is correlated with a favorable clinical outcome,5 although later studies failed to confirm these observations.2,4 In contrast to other B-cell NHL (B-NHL)–associated translocations, theBCL6 translocation is unique in that it can involve not onlyIg genes but also a number of non-Ig loci as partners.9 As the result of the translocation, many types of regulatory sequences on each partner locus substitute for the 5′-untranslated region of the BCL6, and the rearrangedBCL6 is presumed to be under control of the replaced promoter activity.9-11
We have developed the long-distance–polymerase chain reaction (LD-PCR) method to detect Ig oncogene fusion genes.12 On the other hand, the long-distance inverse–PCR (LDI-PCR) method allowed us to readily clone non-Ig partner loci.13 This study was to determine whether the diverse partners of BCL6translocations, which have been identified by these new PCR techniques, are correlated with pretreatment clinical features and treatment outcome of patients with DLBCL.
Study design
Patient characteristics
The characteristics of 203 patients with DLBCL were described in an earlier report.14 Initial pathological diagnosis was made by local pathologists, including those of our institution, and relevant cases were reviewed (H.Y.). Subclassification of DLBCL was not performed. The lymphoma tissues were subjected to immunohistochemical analysis, surface immunophenotyping, and gene rearrangement analysis using a probe for the J region of the heavy chain gene to determine the B-cell origin of lymphoma cells. Of 199 patients, 178 patients in whom treatment details were available were treated with combination chemotherapy including doxorubicin or other anthracyclines.
Molecular analyses
Rearrangement of the BCL6 gene was determined by Southern blot analysis using a probe representing the major translocation cluster (MTC) region.4 LD- and LDI-PCR for detecting Ig/BCL6 and non-Ig/BCL6 fusions were described in our previous papers.12 13 The PCR products were cloned into the pCR2.1 plasmid (Invitrogen, San Diego, CA), and isolation of DNA was performed according to the established methods. Nucleotide sequencing was performed with a BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA), and the sequencing reactions were resolved on an ABI 310 automated sequencer (PE Applied Biosystems). The GenBank sequence database at the National Center of Biotechnology Information was accessed on the Internet. Physical mapping of the isolated clones was performed with the GeneBridge 4 Radiation Hybrid Panel, RH02 (Research Genetics, Huntsville, AL), and the data vectors were submitted to the WICGR Mapping Service (http://www.genome.wi.mit.edu).
Statistical analyses
Correlations between the BCL6 rearrangement and prognostic variables were analyzed with the Fisher exact test. Overall survival was calculated from the date of diagnosis until the patient's death or last follow-up. Survival curves were plotted by the Kaplan-Meier method, and the curves were compared by the log-rank test. The data were analyzed with the Statview statistical software package (Abacus Concepts, Berkeley, CA).
Results and discussion
Diverse partners of the BCL6 translocation
Genomic DNA obtained from involved tissues of the 203 patients with DLBCL was first subjected to Southern blot analysis using a probe for MTC. The results showed that 43 cases (21.2%) had a rearrangement of the BCL6 gene. We next performed LD-PCR and LDI-PCR on 39 patients to determine the partner locus of each rearrangement. Restriction mapping and nucleotide sequencing of the PCR products revealed that the Ig genes were affected as partners in 21 cases; the heavy chain gene, 16 cases; and the λ-light chain gene, 5 cases. In contrast, non-Ig partners affected 15 cases; a deletion of more than a 1-kb segment encompassing the noncoding exon 1, 2 cases; and a point mutation leading to the generation of a restriction site, 1 case. The non-Ig partner loci, where previously identified genes are localized in the vicinity of the breakpoints, included the H4 (histone) gene11 in 2 cases; the transferrin receptor gene15 in 2 cases; and the heat-shock protein 89α gene,PIM1 proto-oncogene, and α-NAC transcriptional coactivator gene16 all in a single case.
Restriction and sequencing analysis of non-Ig partners, whose sequences were not registered in the GenBank sequence database, revealed that an identical locus was involved in 3 cases. PCR screening of the radiation hybrid panel assigned this recurrent locus to band 3q26-27, which was 3-4 cR centromeric to that of BCL6. It remains to be determined whether the BCL6 rearrangement involving this particular locus is the result of translocation between the chromosome 3 pair or inversion occurring within an area includingBCL6. The other chromosomal regions determined by the radiation hybrid panel were 1p32-35 (chromosome 1, short arm), 7p11.2-12, 7p21-22, 14q11-12, and 16p13.3.
Comparison of DLBCL associated either with Ig or non-Ig partners
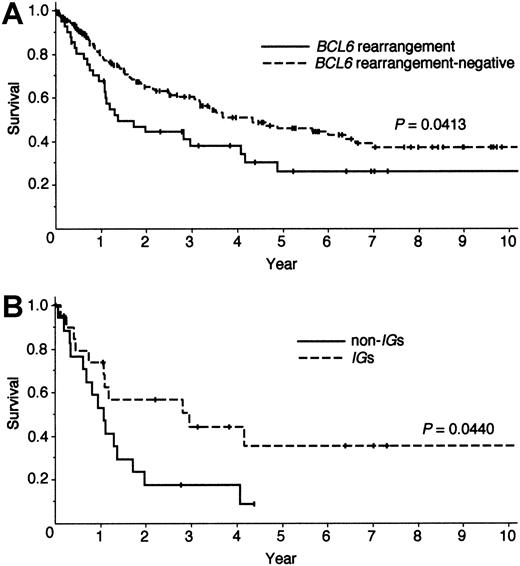
Of the 39 patients with the BCL6 rearrangement, treatment details were available for 37 patients. Although these 37 patients received a regimen containing doxorubicin as the initial chemotherapy, 7 patients first received surgery. Seven patients were further treated by consolidating radiation therapy. As shown in Figure 1A, the patients with theBCL6 rearrangement showed poor clinical outcome compared with the remaining DLBCL patients (P = .0413), thereby contradicting previously published results.5
Survival curves of patients with DLBCL.
(A) Overall survival of patients with DLBCL separated by the presence (n = 43) or absence (n = 160) of the BCL6 rearrangement as determined by Southern blot analysis. (B) Overall survival of patients with the BCL6 rearrangement separated by theIg (n = 21) or non-Ig. (n = 17) partners. Two patients with a deletion of more than a 1-kb segment within theBCL6 were included in the latter group.
Survival curves of patients with DLBCL.
(A) Overall survival of patients with DLBCL separated by the presence (n = 43) or absence (n = 160) of the BCL6 rearrangement as determined by Southern blot analysis. (B) Overall survival of patients with the BCL6 rearrangement separated by theIg (n = 21) or non-Ig. (n = 17) partners. Two patients with a deletion of more than a 1-kb segment within theBCL6 were included in the latter group.
We compared pretreatment clinical features between the 21 patients associated with Ig partners and the 17 patients with non-Ig partners including 2 patients with a deletion. There was no significant difference between the 2 groups in widely used prognostic variables including those of the international prognostic index, “B” symptoms, bone marrow involvement, and bulky mass. Additional molecular lesions, which sometimes coexist with theBCL6 rearrangement,4 9 were equally observed in both groups.
To determine whether the partner locus influenced prognosis, the survival of the patients with the BCL6 rearrangement was stratified according to Ig or non-Ig partners (Figure 1B). The result showed that the overall survival of the non-Ig group was significantly inferior to that of theIg group (P = .0440), and the estimated 2-year overall survival of the 2 groups was 17.6% vs. 58.3% (P = .005). Although there was no difference in the response rates to initial treatments in the Ig group versus the non-Ig group (75.0% vs 76.5%,P > .9999), 14 patients of the non-Ig group died within 2 years, whereas 5 patients of the Ig group are currently enjoying long disease-free survival. Of interest, the survival curve of the Ig partner group overlapped that of patients lacking the BCL6 rearrangement.
Diverse partners of BCL6 translocation could affect clinical behavior of DLBCL
We recently showed that the non-Ig/BCL6 translocation results in replacement of its 5′-noncoding region with heterogeneous promoters, which are activated by a variety of stimuli including cell cycle control, changes in the physical environment, and the response to cytokines.13 Therefore, it is possible that non-Ig partners have influence on the pattern ofBCL6 deregulation in a different fashion from Igs and thereby affect the clinical behavior of DLBCL patients carrying the corresponding BCL6 translocation. Table1 summarizes 3 DLBCL patients with theBCL6 translocation involving the 3q26-27 locus. It is apparent that these patients had homogeneous clinical features, ie, they presented with an advanced disease, a high lactate dehydrogenase level, and a resistance to chemo-radiotherapy. This observation suggested that DLBCL with the BCL6 translocation could be subclassified according to the individual partner locus and/or gene, although the responsible gene of the 3q26-27 locus is currently unknown. Our study suggests that the non-Ig/BCL6fusion is a poor prognostic indicator of DLBCL, but additional studies of larger cohorts of uniformly treated patients, performed in a prospective fashion, will be required to confirm our findings.
Clinical features of 3 DLBCL cases with BCL6 translocation affecting the 3q26-27 locus
| Case no. . | Age (y)/ sex . | Stage/PS . | Involved organs at presentation . | LDH (IU) . | Treatment . | Outcome . | Survival (d) . |
|---|---|---|---|---|---|---|---|
| 90 | 70/M | III/1 | Lymph nodes | 735 | CHOP, radiation | CR-R-PD | 630 |
| 880 | 59/M | IV/1 | Lymph nodes, spleen, bone marrow, liver | 1950 | LSG4* | PD | 30 |
| 910 | 35/F | IVE/3 | Lymph nodes, breasts, ovary, uterus | 737 | Mastectomy, CHOP, high-dose chemotherapy | PR-R-PD | 501 |
| Case no. . | Age (y)/ sex . | Stage/PS . | Involved organs at presentation . | LDH (IU) . | Treatment . | Outcome . | Survival (d) . |
|---|---|---|---|---|---|---|---|
| 90 | 70/M | III/1 | Lymph nodes | 735 | CHOP, radiation | CR-R-PD | 630 |
| 880 | 59/M | IV/1 | Lymph nodes, spleen, bone marrow, liver | 1950 | LSG4* | PD | 30 |
| 910 | 35/F | IVE/3 | Lymph nodes, breasts, ovary, uterus | 737 | Mastectomy, CHOP, high-dose chemotherapy | PR-R-PD | 501 |
PS indicates performance status; LDH, lactic dehydrogenase; M, male; F, female. CHOP indicates cyclophosphamide, doxorubicin, vincristine, and prednisone. CR indicates complete remission; PR, partial remission; R, relapse; and PD, progressive disease. Details of the clinical features of case no. 910 were described previously.17
LSG4 is the multidrug regimen proposed by the Japan Lymphoma Study Group.
Supported by grants-in-aid 7-29 and 9-10 from the Ministry of Health and Welfare, Japan, and grant 11670994 from the Ministry of Education, Science, Sports, and Culture of Japan, Tokyo, Japan. T.A. is a fellow in Cancer Research of the Japan Society for the Promotion of Science, Tokyo, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hitoshi Ohno, First Division, Department of Internal Medicine, Faculty of Medicine, Kyoto University, 54-Shogoin-Kawaramachi, Sakyo-ku, Kyoto 606-8507, Japan; e-mail:hohno@kuhp.kyoto-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal