Abstract
Genetic alteration is considered a probable cause of malignant lymphoma. Folate and methionine metabolism play essential roles in DNA synthesis and DNA methylation, and their metabolic pathways might thus affect disease susceptibility. In the present study, 2 polymorphisms were evaluated for a folate metabolic enzyme, methylenetetrahydrofolate reductase (MTHFR), and one was evaluated for methionine synthase (MS). The 2 polymorphisms, MTHFR677 C→T and MTHFR1298 A→C, are reported to reduce the enzyme activity, which causes intracellular accumulation of 5,10-methylenetetrahydrofolate and results in a reduced incidence of DNA double-strand breakage. The MS2756 A→G polymorphism also reduces the enzyme activity and results in the hypomethylation of DNA. To evaluate the association between malignant lymphoma susceptibility and these polymorphisms, hospital-based case-control study was conducted in Aichi Cancer Center. Ninety-eight patients with histologically confirmed lymphoma and 243 control subjects without cancer were evaluated. Unconditional logistic regression analyses revealed a higher susceptibility with the MTHFR677 CC and the MTHFR1298 AA genotypes (odds ratio, 2.26; 95% confidence interval, 1.26-4.02) when those harboring at least one variant allele in either polymorphism of MTHFR were defined as the reference. For the MS polymorphism, the MS2756 GG genotype also showed a higher susceptibility (odds ratio, 3.83; 95% CI, 1.21-12.1) than those with MS2756 AA or AG types. The significance was not altered when these 3 polymorphisms were evaluated in combination, and the results suggest that folate and methionine metabolism play important roles in the occurrence of malignant lymphomas. Further studies to confirm the association and detailed biologic mechanisms are now required.
Introduction
Biologic mechanisms underlying the genesis of lymphoid malignancies remain to be clarified in detail. However, accumulated evidence suggests that certain genetic events during cell differentiation, such as chromosomal translocation,1,2play an important role. Methylation status of various oncogenes or tumor suppressor genes may induce the selective growth transformation of cells or its inhibition.3 Regardless, a single genetic event is insufficient to explain carcinogenesis of lymphoid tissue, as supported by the transgenic mouse experiment.4
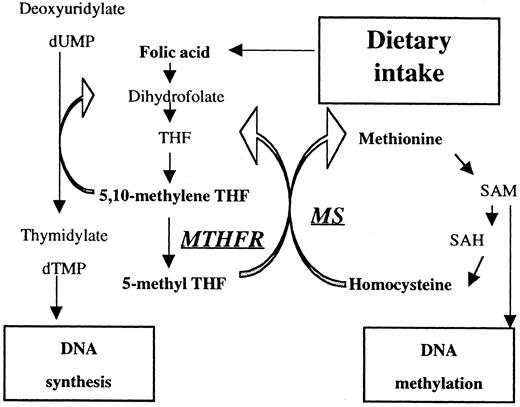
Folic acid is an important nutrient required for DNA synthesis, and the related methionine metabolic pathway is necessary for DNA methylation (Figure 1). An antifolic acid agent, methotrexate, has proven to be an effective chemotherapeutic drug for lymphoid malignancies, giving insight into the association between folate metabolism and the carcinogenesis of these malignancies.
Overview of folate- and methionine-metabolizing pathways.
MTHFR catalyzes the reduction of 5,10-methylene THF to 5-methyl THF. Reduced activity of MTHFR results in the accumulation of 5,10-methylene THF, which accelerates methylation of dUMP to dTMP. MS catalyzes the transfer of the methyl base from 5-methyl THF to homocysteine. Reduced activity of MS leads to hypomethylation of DNA.
Overview of folate- and methionine-metabolizing pathways.
MTHFR catalyzes the reduction of 5,10-methylene THF to 5-methyl THF. Reduced activity of MTHFR results in the accumulation of 5,10-methylene THF, which accelerates methylation of dUMP to dTMP. MS catalyzes the transfer of the methyl base from 5-methyl THF to homocysteine. Reduced activity of MS leads to hypomethylation of DNA.
Methylenetetrahydrofolate reductase (MTHFR) catalyzes the reduction of 5,10-methylenetetrahydrofolate (methylene THF) to 5-methyltetrahydrofolate (5-methyl THF),5 the predominant circulatory form of folate and carbon donor for the remethylation of homocysteine to methionine (Figure 1). The MTHFR gene, which is located on chromosome 1p36,6 is reported to have 2 polymorphisms involving nucleotides 677 (C→T; alanine→valine) and 1298 (A→C; glutamate→alanine). Both polymorphisms lead to reduced MTHFR activity7,8 and result in the accumulation of 5,10-methylene THF. This in turn reduces the chance for the misincorporation of uracil into DNA, a cause of double-strand breaks during uracil excision repair.9,10 Neural tube defect,8,11 coronary artery disease,7,12,13cerebrovascular disease,14,15 venous thrombotic disease,16,17 colorectal cancer,18,19and endometrial cancer20 are reported to be associated with these polymorphisms. Skibola et al21 reported a link between acute lymphoblastic leukemia and 2 polymorphisms ofMTHFR and concluded there is lower susceptibility with either mutant.
Methionine synthase (MS) catalyzes the transfer of methyl base from 5-methyl THF to homocysteine (Figure 1). Its gene is located on 1q43,22 producing methionine and tetrahydrofolate. It is reported to have a polymorphism in 2756 A to G (glycine→aspartic acid), resulting in a lower enzyme activity,22 and it is thought to result in homocysteine elevation and DNA hypomethylation. Moderate associations of the polymorphism with methionine level23 and colorectal cancer24 have been reported. In the present case-control study, we examined the association of the folate metabolic polymorphisms (MTHFR677, MTHFR1298) and the methylation-related polymorphism (MS2756) with the development of malignant lymphoma.
Patients, materials, and methods
Study population and sample collection
Recruitment was made from among patients at Aichi Cancer Center who received histologic confirmation of malignant lymphoma between October 1986 and February 2000. Those with a history of other types of malignancy were excluded. Control subjects were outpatients without any history of cancer who underwent physical examination and gastroscopy at Aichi Cancer Center from March to December 1999. All were disease free except several patients with digestive ulcers. Chronic diseases, such as cardiovascular and cerebrovascular diseases, and diabetes are not treated at our hospital. All patients and control subjects were Japanese. Subjects who provided written informed consent for participation in this study were asked to complete a self-administered questionnaire and to provide blood from a peripheral vein. This study was approved by the Institutional Review Board of Aichi Cancer Center.
Genotype analyses of the MTHFR677, MTHFR1298, and MS2756
DNA of each subject was extracted from the buffy coat fraction with a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA).
Before genotyping of patients and controls, cancer cell lines (HL60, NOMO-1, CMK, K562, Jurkat, SUDHL1, Hut102, NALL-1, SUDHL6, and Daudi) were examined to confirm patterns of polymorphism for possible use as controls. Three cell lines were selected—K562 for homozygous wild type (CC) of MTHFR677 and heterozygous type (AG) of MS2756, HL60 for homozygous mutant type (TT) of MTHFR677 and homozygous wild type (AA) of MTHFR1298, and Jurkat for homozygous mutant type (CC) of MTHFR1298 and homozygous wild type (AA) of MS 2756. No homozygous mutant was observed for MS2756.
Genotyping was performed according to previously described methods for MTHFR6777 and MTHFR129821 of theMTHFR gene and MS275623 25 of the MSgene polymorphisms in detail. For the 677 C→T polymorphism, extracted DNA was amplified with the forward primer 5′-TGA AGG AGA AGG TGT CTG CGG GA-3′ and the reverse primer 5′-AGG ACG GTG CGG TGA GAG TG-3′. Polymerase chain reaction (PCR) thermal cycling conditions were 2-minute denaturation at 94°C, then 40 cycles of 94°C for 30 seconds, 62°C for 30 seconds, and 72°C for 30 seconds. This was followed by 7-minute extension at 72°C. Amplified 198-bp PCR products were digested with HinfI (Boehringer Mannheim, Mannheim, Germany) and were visualized under electrophoresis on 4% agarose gel with ethidium bromide. The C allele produced a 198-bp band, and the T allele produced 175- and 23-bp fragments. For the MTHFR1298 A→C polymorphism, DNA was amplified with the forward primer 5′-CTT TGG GGA GCT GAA GGA CTA CTA C-3′ and the reverse primer 5′-CAC TTT GTG ACC ATT CCG GTT TG-3′. PCR conditions were 2-minute denaturation at 92°C followed by 35 cycles of 92°C for 1 minute, 60°C for 1 minute, and 72°C for 30 seconds, with 7-minute extension at 72°C. With 4% agarose gel visualization, MboII (New England Biolab, Beverly, MA) digestion produced 56-, 31-, 30-, 28-, and 18-bp bands for the A allele and 84-, 31-, 30-, and 18-bp bands for the C allele. For the MS 2756 A→G polymorphism, DNA was amplified with the forward primer 5′-TGT TCC AGA CAG TTA GAT GAA AAT C-3′ and the reverse primer 5′-GAT CCA AAG CCT TTT ACA CTC CTC-3′. PCR thermal cycling conditions were 2-minute denaturation at 95°C followed by 35 cycles at 95°C for 1 minute, 60°C for 1.5 minutes, and 72°C for 1 minute, with a 7-minute extension at 72°C. PCR products were digested withHaeIII (Boehringer Mannheim), resulting in a 211-bp band for the A allele and 131- and 80-bp fragments for the G allele, after 4% agarose gel electrophoresis. Heterozygotes produced bands for each allele.
Statistical analysis
All statistical analyses in this study were performed using STATA (College Station, TX) statistical software. Accordance with the Hardy-Weinberg equilibrium, which indicates an absence of discrepancy between genotype and allele frequency, was checked for control subjects using χ2 test. Odds ratios (OR) and 95% confidence intervals (95% CI) were adjusted for sex and age as 3 binary variables for 4 age categories (younger than 45, 45 to 54, 55 to 64, and 65 or older) using an unconditional logistic regression model. Adjustment for multiple comparison was not performed because the analyses were conducted in an exploratory context, which requires careful interpretation of any P values.
Results
Study population
Ninety-eight patients (age range, 20-83 years; mean age, 54.5 years; male, 56.7%) and 243 control subjects (age range, 39-69 years; mean age, 56.8 years; male, 49.0%) were recruited. Histologic types were diffuse large B-cell lymphoma (n = 33), follicular lymphoma (n = 25), MALT lymphoma (n = 12), peripheral T-cell lymphoma (n = 6), Hodgkin disease (n = 5), cutaneous T-cell lymphoma (n = 4), mantle cell lymphoma (n = 3), nodal marginal zone lymphoma (n = 2), lymphoblastic lymphoma (n = 2), angioimmunoblastic T-cell lymphoma (n = 1), anaplastic large cell lymphoma (n = 1), low-grade lymphoma, and not otherwise specified (n = 4).
Genotyping for MTHFR677, MTHFR1298, and MS2756
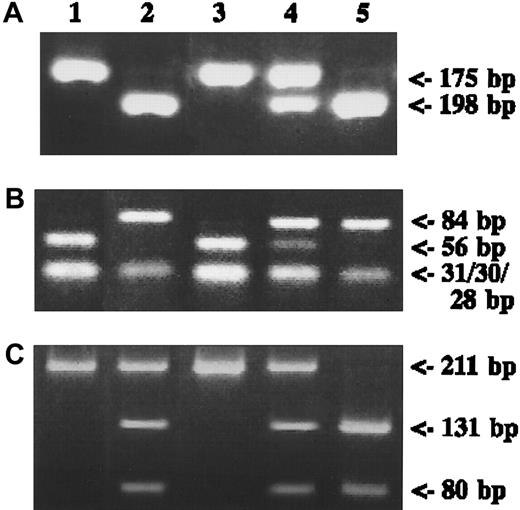
Representative results are shown in Figure2. Among the 98 adult patients analyzed, the frequency of the MTHFR677 mutated-allele was 34.2% for patients and 40.7% for controls. Frequencies of the MTHFR677CC, MTHFR677CT, and MTHFR677TT genotypes were 45.9%, 39.8%, and 14.3%, respectively, for patients and 33.3%, 51.9%, and 14.8% for controls (χ2test, P = .078). For MTHFR1298, the mutated-allele frequency was 18.6% for patients and 19.1% for controls. Frequencies of MTHFR1298AA, MTHFR1298AC, and MTHFR1298CC genotypes were 63.3%, 34.7%, and 1.0%, respectively, for patients and 65.4%, 30.8%, and 3.7% for controls (χ2 test, P = .352). For MS2756, the mutated-allele frequency was 20.1% for patients and 19.1% for controls. Frequencies of the MS2756AA, MS2756AG, and MS2756GG genotypes were 64.3%, 26.5%, and 7.2%, respectively, for patients and 64.2%, 33.3%, and 2.5% for controls (χ2 test,P = .080). The χ2 test for the Hardy-Weinberg equilibrium with each polymorphism was not statistically significant (MTHFR677, P = .29; MTHFR1298,P = 1.00; and MS2756, P = .30).
Polymorphisms of
MTHFR and MS genes. (A) Genomic DNA from each control subject was amplified with MTHFR677 primers digested with HinfI and was run on a 4% agarose gel. (B) Results for MTHFR1298 with MboII digestion. (C) Results for MS2756 with HaeIII digestion. DNA fragments stained with ethidium bromide are shown. Lane 1, positive control for wild type; lane 2, positive control for mutant type, except for heterozygote in panel C; lane 3, wild type; lane 4, heterozygote; lane 5, mutant.
Polymorphisms of
MTHFR and MS genes. (A) Genomic DNA from each control subject was amplified with MTHFR677 primers digested with HinfI and was run on a 4% agarose gel. (B) Results for MTHFR1298 with MboII digestion. (C) Results for MS2756 with HaeIII digestion. DNA fragments stained with ethidium bromide are shown. Lane 1, positive control for wild type; lane 2, positive control for mutant type, except for heterozygote in panel C; lane 3, wild type; lane 4, heterozygote; lane 5, mutant.
Risk estimation for genotypes by the unconditional logistic model
Table 1 shows the frequency of genotypes for patients and controls and the sex- and age-adjusted OR for each polymorphism. When the MTHFR677 CC genotype was defined as the reference, the MTHFR677 CT/TT genotype showed a reduced OR (0.64; 95% CI, 0.39-1.05). When the MTHFR1298 AA genotype was defined as the reference, the adjusted OR for the MTHFR AC/CC genotype was 1.14 (0.69-1.89). When the MS 2756 AA/AG genotype was defined as the reference, the MS2756 GG genotype showed a higher adjusted OR (3.83; 95% CI, 1.12-12.1; P = .023).
Number of patients and controls, sex- and age-adjusted odds ratios and 95% confidence intervals, for methylenetetrahydrofolate reductase 677 (MTHFR677), MTHFR1298, and methionine synthase 2956
| Genotype . | Patients n = 98 . | Controls n = 243 . | OR . | 95% CI . | P value . |
|---|---|---|---|---|---|
| MTHFR 677 | |||||
| CC | 45 (45.9%) | 81 (33.3%) | 1.00 | Reference | |
| CT | 39 (39.8%) | 126 (51.9%) | 0.62 | 0.37-1.05 | .366 |
| TT | 14 (14.3%) | 36 (14.8%) | 0.71 | 0.34-1.49 | .076 |
| CT/TT | 53 (54.1%) | 162 (66.7%) | 0.64 | 0.39-1.05 | .078 |
| MTHFR 1298* | |||||
| AA | 62 (63.3%) | 159 (65.4%) | 1.00 | Reference | |
| AC | 34 (34.7%) | 75 (30.8%) | 1.25 | 0.75-2.10 | .391 |
| CC | 1 (1.0%) | 9 (3.7%) | 0.30 | 0.38-2.45 | .233 |
| AC/CC | 35 (35.7%) | 84 (33.8%) | 1.14 | 0.69-1.89 | .611 |
| MS2756† | |||||
| AA | 63 (64.3%) | 156 (64.2%) | 1.00 | Reference | |
| AG | 26 (26.5%) | 81 (33.3%) | 0.81 | 0.47-1.41 | .461 |
| GG | 7 (7.2%) | 6 (2.5%) | 3.59 | 1.11-11.5 | .032 |
| Genotype . | Patients n = 98 . | Controls n = 243 . | OR . | 95% CI . | P value . |
|---|---|---|---|---|---|
| MTHFR 677 | |||||
| CC | 45 (45.9%) | 81 (33.3%) | 1.00 | Reference | |
| CT | 39 (39.8%) | 126 (51.9%) | 0.62 | 0.37-1.05 | .366 |
| TT | 14 (14.3%) | 36 (14.8%) | 0.71 | 0.34-1.49 | .076 |
| CT/TT | 53 (54.1%) | 162 (66.7%) | 0.64 | 0.39-1.05 | .078 |
| MTHFR 1298* | |||||
| AA | 62 (63.3%) | 159 (65.4%) | 1.00 | Reference | |
| AC | 34 (34.7%) | 75 (30.8%) | 1.25 | 0.75-2.10 | .391 |
| CC | 1 (1.0%) | 9 (3.7%) | 0.30 | 0.38-2.45 | .233 |
| AC/CC | 35 (35.7%) | 84 (33.8%) | 1.14 | 0.69-1.89 | .611 |
| MS2756† | |||||
| AA | 63 (64.3%) | 156 (64.2%) | 1.00 | Reference | |
| AG | 26 (26.5%) | 81 (33.3%) | 0.81 | 0.47-1.41 | .461 |
| GG | 7 (7.2%) | 6 (2.5%) | 3.59 | 1.11-11.5 | .032 |
OR indicates odds ratio; CI, confidence interval; MS, methionine synthase; PCR, polymerase chain reaction.
One patient (1.0%) was excluded from analysis because DNA was not amplified by PCR.
Two patients (2.0%) were excluded from analysis because DNA was not amplified by PCR.
Table 2 shows combined results for the MTHFR677 and MTHFR1298 polymorphisms. Risk estimation showed lower than unity for each combination of alleles, but statistical significance could not be assessed because of the small number of patients. When the MTHFR677CC/1298AA genotype was defined as the reference, the adjusted OR for the other genotypes combined showed markedly reduced risk (OR, 0.45; 95% CI, 0.25-0.80; P = .007). This means that those with full MTHFR enzyme activity had approximately 2 times higher susceptibility than others with at least one mutant allele.
Number of patients and controls, sex- and age-adjusted odd ratios and 95% confidence intervals for methylenetetrahydrofolate reductase 677 (MTHFR677) and MTHFR1298
| MTHFR677 . | MTHFR1298 . | Patients n = 97* . | Controls n = 243 . | OR . | 95% CI . | P value . |
|---|---|---|---|---|---|---|
| CC | AA | 29 (29.9%) | 37 (15.2%) | 1.00 | Reference | |
| CT/TT | AC/CC | 68 (70.1%) | 206 (84.8%) | 0.45 | 0.25-0.80 | .007 |
| CC | AC | 14 (14.4%) | 35 (14.4%) | 0.49 | 0.22-1.12 | |
| CC | CC | 1 (1.0%) | 9 (3.7%) | 0.14 | 0.02-1.17 | |
| CT | AA | 20 (20.6%) | 86 (35.4%) | 0.30 | 0.15-0.61 | |
| CT | AC | 19 (19.6%) | 40 (16.5%) | 0.72 | 0.34-1.52 | |
| CT | CC | 0 | 0 | |||
| TT | AA | 13 (13.4%) | 36 (14.8%) | 0.46 | 0.20-1.05 | |
| TT | AC | 1 (1.0%) | 0 | NE | ||
| TT | CC | 0 | 0 |
| MTHFR677 . | MTHFR1298 . | Patients n = 97* . | Controls n = 243 . | OR . | 95% CI . | P value . |
|---|---|---|---|---|---|---|
| CC | AA | 29 (29.9%) | 37 (15.2%) | 1.00 | Reference | |
| CT/TT | AC/CC | 68 (70.1%) | 206 (84.8%) | 0.45 | 0.25-0.80 | .007 |
| CC | AC | 14 (14.4%) | 35 (14.4%) | 0.49 | 0.22-1.12 | |
| CC | CC | 1 (1.0%) | 9 (3.7%) | 0.14 | 0.02-1.17 | |
| CT | AA | 20 (20.6%) | 86 (35.4%) | 0.30 | 0.15-0.61 | |
| CT | AC | 19 (19.6%) | 40 (16.5%) | 0.72 | 0.34-1.52 | |
| CT | CC | 0 | 0 | |||
| TT | AA | 13 (13.4%) | 36 (14.8%) | 0.46 | 0.20-1.05 | |
| TT | AC | 1 (1.0%) | 0 | NE | ||
| TT | CC | 0 | 0 |
NE, not estimated; for other abbreviations, see Table 1.
One patient was excluded because DNA was not amplified by PCR.
Table 3 shows the results of analysis of the 2 genes, MTHFR and MS, in combination. In this analysis, the patients having MTHFR677 CT/TT with MTHFR1298 AC/CC and MS2756 AA/AG were redefined as the reference group (group A) because they were expected to have the lowest susceptibility. Adjusted OR for the group at risk was 2.51 (95% CI, 1.45-4.37;P = .001), and the ORs for MTHFR677CT/TT with MTHFR1298AC/CC and MS2756 GG (group B) and MTHFR677CC/1298AA with MS2756 AA/AG (group C) were 3.86 (95% CI, 1.16-12.8;P = .028) and 2.32 (95% CI, 1.28-4.18;P = .005, respectively). An OR for MTHFR677CC/1298AA with the MS2756 GG genotype, which might have had the highest susceptibility, could not be estimated because none of the 243 controls had this genotype.
Number of patients and controls, sex- and age-adjusted odd ratios and 95% confidence intervals for combination status of methylenetetrahydrofolate reductase and methionine synthase
| MTHFR 677 and 1298 . | MS2756 . | Patients n = 953-150 . | Controls n = 243 . | OR . | 95% CI . | P value . |
|---|---|---|---|---|---|---|
| CT/TT and AC/CC (Group A) | AA/AG | 61 (63.5%) | 200 (82.3%) | 1.00 | Reference | |
| (Groups B, C) | 34 (34.%) | 43 (17.7%) | 2.51 | 1.45-4.37 | .001 | |
| CT/TT and AC/CC (Group B) | GG | 6 (6.3%) | 6 (2.5%) | 3.86 | 1.16-12.8 | |
| CC and AA (Group C) | AA/AG | 28 (28.6%) | 37 (15.2%) | 2.32 | 1.28-4.18 |
| MTHFR 677 and 1298 . | MS2756 . | Patients n = 953-150 . | Controls n = 243 . | OR . | 95% CI . | P value . |
|---|---|---|---|---|---|---|
| CT/TT and AC/CC (Group A) | AA/AG | 61 (63.5%) | 200 (82.3%) | 1.00 | Reference | |
| (Groups B, C) | 34 (34.%) | 43 (17.7%) | 2.51 | 1.45-4.37 | .001 | |
| CT/TT and AC/CC (Group B) | GG | 6 (6.3%) | 6 (2.5%) | 3.86 | 1.16-12.8 | |
| CC and AA (Group C) | AA/AG | 28 (28.6%) | 37 (15.2%) | 2.32 | 1.28-4.18 |
MTHFR indicates methylenetetrahydrofolate reductase; for other abbreviations, see Table 1.
Patients were genotyped for all three polymorphisms. Three patients were excluded.
We performed subgroup analyses for diffuse large B-cell lymphoma and follicular lymphoma, which comprise 33.7% and 25.5% of patients, respectively. For diffuse large B-cell lymphoma, the MTHFR677 CT/TT and MTHFR1298 AC/CC type showed a reduced risk (OR, 0.29; 95% CI, 0.14-0.64; P = .002) compared with the MTHFR677 CC and MTHFR1298 AA types, whereas the MS2756 GG genotype showed a higher risk (OR, 3.59; 95% CI, 0.81-15.9; P = .093). On the other hand, MTHFR mutant allele carriers showed risk reduction, though it was not statistically significant (OR, 0.52; 95% CI, 0.19-1.43;P = .207), and the MS2756 GG type showed significant risk elevation (OR, 6.43; 95% CI, 1.45-28.5; P = .014) for follicular lymphoma.
Discussion
In the present study, 98 patients and 243 controls were enrolled. The statistical power for this sample was more than 60% for an OR of 2 or 0.5 under a 2-sided significance level of 0.05, when a genotype frequency among the controls was between 30% and 70%. It was more than 95% for an OR of 3 or 0.33 under the same conditions. Subjects with both MTHFR677 and MTHFR1298 wild types (MTHFR677CC with MTHFR1298AA) had approximately 2 times higher susceptibility than those with other types (OR, 2.26; 95% CI, 1.26-4.02). On the other hand, mutant-type subjects for MS2756 (MS2756 GG) had more than 3 times higher susceptibility than those with other types (OR, 3.83; 95% CI, 1.12-12.1). Analyses in combination with MTHFR677, MTHFR1298, and MS2756 also showed a positive association, revealing the lowest susceptibility for MTHFR677CT/TT and MTHFR1298AC/CC with the MS2756AA/AG genotype. Subgroup analyses showed a similar trend in overall analyses in diffuse large B-cell lymphoma and follicular lymphoma, though the associations with specific histologic subtype remain to be clarified.
Sampling of control subjects is very important in case-control studies. In this study, outpatients without any malignancies (most were free from any kind of disease) were adopted as controls, and the genotype frequencies of the 3 polymorphisms were in accordance with the Hardy-Weinberg laws of equilibrium, indicating that no selective mechanisms for a specific genotype of these polymorphisms existed among the controls. For MTHFR677, the frequency of the T-containing allele was slightly lower in this study than in a previous study in Japan12 but was similar to that for other populations.24,26 For MTHFR1298, there has been no report on frequency in the Japanese population, but results observed were relatively low compared to results of other investigations.8,21,27 It is likely that discrepancies in allele frequency result from ethnic or regional differences. For the MS2756 polymorphism, the allele frequency observed in this study was similar to that in previous studies.22-25,28 29
Because these enzymes were active in related metabolic pathways, their polymorphisms were evaluated in a combined setting. Results of combined analysis of MTHFR677 and MTHFR1298 illustrated a clear association between folate metabolism and susceptibility to lymphoid malignancies. Folate is an essential nutrient for DNA synthesis, and mutations of MTHFR677 and MTHFR1298 reduce enzyme activity. The resultant inhibition of the methyl THF pathway leads to increased levels of methylene THF, and this elevation accelerates the methylation of uridylate to thymidylate. Uracil is normally only an RNA base, but it is incorrectly incorporated into DNA if the methylation of uridylate to thymidylate is insufficient during DNA synthesis.9 Misincorporated uracil is excised by uracil DNA glycosylase, and this can generate transient single-strand breaks.10 Higher rates of occurrence of misincorporation raise the incidence of 2 closely spaced uracils on opposite strands, and excision repair of these close sites may induce double-strand breakage of DNA. Approximately 1,000 times higher rates of misincorporation of uracil were observed in an experimental model of folate deficiency, and this led to 50 times higher rates of double-strand breakage,9 a possible explanation for genetic instability and occurrence of malignant disease.30
Our results for the MS2756 polymorphism provided the first indication, to our knowledge, that the hypomethylation of DNA is associated with a higher susceptibility to malignant lymphoma. Basically, hypermethylation of specific genes causes lower expression of the coding region. Resultant inactivation of 1 or 2 alleles of a tumor-suppressor gene—introduced during development or aging or by carcinogens or chemotherapeutic drugs—could lead to the formation of cells with increased carcinogenic potential.3Growth-inducing genes such as oncogenes, in contrast, may be overexpressed when they are hypomethylated.3 Studies point to associations between lymphoid malignancy and methylation status of specific genes such as p53,31p15,31,32 and p16.31-33Most of them have shown a higher frequency of hypermethylation in tumor-suppressor genes in patients with lymphoma, but our results raise the possibility that the hypomethylation of growth-inducing genes may also play an important role in lymphomagenesis. The relation between methylation status and polymorphism of MS requires further investigation before a conclusion can be drawn.
Our results for MTHFR polymorphisms are similar to those published for other types of malignacy.18,20,21 Genetic instability caused by altered folate metabolism is in line with epidemiologic evidence of higher cancer susceptibility in populations with higher chromosomal aberrations.34 35
This is the first report of higher susceptibility to malignant lymphoma with the mutant form of MS polymorphism. However, the opposite result—lower susceptibility to colorectal cancer with the mutant allele—has been reported.24 The reason for this discrepancy remains unclear, but the pattern of methylation in cancer-related genes and the resultant cell transformation might be different in each cancer type.3
A gene-dose effect was not observed in this study. Most of the reported associations with diseases or biomarkers for these polymorphisms did not show this effect,11,12,14,16,17,19,20,23,24 26suggesting that disease susceptibility is not determined in a gene-dose manner. The threshold of susceptibility may differ among gene polymorphisms, depending on the biologic mechanism. In line with the grouping adopted by studies, we classified the genotypes used in this study into CC versus CT/TT for MTHFR677, AA versus AC/CC for MTHFR1298, and AA/AG versus GG for MS2756.
For cancer prevention, higher amounts of folic acid could reduce the risk for lymphoid malignancy by averting uracil misincorporation and hypomethylation. Neural tube defects are reported to be associated with MTHFR polymorphisms, and folic acid supplementation reduces the risk for them.36 In a report concerning colorectal cancer, an insufficient plasma folate level is found to negate the protective effect of MTHFR677.18 Greater protection from folate supplementation might be obtained in populations having variant alleles for MTHFR677 or MTHFR1298, but this speculation warrants further investigation. The relation between DNA methylation status and folate supplementation has yet to be evaluated in detail.
In conclusion, the present study provided evidence that MTHFR mutant alleles are associated with lower susceptibility to malignant lymphoma and that the MS mutant allele is associated with higher susceptibility to it. This suggests that folate and methionine metabolism play important roles in their genesis. Further studies to confirm the association of the polymorphisms with malignant lymphoma risk and to investigate the detailed biologic mechanisms are required.
We thank Ms Yohko Kurobe, Ms Keiko Asai, and Ms Hiroko Fujikura for their technical assistance.
Supported in part by a Grant-in-Aid for Scientific Research on Priority Area (C) in 2000-2003 from the Ministry of Education, Science, Sports and Culture.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Keitaro Matsuo, Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, 1-1 Kanokoden Chikusa-ku, Nagoya 464-8681, Japan; e-mail:kmatsuo@aichigw.aichi-cc.pref.aichi.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal