Abstract
Adhesion of mature Plasmodium falciparum parasitized erythrocytes to microvascular endothelial cells or to placenta contributes directly to the virulence and severe pathology of P falciparum malaria. Whereas CD36 is the major endothelial receptor for microvasculature sequestration, infected erythrocytes adhering in the placenta bind chondroitin sulfate A (CSA) but not CD36. Binding to both receptors is mediated by different members of the large and diverse protein family P falciparum erythrocyte membrane protein-1 (PfEMP-1) and involves different regions of the molecule. The PfEMP-1–binding domain for CD36 resides in the cysteine-rich interdomain region 1 (CIDR-1). To explore why CSA-binding parasites do not bind CD36, CIDR-1 domains from CD36- or CSA-binding parasites were expressed in mammalian cells and tested for adhesion. Although CIDR-1 domains from CD36-adherent strains strongly bound CD36, those from CSA-adherent parasites did not. The CIDR-1 domain has also been reported to bind CSA. However, none of the CIDR-1 domains tested bound CSA. Chimeric proteins between CIDR-1 domains that bind or do not bind CD36 and mutagenesis experiments revealed that modifications in the minimal CD36-binding region (M2 region) are responsible for the inability of CSA-selected parasites to bind CD36. One of these modifications, mapped to a 3–amino acid substitution in the M2 region, ablated binding in one variant and largely reduced binding of another. These findings provide a molecular explanation for the inability of placental sequestered parasites to bind CD36 and provide additional insight into critical residues for the CIDR-1/CD36 interaction.
Introduction
Of the 4 species of Plasmodium that naturally infect humans, Plasmodium falciparum is responsible for most disease and nearly all the mortality.1 One important survival mechanism of P falciparum parasites is the sequestration of early-stage gametocyte and asexual-stage parasitized erythrocytes (PEs) from blood circulation by attachment to host endothelium or placenta.2-6 Sequestration allows the parasite to avoid spleen-dependent killing mechanisms and enhances transmission to mosquitoes.5,7 Unfortunately, sequestration contributes directly to P falciparum pathogenesis. Adherence of PEs to brain microvasculature or to placenta is associated with development of cerebral malaria or increased fetal and newborn mortality, respectively.6 8-10
Sequestration is mediated by specific ligand-receptor interactions.2-4,11 A large number of host receptors have been implicated in this phenomenon, including CD36,12intercellular cell adhesion molecule-1 (ICAM-1),13vascular cell adhesion molecule,14 thrombospondin (TSP),15 chondroitin sulfate A (CSA),16,17platelet/endothelial cell adhesion molecule (PECAM),18E-selectin,14 P-selectin,19 and hyaluronic acid.20 Whereas most isolates bind CD36, binding to other receptors is more variable.21-24 Thus, CD36 is considered the major endothelial receptor for sequestration. However, not all parasites bind CD36. A striking exception consists of parasites isolated from human placenta that bind CSA and hyaluronic acid instead.20 25 Interestingly, CSA binding is a rare adherence property of parasites that infect nonpregnant individuals. Therefore, the dichotomy between parasite adhesion to CD36 and to CSA may control the tissue distribution of infected erythrocytes and account for malaria pathogenesis during pregnancy.
Both receptors, CD36 and CSA, are recognized by members of a large, highly diverse gene family known as var genes that encodeP falciparum erythrocyte membrane protein-1 (PfEMP-1).26-28 PfEMP-1 is expressed on the erythrocyte surface and mediates binding to several adhesion receptors, including CD36, TSP, ICAM-1, PECAM, and CSA.29-34 PfEMP-1 possesses 2 adhesive modules: the cysteine-rich interdomain regions (CIDRs) and the Duffy binding-like (DBL) domains.35 The CIDR-1 domain, located after the first DBL domain, was described as the binding domain for CD36,33 whereas the DBLβ-C2 and DBLγ bind, respectively, to ICAM-1 and CSA.29,31 Interestingly, the CIDR-1 domain from CSA-adherent parasites is not significantly different from the CIDR-1 domains that bind CD36. No gross changes are found in the minimal CD36-binding region of CIDR-1, and all cysteine residues critical for CD36 binding are conserved.33 35Thus, small modifications in the CIDR-1 sequence may change its ability to bind CD36.
In the present paper, we explore the molecular basis for the lack of CD36 binding by CSA-adherent parasites and identify residues important for the CIDR-1/CD36 interaction. For this purpose, CIDR-1 domains were expressed from PfEMP-1 of CD36-adherent and CD36-nonadherent PEs and tested for binding to CD36. In addition, chimeric proteins and site-directed mutagenesis of CIDR-1 were used to identify residues that are critical for CD36 binding. Our findings suggest that modifications in the CIDR-1 domain of PfEMP-1 are responsible for the inability of placental sequestered parasites to bind CD36.
Materials and methods
Construction of recombinant plasmids for surface expression in mammalian cell lines
Constructs were amplified from genomic DNA by polymerase chain reaction (PCR) and cloned into the pSRα5 vector (Affymax Research Institute, Santa Clara, CA) between theBamHI/NotI and EcoRI sites for expression in mammalian cells. The pSRα5 vector supplies a signal sequence and a glycosylphosoinositol anchor for cell surface expression. The following domains were used for this study: Malayan Camp (MC) CIDR-1 (residues 395-852, GenBank accession no.AAB60251); A4 CIDR-1 (residues 401-846, accession no. L42244); A4tres CIDR-1 (residues 375-724, accession no. AF193424); FCR3-CSA CIDR-1 (residues 418-824, accession no. AJ133811); ItG2-CS2 CIDR-1 (residues 421-920, accession no. AF134154); 3D7 CIDR-1 (residues 397-824, accession no. AL010226); FVO CIDR-1 (residues 1-480, accession no. AF286005); MC M2 (residues 576-754, accession no.AAB60251); and FVO M2 (residues 180-354, accession no.AF286005).
Construction of chimeric proteins
To allow easy exchange of domains between the MC CIDR-1 and the FCR3-CSA CIDR-1, new restriction sites (MluI,HindIII, and NdeI) were introduced at the desired locations. As a result, several residues were modified, as indicated (Figure 2). Chimeric constructs were created by means of PCR amplification, digestion with specific restriction enzymes, and ligation into the pSRα5 vector.
Site-directed mutagenesis
The MC CIDR-1 C13 clone (MC1 + MC2a + MC2b + MC3) and ItG2-CS2 (DIE [single-letter amino acid codes]) CIDR-1 subcloned into the Puc18 between the BamHI and EcoRI sites were used as a template for mutagenesis. The mutations were performed by means of the Quikchange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). All mutants were generated by means of overlapping forward (F) and reverse (R) primers containing the desired residue change, and the modifications introduced are indicated in lowercase letters. The primers used (identified with single-letter amino acid codes) were as follows:
S203T(F) 5′-GATATGTTAATTGATaCTATTAAATGGAGAG-3′; S203T(R) 5′-CTCTCCATTTAATAGtATCAATTAACATATC-5′; R207N(F) 5′-GATTCTATTAAATGGAacGACGAACATGGTAGG-3′; R207N(R) 5′-CCTACCACGTTCGTCgtTCCATTTAATAGAATC-3′; K218T(F) 5′-GTATAAATAAAGATAcGGGAAAAACATGTATA-3′; K218T(R) 5′-TATACATGTTTTTCCCgTATCTTTATTTATAC-3′; K235R(F) 5′-GTATATGTTTCCAAAgATGGGTTGAACAA-3′; K235R(R) 5′-TTGTTCAACCCATcTTTGGAAACATATAC-3′; K246S(F) 5′-ACCGAATGGGGGtcAATAAAAAAGCTTT-3′; K246S(R) 5′-AAAGCTTTTTTATTgaCCCCCATTCGGT-3′; C213-C222(F) 5′-GACGAACATGGTAGGTGTATAAATAAtacTAcGacAcAcTGTATAAAAGGATGTAACAAAAAATG-3′; C213-C222(R) 5′-CATTTTTTGTTACATCCTTTTATACAgTgTgtCgTAgtaTTATTTATACACCTACCATGTTCGTC-3′; MCCIDR-1 EIK → GHR (F) 5′-CTTATGGGGATGCAAATGgAcacAgACGTATTGAGGCACTGTTGG-3′; MCCIDR-1 EIK → GHR (R) 5′-CCAACAGTGCCTCAATACGTcTgtgTcCATTTGCATCCCCATAAG -3′; CS2CIDR-1 DIE → GHR(F) 5′-GAGGGTTATGGGAATGCAAAAGGACATAGACGTATTGAGGCACTGTTGAAAG-3′; CS2CIDR-1 DIE → GHR(R) 5′-CTTTCAACAGTGCCTCAATACGTCTATGTCCTTTTGCATTCCCATAACCCTC-3′.
Sequence analysis
Multiple sequence alignments were obtained by means of Clustalw software at: http://www.ebi.ac.uk/clustalw/. Consensus residues were identified by means of the Consensus program at:http://www.bork.embl-heidelberg.de/Alignment/consensus.html.
Mammalian cell cultures and transfection
African green monkey kidney fibroblast-like cell line (COS-7), Chinese hamster ovary K1 cells (CHO-K1), and the derived Chinese hamster ovary PgsA-745 cells (CHO-745) deficient in glycosaminoglycans were obtained from the American Type Culture Collection (Manassas, VA). The cells were grown in RPMI 1640 medium (Life Technologies, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal calf serum (Life Technologies). These cells were used for transient or stable expression of the various CIDR-1 constructs. For transient expression, fresh monolayers of COS-7 cells were seeded onto coverslips and grown overnight. The next day, cells were transfected with 2.5 μg plasmid DNA by means of the superfect transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's recommendations. Cells were analyzed for surface expression by immunofluorescence 48 hours later with mAb 179 (Affymax Research Institute), which recognizes an epitope tag at the carboxy terminal of the insert.31,36Cell lines stably expressing CIDR-1 domains were transfected and selected as described elsewhere.29,31 36
Binding assays with Dynal beads coated with recombinant CD36
Binding assays with CD36-coated magnetic beads were performed as described.31,36 In brief, 107 sheep anti–murine immunoglobulin G magnetic beads (Dynal, Oslo, Norway) were coated with 1.0 μg anti-FLAG M1 (Sigma, St Louis, MO) and used to immobilize soluble CD36 (Affymax Research Institute). Transfected cells grown on coverslips were overlaid with 45 μL RPMI binding medium (RPMI 1640, 25 mM Hepes, and 1% bovine serum albumin, pH 6.8) containing 1 500 000 CD36-coated beads and incubated 1 hour at 37°C in a humidified chamber. After incubation, the coverslips were flipped cell-side down onto a stand in 24-well plates filled with 1 mL RPMI binding medium and incubated for 5 minutes to allow unbound beads to settle by gravity and then fixed for 2 hours at room temperature in phosphate buffered saline containing 2% paraformaldehyde (Electron Microscopy Sciences, Ft Washington, PA). Fixed coverslips were processed for immunofluorescence as previously described.31 36 One hundred cells expressing the recombinant protein, as determined by immunofluorescence, were checked for the presence of CD36-coated Dynal beads on their surface. Cells with 4 or more beads attached were considered positive for binding. In some assays, the total number of beads associated with 100 binding positive cells was determined.
Results
The CIDR-1 domain from CD36-adherent PEs but not from CSA-adherent PEs bind CD36
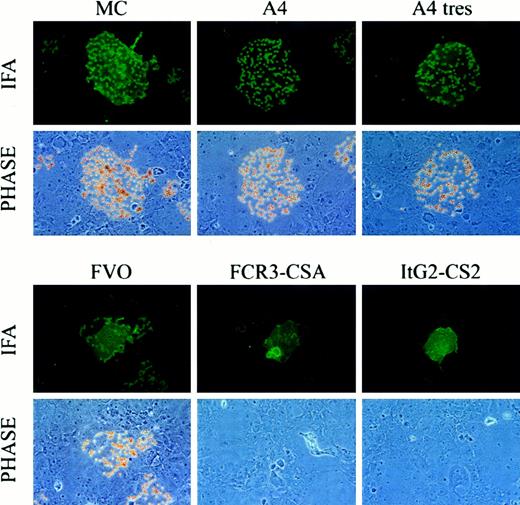
CIDR-1 domains from several laboratory strains were expressed on the surface of COS-7 cells and tested for binding to CD36. Four CIDR-1 domains (MC, A4, A4tres, and FVO) from CD36-adherent parasites strongly bound CD36 (Table 1; Figure1). For each domain, the percentage of transfected cells that were determined positive for binding to CD36 (cells with 4 or more CD36-coated beads) was above 90% (Table 1 and Figure 1).
Binding characteristics of different cysteine-rich interdomain region 1 to CD36
| Parasite phenotype . | Construct expressed . | Positive cells (%) . |
|---|---|---|
| CD36+/CSA− | MC CIDR-1 | 99 ± 1 |
| A4 CIDR-1 | 97 ± 1 | |
| A4 tres CIDR-1 | 89 ± 3 | |
| FVO CIDR-1 | 93 ± 2 | |
| MC M2 | 92 ± 5.5 | |
| FVO M2 | 46 ± 7.5 | |
| CD36−/CSA+ | FCR3-CSA CIDR-1 | 1 ± 1 |
| ItG2-CS2 CIDR-1 | 1 ± 1 | |
| Unknown | 3D7AL010226 | 0 ± 0 |
| Parasite phenotype . | Construct expressed . | Positive cells (%) . |
|---|---|---|
| CD36+/CSA− | MC CIDR-1 | 99 ± 1 |
| A4 CIDR-1 | 97 ± 1 | |
| A4 tres CIDR-1 | 89 ± 3 | |
| FVO CIDR-1 | 93 ± 2 | |
| MC M2 | 92 ± 5.5 | |
| FVO M2 | 46 ± 7.5 | |
| CD36−/CSA+ | FCR3-CSA CIDR-1 | 1 ± 1 |
| ItG2-CS2 CIDR-1 | 1 ± 1 | |
| Unknown | 3D7AL010226 | 0 ± 0 |
CIDR-1 and M2 constructs from CD36 (CD36+/CSA1−) and CSA (CD36−/CSA+) adherent parasites were transfected into COS-7 cells and tested for binding to CD36-coated Dynal beads.
One hundred cells expressing the recombinant protein, determined by immunofluorescence, were checked for the presence of CD36-coated Dynal beads on their surface. Cells with 4 or more beads attached were considered positive for binding. Results are expressed as the mean and SD of 3 independent experiments.
MC indicates Malayan Camp; CIDR-1, cysteine-rich interdomain region 1.
CD36 binding of CIDR-1 recombinant proteins.
CIDR-1 recombinant proteins from CD36-adherent parasites, but not CSA-adherent parasites, bind CD36. Soluble CD36 was immobilized to beads coated with M1 anti-FLAG mAb. Coated beads were incubated with COS-7 cells grown on coverslips and transfected with different constructs. After washing and fixation, the colocalization of beads and transfected cells was quantified by means of a specific monoclonal antibody against an epitope tag incorporated carboxy terminal to the cell-surface–expressed protein. The degree of bead association was examined under immunofluorescence and phase microscopy. CD36-coated beads specifically bound COS-7 cells expressing CIDR-1 from CD36-adherent strains (MC, A4, A4tres, and FVO) but not CIDR-1 from CSA-adherent strains (FCR3-CSA and ItG2-CS2).
CD36 binding of CIDR-1 recombinant proteins.
CIDR-1 recombinant proteins from CD36-adherent parasites, but not CSA-adherent parasites, bind CD36. Soluble CD36 was immobilized to beads coated with M1 anti-FLAG mAb. Coated beads were incubated with COS-7 cells grown on coverslips and transfected with different constructs. After washing and fixation, the colocalization of beads and transfected cells was quantified by means of a specific monoclonal antibody against an epitope tag incorporated carboxy terminal to the cell-surface–expressed protein. The degree of bead association was examined under immunofluorescence and phase microscopy. CD36-coated beads specifically bound COS-7 cells expressing CIDR-1 from CD36-adherent strains (MC, A4, A4tres, and FVO) but not CIDR-1 from CSA-adherent strains (FCR3-CSA and ItG2-CS2).
In contrast, the CIDR-1 domain of the CSA-binding PEs, (FCR3-CSA and ItG2-CS2) did not bind CD36. Furthermore, a third CIDR-1 domain from parasite strain 3D7 (accession no. AL010226) chosen from the GenBank database for its similarity (40% amino acid identity) to the FCR3-CSA CIDR-1 also failed to bind to CD36. These constructs, except 3D7, were also tested for binding in CHO-K1 or CHO-745 stable transfected cell lines and showed the same pattern of binding (data not shown). These data from the in vitro expression of CIDR-1 domains in mammalian cells provide a molecular basis for the lack of adhesion to CD36 in CSA-adherent parasites and demonstrate that it results from a non–CD36-binding CIDR-1 domain.
It has been suggested that the CIDR-1 of CSA-adherent PEs participates in the binding to CSA.37,38 However, none of the CIDR-1 domains expressed in CHO-745 cells tested in this study demonstrated any binding to CSA (data not shown), which is consistent with our previous results demonstrating the lack of binding of the FCR3-CSA CIDR-1.29
The inability of the FCR3-CSA PfEMP-1 to bind CD36 is linked to the minimal CD36-binding region
By means of bacterially expressed recombinant proteins, the minimal CD36-binding domain in the MC CIDR-1 was shown to reside in a 179–amino acid fragment (named M2 in this study) starting near the center of the domain.33 We have now extended this observation to show that the M2 region from the MC CIDR-1 and the equivalent region from the FVO CIDR-1 expressed in COS-7 cells bound CD36 (Table 1). These results support the claim that this region is critical for PfEMP-1 adhesion to CD36.
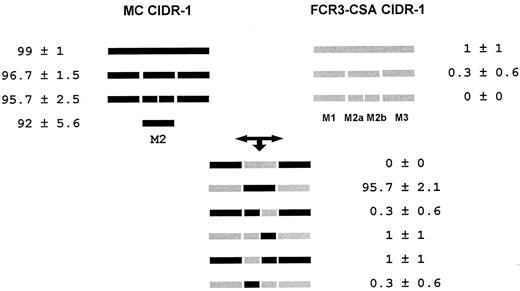
The lack of CD36 binding domains of CSA-binding parasites in CIDR-1 can arise from changes in the minimal CD36-binding region (M2) or from inhibitory effects of the surrounding regions of CIDR1. To explore these possibilities, the CIDR-1 was divided into 3 parts: M1, M2, and M3 (Figure 2). These regions were exchanged between the MC CIDR-1 that binds CD36 and the FCR3-CSA CIDR-1 that does not (Figures 2,3). To generate chimeras, MluI and NdeI restriction sites were introduced into both the MC and the FCR3-CSA CIDR-1; these resulted in amino acid modifications at each site (Figure 2). None of these modifications changed the binding properties of the MC and FCR3-CSA CIDR-1 (Figure 3). Chimeras consisting of the M2 region of MC flanked by the M1 and M3 of FCR3-CSA readily bound to CD36. In contrast, the FCR3-CSA M2 did not bind CD36 even when bordered by the M1 and M3 regions of MC (Figure 3). Thus, the minimal CD36-binding region (M2) and not the surrounding regions of the FCR3-CSA CIDR-1 (M1 and M3) appears to be linked to its non–CD36-binding property.
Alignment of MC and FCR3-CSA CIDR-1.
Alignment was performed by means of Clustalw software at:http://www.ebi.ac.uk/clustalw/. * indicates identical or conserved residues in all sequences in the alignment; :, conserved substitutions; and ., semiconserved substitutions. CIDR-1 domains have been divided into the M1, M2, and M3 regions, in which the M2 encompasses the minimal CD36-binding domain from theMCvar-1 CIDR-1. To exchange the regions of MC and FCR3-CSA CIDR-1, MluI, HindIII, andNdeI restriction sites were introduced. These modifications changed some amino acids at each site, as indicated under the sequences.
Alignment of MC and FCR3-CSA CIDR-1.
Alignment was performed by means of Clustalw software at:http://www.ebi.ac.uk/clustalw/. * indicates identical or conserved residues in all sequences in the alignment; :, conserved substitutions; and ., semiconserved substitutions. CIDR-1 domains have been divided into the M1, M2, and M3 regions, in which the M2 encompasses the minimal CD36-binding domain from theMCvar-1 CIDR-1. To exchange the regions of MC and FCR3-CSA CIDR-1, MluI, HindIII, andNdeI restriction sites were introduced. These modifications changed some amino acids at each site, as indicated under the sequences.
Schematic representation and binding properties of the MC and FCR3-CSA CIDR-1 chimeras.
MC and FCR3-CSA CIDR-1 sequences that were re-engineered with restriction sites and various chimeras between them were tested for binding to CD36. The various constructs were transfected into COS-7 cells and tested for binding to CD36-coated Dynal beads as in Figure 1. The percentage of cells with 4 or more beads attached is given. In each assay, 100 cells expressing the recombinant protein, as determined by immunofluorescence, were examined. Results are expressed as the mean and SD of 3 independent experiments.
Schematic representation and binding properties of the MC and FCR3-CSA CIDR-1 chimeras.
MC and FCR3-CSA CIDR-1 sequences that were re-engineered with restriction sites and various chimeras between them were tested for binding to CD36. The various constructs were transfected into COS-7 cells and tested for binding to CD36-coated Dynal beads as in Figure 1. The percentage of cells with 4 or more beads attached is given. In each assay, 100 cells expressing the recombinant protein, as determined by immunofluorescence, were examined. Results are expressed as the mean and SD of 3 independent experiments.
In an attempt to restore CD36 binding and to localize residues responsible for the lack of binding, the M2 region of both proteins were divided into 2 parts: M2a and M2b (residues 182-248 and 249-360 of the MC CIDR-1, respectively). For this purpose, a new restriction site (HindIII) was introduced that changed 2 residues in the MC179 (DH to KL [single letter amino acid codes]) but introduced no changes in the CSA sequence (Figure 2). This modification in the M2 region of the MC CIDR-1 had no effect on the binding to CD36. Chimeras of all possible combinations were constructed and tested for binding to CD36 (Figure 3). No binding to CD36 was observed with any construct that contained one or more fragments of the FCR3-CSA M2 region (Figure3). Thus, it is probable that several residues located in both parts (M2a and M2b) of the minimal CD36-binding domain are responsible for the inability of the FCR3-CSA CIDR-1 to bind CD36 or that there is some incompatibility between the M2a and M2b regions of the 2 proteins in forming the CD36-binding pocket.
Mutagenesis of conserved residues in the MC M2 region
To define residues in the FCR3-CSA M2 region that are responsible for the lack of binding, we compared the M2 sequence in the CIDR-1 domains that bind and that do not bind CD36 (Figure4). The M2b region (residues 249-360 of the MC CIDR-1) is not highly conserved among the various CIDR-1 domains. By comparison, many residues in the M2a part (residues 182-248 of the MC CIDR-1) are well conserved among binding and nonbinding CIDR-1 domains (Figure 4). However, 5 residues (marked by an asterisk in Figure 4) were conserved among CD36-binding M2a regions but modified or missing in the FCR3-CSA M2a region. Furthermore, a positively charged region in CD36-binding M2a regions (between the first and second cysteines) did not contain charged residues in the FCR3-CSA and 3D7 M2 regions (Figure 4). To investigate the possible involvement of these residues in binding, site-directed mutagenesis was used to substitute FCR3-CSA residues into the MC CIDR-1 (Figure 4). All together, 5 single-residue mutants of the MC M2 region (S203T, R207N, K218T, K235R, and K246S) and a mutant in which the region between the first and second cysteines (C213 and C222) was changed to the corresponding residues of FCR3-CSA were tested for CD36 binding. None of these modifications had any effect on binding (Table2). Thus, these residues are not critical for the interaction of the MC CIDR-1 with CD36.
Multiple alignments of PfEMP-1 M2 domains.
Multiple sequence alignments were obtained by means of Clustalw software at: http://www.ebi.ac.uk/clustalw/. The amino acid numbers correspond to the amino acid numbers of the full-length CIDR-1. Dashes (–) in the multiple alignment indicate gaps that were introduced to maintain alignment. Consensus residues were identified with the Consensus program at:http://www.bork.embl-heidelberg.de/Alignment/consensus.html. The 80% amino acid consensus sequence of the 5 CD36-binding CIDR-1 domains (MC, FVO, A4, A4 tres, and ItG2 [DIE]) and the 100% amino acid consensus sequence of 2 non–CD36-binding CIDR-1 domains (3D7 AL010226and FCR3-CSA) are indicated below the alignments. Identical residues are identified according to single-letter amino acid codes, and conserved cysteines are boxed. The codes for residues that have similar character are as follows: c indicates charge (D, E, H, K, R); + indicates positive (H, K, R); h indicates hydrophobic (A, C, F, I, L, M, V, W, Y); p indicates polar (C, D, E, H, K, N, Q, R, S, T); s indicates small (A, C, D, G, N, P, S, T, V); u indicates tiny (A, G, S); b indicates big (E, K, R, I, L, N, S, Y, W). Residues or regions in the MC CIDR-1 mutated for the FCR3-CSA sequence are marked by an asterisk or a line, respectively.
Multiple alignments of PfEMP-1 M2 domains.
Multiple sequence alignments were obtained by means of Clustalw software at: http://www.ebi.ac.uk/clustalw/. The amino acid numbers correspond to the amino acid numbers of the full-length CIDR-1. Dashes (–) in the multiple alignment indicate gaps that were introduced to maintain alignment. Consensus residues were identified with the Consensus program at:http://www.bork.embl-heidelberg.de/Alignment/consensus.html. The 80% amino acid consensus sequence of the 5 CD36-binding CIDR-1 domains (MC, FVO, A4, A4 tres, and ItG2 [DIE]) and the 100% amino acid consensus sequence of 2 non–CD36-binding CIDR-1 domains (3D7 AL010226and FCR3-CSA) are indicated below the alignments. Identical residues are identified according to single-letter amino acid codes, and conserved cysteines are boxed. The codes for residues that have similar character are as follows: c indicates charge (D, E, H, K, R); + indicates positive (H, K, R); h indicates hydrophobic (A, C, F, I, L, M, V, W, Y); p indicates polar (C, D, E, H, K, N, Q, R, S, T); s indicates small (A, C, D, G, N, P, S, T, V); u indicates tiny (A, G, S); b indicates big (E, K, R, I, L, N, S, Y, W). Residues or regions in the MC CIDR-1 mutated for the FCR3-CSA sequence are marked by an asterisk or a line, respectively.
Binding characteristics of different Malayan Camp cysteine-rich interdomain region 1 mutants
| MC CIDR-1 mutant expressed* . | Positive cells (%)† . |
|---|---|
| MC CIDR-1 | 97.3 ± 1.5 |
| MC CIDR-1 S203T | 97 ± 1.7 |
| MC CIDR-1 R207N | 96 ± 3 |
| MC CIDR-1 K218T | 96.6 ± 2.5 |
| MC CIDR-1 K235R | 94.6 ± 2.5 |
| MC CIDR-1 K246S | 97.6 ± 0.6 |
| MC CIDR-1 C213-C222 | 96 ± 2.6 |
| MC CIDR-1 mutant expressed* . | Positive cells (%)† . |
|---|---|
| MC CIDR-1 | 97.3 ± 1.5 |
| MC CIDR-1 S203T | 97 ± 1.7 |
| MC CIDR-1 R207N | 96 ± 3 |
| MC CIDR-1 K218T | 96.6 ± 2.5 |
| MC CIDR-1 K235R | 94.6 ± 2.5 |
| MC CIDR-1 K246S | 97.6 ± 0.6 |
| MC CIDR-1 C213-C222 | 96 ± 2.6 |
A 3–amino acid substitution in the M2 region is responsible for the inability of ItG2-CS2 CIDR-1 to bind CD36
The ItG2-CS2 parasite binds CSA.30 When we attempted to amplify the published ItG2-CS2 var sequence (accession no. AF134154) using genomic DNA from the parental P falciparum line (ItG2) that was maintained at the National Institutes of Health (NIH), we found that the DBL2γ had an identical sequence to the published one but that the CIDR-1 region had a small difference. The ItG2-CS2 and the ItG2 (NIH) sequences were GCAAAAGGACATAGACGT and GCAAAGGACATAGAACGT, respectively. These changes result in a 3–amino acid difference: AKGHRR to AKDIER (residues 340-342 of the ItG2-CS2 CIDR-1; Figure 4). We were unable to detect anyvar gene in the parental line DNA with a CIDR-1 sequence identical to the CS2–CIDR-1 (unpublished observations, September 2000) and assume that the CS2–CIDR-1 sequence resulted from a mutation that occurred during the in vitro cultivation and selection of this clone. The CIDR-1 with the GHR sequence (ItG2-CS2 [GHR] CIDR-1) did not bind CD36 (Table 1, Figure 1), which is consistent with the non–CD36-adherence phenotype of CS2 PEs.30 The CIDR-1 with the DIE mutation (ItG2 [DIE] CIDR-1), however, readily bound CD36 (Table 3). Thus, a 3–amino acid change from DIE to GHR in the M2 region of ItG2-CS2 ablated its binding to CD36. To test if this area was critical for CD36 binding in other CIDR-1 domains, we introduced the same mutation into the MC CIDR-1 by converting an EIK sequence to GHR (residues 289-291 of the MC CIDR-1; Figure 4). This substitution reduced the percentage of positive cells (transfected cells with 4 or more beads) by 55% and the total number of beads present on 100 binding positive cells by 64% (Table 3). No difference in the level of expression between MC-EIK and MC-GHR was observed by immunofluorescence. Therefore, unlike CS2, the modification did not totally eliminate CD36 binding; this leads to the conclusion that these residues play a significant role in the CIDR1-CD36 interaction and that their specific contribution is influenced by the surrounding sequence. Thus, these residues have varied effects on the structure and function of CIDR-1 domains in accordance with their specific sequence.
Effect of the GHR mutation on CD36-binding characteristics
| Construct expressed . | Positive cells (%)3-150 . | No. beads per 100 positive cells3-151 . |
|---|---|---|
| ItG2 (DIE) CIDR-1 | 91.7 ± 3.1 | 5730 ± 128 |
| ItG2-CS2 (GHR) CIDR-1 | 1 ± 1 | — |
| MC CIDR-1 | 97.3 ± 1.5 | 7990 ± 263 |
| MC (GHR) CIDR-1 | 43.7 ± 4.5 | 2869 ± 188 |
| Construct expressed . | Positive cells (%)3-150 . | No. beads per 100 positive cells3-151 . |
|---|---|---|
| ItG2 (DIE) CIDR-1 | 91.7 ± 3.1 | 5730 ± 128 |
| ItG2-CS2 (GHR) CIDR-1 | 1 ± 1 | — |
| MC CIDR-1 | 97.3 ± 1.5 | 7990 ± 263 |
| MC (GHR) CIDR-1 | 43.7 ± 4.5 | 2869 ± 188 |
CIDR-1 constructs were transfected into COS-7 cells and tested for binding to CD36-coated Dynal beads.
For abbreviations, see Table 1.
See footnote (marked by an asterisk) to Table 1.
The number of beads was counted on 100 cells determined positive for binding to CD36-coated Dynal beads. Results are expressed as the mean and SD of 3 independent experiments.
Discussion
PfEMP-1 expressed on the surface of PEs mediates adhesion to a variety of host receptors.39 Among these receptors are CD36 and CSA, which play a critical role in PE adhesion to endothelial cells and to syncytiotrophoblasts in placenta, respectively.12,17,25,40 PEs present in the placenta bind CSA but not CD36,6,41,42 and PEs with CSA-adherence phenotype are common only in infections of pregnant women.22,41 In contrast, the CD36-binding phenotype is characteristic of PEs of all other infections and is the most common adhesion phenotype.21-24 Previously, CIDR-1 domains from 3 different parasites expressed either in bacterial cells (MC) or on the surface of COS-7 cells (A4 and A4tres) were shown to bind CD36.31,33 36
In the present study, we show that 2 CIDR-1 domains from parasites that adhere to CSA and do not adhere to CD36 (FCR3-CSA and ItG2-CS2 lines) failed to bind CD36, although several CIDR-1 domains from CD36-adherent parasites readily bound CD36. Thus, PfEMP-1 appears to be essential for parasite adhesion to CD36. In addition, these findings suggest that other molecules proposed as being involved in adhesion to CD36, pfalhesin,43 sequestrin,44 and CLAG9,45 cannot compensate for a non–CD36-binding CIDR-1.
The defects in CD36 binding were mapped to the originally described CIDR-1 minimal CD36-binding domain (the M2 region).33 With the use of the FCR3-CSA PfEMP-1, both parts, M2a and M2b, were implicated in binding. Although the FCR3-CSA M2a region is highly similar to the same region in CD36-binding CIDR-1 domains (Figure 4), it could not substitute for the MC sequence, and several mutations in this region did not convert the MC CIDR-1 to a non–CD36-adherent one.
The importance of the CIDR-1 M2 region to CD36 binding was confirmed by testing a second CSA-adherent PfEMP-1 cloned from the ItG-CS2 parasite. With the exception of 3 amino acids, the CIDR-1 from the CS2 clone of the P falciparum ItG2 line was identical to the sequence cloned from the parental ItG2 line maintained at NIH. However, these changes, located in the M2b region of CIDR-1, converted the CIDR-1 domain from a CD36 binder (ItG2-DIE) to a nonbinder (ItG2CS2-GHR). Surprisingly, these results focus attention on the less conserved M2b region as critically important for CD36 binding. A similar change made to the MC CIDR-1 (EIK to GHR substitution) strongly reduced, but did not completely abolish, binding to CD36. Although this 3–amino acid motif is conserved in many var genes,33 it is not present in all CD36-binding CIDR-1 domains (Figure 4). Thus, these 3 residues in the M2b region have an important role in CD36 binding, but their effect is influenced by the surrounding protein sequence; this is consistent with the idea that CD36 binding is influenced by residues throughout the M2 region.
It is striking that the M2 region has such conserved function for CD36 binding despite the sequence variation that occurs within it. It is not clear whether amino acid changes in CSA-adherent parasites affect CIDR-1 structure or whether contact residues are substituted, abolishing the parasites' ability to adhere to CD36.
An attractive idea to explain the apparent mutually exclusive binding of parasites to CD36 and CSA is that the same region of PfEMP-1 is involved in the binding to both receptors. Ample evidence, including this study, has demonstrated that the PfEMP-1 CIDR-1 domain binds CD36.31,33,34,36 It has also been reported that some CIDR-1 or fragments of CIDR-1 domains bind CSA.30,37,38 In these studies, the bacterially expressed recombinant proteins lacked conserved cysteine residues within the M2 region, which was shown to be critical for correct folding for CD36 binding.33 Moreover, the fragments used by Reeder et al37 also bound chondroitin sulfate C (CSC), a form of chondroitin sulfate that does not support PE adhesion. In addition, the parasite culture from which Degen et al38 expressed CIDR-1 fragments did not bind CSA. Thus, the reported interaction between CIDR-1 and CSA may not be biologically relevant. The findings of a previous publication29 and the fact that none of the complete CIDR-1 expressed in this study (including ItG2-CS2) bound CSA do not support this idea. Our study leaves open the question of why there is a dichotomy in parasite binding to CD36 and CSA; it only provides an answer as to why CSA-adherent parasites do not bind CD36. An intriguing possibility is that the CIDR-1 of placental binding parasites has been modified to bind hyaluronic acid20 or an undiscovered coreceptor in placenta.
According to the current paradigm, malaria during pregnancy occurs when mothers become infected with parasites that cannot sequester in microvasculature because they do not bind CD36, but sequester in the placenta because they bind CSA.25 Thus, pregnant women offer a specialized niche for a particular binding subset of parasites to survive. Regardless of the specific combination of residues that leads to a non–CD36-binding CIDR-1, our findings clearly demonstrate that CSA-binding PEs fail to bind CD36 owing to a dysfunctional CIDR-1 domain.
In conclusion, the mapping of the M2 region as responsible for the inability of CSA-selected parasites to bind CD36 and the identification of important residues for the CIDR-1/CD36 interaction enhance the understanding of PE adhesion to CD36 and may contribute to the development of antiadherence therapeutics.
We thank Artur Scherf and Pierre Buffet for providing the FCR3-CSA CIDR-1 construct.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dror I. Baruch, National Institutes of Health, National Institute of Allergy and Infectious Diseases, Bldg 4, Rm B1-37, 4 Center Dr, MSC 0425, Bethesda, MD 20892; e-mail:dbaruch@atlas.niaid.nih.gov.


![Fig. 4. Multiple alignments of PfEMP-1 M2 domains. / Multiple sequence alignments were obtained by means of Clustalw software at: http://www.ebi.ac.uk/clustalw/. The amino acid numbers correspond to the amino acid numbers of the full-length CIDR-1. Dashes (–) in the multiple alignment indicate gaps that were introduced to maintain alignment. Consensus residues were identified with the Consensus program at:http://www.bork.embl-heidelberg.de/Alignment/consensus.html. The 80% amino acid consensus sequence of the 5 CD36-binding CIDR-1 domains (MC, FVO, A4, A4 tres, and ItG2 [DIE]) and the 100% amino acid consensus sequence of 2 non–CD36-binding CIDR-1 domains (3D7 AL010226and FCR3-CSA) are indicated below the alignments. Identical residues are identified according to single-letter amino acid codes, and conserved cysteines are boxed. The codes for residues that have similar character are as follows: c indicates charge (D, E, H, K, R); + indicates positive (H, K, R); h indicates hydrophobic (A, C, F, I, L, M, V, W, Y); p indicates polar (C, D, E, H, K, N, Q, R, S, T); s indicates small (A, C, D, G, N, P, S, T, V); u indicates tiny (A, G, S); b indicates big (E, K, R, I, L, N, S, Y, W). Residues or regions in the MC CIDR-1 mutated for the FCR3-CSA sequence are marked by an asterisk or a line, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/10/10.1182_blood.v97.10.3268/6/m_h81011037004.jpeg?Expires=1763528015&Signature=DwCnM6FGdQHt2zSC~cmZtzGLj-ciEAxak4bnhwxhzWnr-BuQXKP9c6NDVARUid9G5vpKw33noWeTCXN42j031jlmC3wGWc1GHQ8LGs7uP5JWV4fZweckOcRIhKPdWIUZA3jXoEtWNAN8mLFwPnAXKsmpl-isl1nrbJFU6uruJB-ome-4LFmwT11HqyP~KdhdUAHzlrBiEsu--kJ7u0e1aKQ1EE1f7DqD4eqcJ6LnQ-g7NWx~kO8oqSTPjeOXpc8azFOq9n8jOqm8ZneJd4cF3tl-we7cBqdYJWH0Hrujdx~SNTTJb5A0L-bMS3DgjRgOb3-S45n~ey7BBU-jaKmGOA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal