For the management of chronic myeloid leukemia (CML), prediction or early determination of the response to interferon-alpha (IFN-α) treatment is important for identifying nonresponder patients to whom alternative therapy may be proposed. In this study, the levels of expression of both BCR-ABL and subunit 2c of IFN-α receptor (IFN-αR2c) genes were analyzed at diagnosis in 74 patients with chronic phase CML treated with an IFN-α monotherapy. By using blood samples, real-time quantitative polymerase chain reaction was performed to quantify BCR-ABL, IFN-αR2c, and G6PDH mRNA as external control. The results were compared with hematologic and cytogenetic responses to IFN-α. A wide variation in the BCR-ABL/G6PDH ratio was observed at diagnosis (median, 6.68%; range, 0.18%-41.31%), but no significant association with response to IFN-α was observed. In contrast, the variation of IFN-αR2c/G6PDH ratio at diagnosis was significantly associated with the achievement of major cytogenetic response (MCR; 34% or lower Ph+metaphases). Median values of IFN-αR2c/G6PDH ratio for patients achieving MCR and for those who did not achieve it were 110.75% (range, 9.47%-612.30%) and 64.42% (range, 5.96%-425.40%), respectively (P = .037). In addition, this novel molecular factor, combined with the achievement of complete hematologic response at 3 months, makes it possible to predict MCR achievement with high probability by Kaplan-Meier analysis (91% ± 17% at 24 months; P = .0001).
Introduction
Chronic myeloid leukemia (CML) constitutes a clonal myeloproliferative disorder characterized in more than 95% of patients by the reciprocal translocation between chromosomes 9 and 22, t(9;22)(q34;q11), which is referred to as the Philadelphia translocation (Ph).1,2 Molecular studies have demonstrated that the rearrangement disrupts the normal ABL andBCR genes. The resultant BCR-ABL fusion gene is, in most patients, transcribed to an 8.6-kb chimeric mRNA and encodes a 210-kd hybrid protein.3 Most patients have breakpoints that result in fusion mRNA in which either the b2 or the b3 major bcr (M-bcr) exon is fused to the ABL exon a2 to form the b2a2 and b3a2 transcripts. In a minority of patients, this breakpoint may occur 5′ from M-bcr with a minor bcr (m-bcr; 190-kd protein) or 3′ from M-bcr with a micro bcr (μ-bcr; 230-kd protein).4
The BCR-ABL hybrid gene plays a key role in the pathogenesis of the chronic phase of CML.5 This gene and its products are specific to leukemia cells and can therefore be used as sensitive markers of the disease through molecular biology techniques.6
At present, allogeneic transplantation is the only curative treatment for CML, but fewer than half the patients are eligible for this therapy. Interferon-α (IFN-α) treatment has been shown to increase the overall survival of a subset patients with CML in chronic phase.7 Response patterns of patients with CML to IFN-α are heterogeneous. IFN-α therapy is able to induce a cytogenetic response—that is, to decrease the percentage number of Ph+cells—but in two thirds of the patients this treatment is ineffective.8
Like many cytokines and growth factors, the effect of IFN-α is mediated through its interaction with specific cell surface receptors.9 The functional type 1 interferon receptor has a multichain structure composed of the α subunit (IFN-αR1) and the β subunit (IFN-αR2).10 By alternative splicing of the same gene, 3 different forms of IFN-αR2 mRNA have been reported (IFN-αR2a, IFN-αR2b, and IFN-αR2c mRNA).11Only the IFN-αR2c protein mediates a biologic response when associated with the IFN-αR1 protein.12 13
In this study, real-time quantitative polymerase chain reaction (q-PCR) was used to study at diagnosis the levels of BCR-ABL and IFN-αR2c mRNA in the peripheral blood of 74 patients with CML consecutively treated with IFN-α monotherapy. Using LightCycler technology (Roche Diagnostic, Mannheim, Germany), the level of each mRNA transcript was determined and normalized with the quantification of G6PDH mRNA transcripts.14 The ratios BCR-ABL/G6PDH and IFN-αR2c/G6PDH were used to check retrospectively the correlation with the response to IFN-α treatment. We found that the BCR-ABL mRNA level was highly variable at diagnosis but was not correlated with the response to IFN-α treatment in patients with newly diagnosed CML. In contrast, the expression rate of IFN-αR2c mRNA was significantly associated with the major cytogenetic response (MCR) (ie, 34% or lower of Ph+ metaphases) on IFN-α treatment. We therefore compared the latter with other prognostic factors reported in our previous work, such as the complete hematologic response (CHR) at 3 months.15
Patients and methods
Patients
From 116 patients treated with IFN-α alone, as already reported,15 we analyzed 74 patients from whom we obtained samples at diagnosis. For the other patients, the samples were not available or were damaged. These 74 patients with CML in chronic phase were retrospectively analyzed at diagnosis before the start of any treatment. All patients studied were 100% Ph+ and had no clinical or biologic signs of transformation. As shown in Table1, the patients' clinical characteristics were similar to those previously reported.15 Patients were divided into 3 categories according to Sokal classification: low risk (less than 0.8; n = 36), intermediate risk (0.8-1.2; n = 29), and high risk (1.2 or greater; n = 8).16
Clinical characteristics of patients at diagnosis
| Characteristics . | Median value (range) . |
|---|---|
| Age (y) | 49.7 (9.1-70.1) |
| Sex (M/F) | 45/29 |
| Enlarged spleen (cm) | 5 (1-18)* |
| White blood cell counts (×109/L) | 72.3 (14.4-492) |
| Platelet counts (×109/L) | 320 (110-2970) |
| Peripheral blood blast cells (%) | 1 (0-7) |
| Peripheral basophil (%) | 3 (0-13) |
| Hemoglobin level (g/L) | 126 (67-158) |
| Interval diagnosis–IFN-α (mo) | 1.18 (0-6.30) |
| Characteristics . | Median value (range) . |
|---|---|
| Age (y) | 49.7 (9.1-70.1) |
| Sex (M/F) | 45/29 |
| Enlarged spleen (cm) | 5 (1-18)* |
| White blood cell counts (×109/L) | 72.3 (14.4-492) |
| Platelet counts (×109/L) | 320 (110-2970) |
| Peripheral blood blast cells (%) | 1 (0-7) |
| Peripheral basophil (%) | 3 (0-13) |
| Hemoglobin level (g/L) | 126 (67-158) |
| Interval diagnosis–IFN-α (mo) | 1.18 (0-6.30) |
| Sokal classification* . | No. patients (%) . |
|---|---|
| Low risk | 36 (49) |
| Intermediate risk | 29 (40) |
| High risk | 8 (11) |
| Sokal classification* . | No. patients (%) . |
|---|---|
| Low risk | 36 (49) |
| Intermediate risk | 29 (40) |
| High risk | 8 (11) |
Only 31 patients had enlarged spleens, and one patient had a splenectomy before diagnosis.
Hematologic and cytogenetic responses to IFN-α treatment were evaluated according to the criteria of the Houston group.17 The criterion for CHR was the normalization of the peripheral white blood cell count (WBC) to less than 10 × 109/L with the disappearance of immature circulating cells (blasts, promyelocytes, myelocytes, metamyelocytes), the normalization of platelet count (less than 450 × 109/L), and the disappearance of all signs and symptoms of the disease (in particular, splenomegaly). Cytogenetic response was classified according to the proportion of Ph metaphases (20 or more metaphases studied),18 no response when the Ph chromosome persisted in all metaphases, minor response when it persisted in 35% to 95% of metaphases, and major response (MCR) when it persisted in 34% or fewer metaphases.
Thirty-two patients achieved MCR, and 23 achieved complete cytogenetic response. Fifty-nine patients achieved CHR—25 of them at 3 months. The median follow-up time of the population was 50.8 months (range, 5.3-141.8 months).
Cell lines
Four human cell lines were used in this study: K562 (b3a2 BCR-ABL+), LAMA84 (b3a2 BCR-ABL+), AR230 (e19a2 BCR-ABL+), and HL60 (BCR-ABL−). Cell lines were purchased from cell repository banks (American Tissue Culture Collection, Rockville, MD; European Collection of Cell Cultures, Winchester, United Kingdom; or German Collection of Micro-organisms and Cell Cultures, Braunschweig, Germany). Two cell lines (LAMA84HIGH and AR230HIGH) exhibited an overexpression of BCR-ABL mRNA compared to the parental counterparts and were kindly provided by J. Melo (ICSM; Haematology Department, Hammersmith Hospital, London, United Kingdom). They were all grown in liquid culture medium containing RPMI-1640 and 10% fetal calf serum.
RNA extraction and cDNA synthesis
Total RNA was extracted from 106 to 107cells of either peripheral blood cryopreserved in liquid nitrogen at diagnosis or the different cell lines used. RNA extraction was performed by using the acid guanidinium thiocyanate and phenol-chloroform method19 and a commercially available extraction kit (Trizol; Gibco BRL Life Technologies, Gaithersburg, MD). Briefly, cells were thawed in liquid culture medium containing RPMI-1640 and 10% fetal calf serum. Cell viability was checked, and extraction was performed as described above. cDNA synthesis was performed according to the manufacturer's instructions using random hexamer priming and AMV reverse transcriptase (First Strand cDNA Synthesis Kit for RT-PCR; Roche Diagnostic, Mannheim, Germany). One microgram total RNA was reverse-transcribed and stored at −20°C.
Real-time quantitative polymerase chain reaction
Real-time q-PCR was performed by using the LightCycler Technology (Roche Diagnostic). PCR protocol and oligonucleotides used in the study for BCR-ABL and G6PDH amplifications were reported elsewhere.20
For IFN-αR2c mRNA amplification, 2 μL cDNA was added in 20 μL final volume to 2 μL Mastermix (LightCycler DNA Master hybridization probes; Roche Diagnostic), 1 U heat-labile uracil DNA glycosylase (UDG, Roche Diagnostic), 0.4 μg TaqStart monoclonal antibody (Clontech), MgCl2 solution (5 mM), forward and reverse oligonucleotide primers (0.3 μM), and fluorescent hybridization probes (0.2 and 0.4 μM). The amplification mix was incubated for 5 minutes at room temperature to allow the degradation of specific contaminating PCR products by uracyl DNA glycosylase (UDG). Heat-labile UDG and TaqStart antibody were deactivated by an initial denaturation step for 1 minute at 95°C. The PCR program included 45 cycles (95°C, 1 second, 20°C/s)/(57°C, 10 seconds, 20°C/s)/(72°C, 10 seconds, 2°C/s). Primers (R2A, R2B) and probes (S1R2, S2R2) used for IFN-αR2c mRNA reverse-transcribed amplification were as follows: R2A, 5′AGTGATGAGCAAGCAGTAA; R2B, 5′ AGGTTAGGAAAT GGCCAG; S1R2, 5′CCATAGTGACACTGAAATGGATTGGTTATATA-(fluorescein); and S2R2, 5′(Red640)-GCTTAAGAAATAGCCTCCCCAAAGTCT-phosphate). The estimated size of the IFN-αR2c–amplified fragment matched the calculated size (238 bp) by electrophoresis on 2% ethidium bromide–stained agarose gels.
Detection of the PCR product was based on fluorescence resonance energy transfer between the fluorophores. Cycle threshold (CT) was defined as the cycle number at which a significant increase in the fluorescence signal was first detected. Using serial dilutions of plasmids (10 to 106 molecules per reaction) of external standard (pGD210b3a2, BCR-ABL, pIFN-αR2-2IFN-αR2c, and pGdBBXG6PDH), the standard curve was generated between the CT value and the logarithm of the starting copy number of plasmids. Quantification of the unknown samples was performed automatically by the LightCycler software (version 3.39; Roche Diagnostic) (Figure1). Results were expressed initially as the number of target molecules/2 μL cDNA. To standardize results for variability in RNA and cDNA quantity and quality, we quantified total G6PDH transcripts in each sample as the internal control.14 Normalized levels of BCR-ABL and IFN-αR2c transcripts were calculated as the ratio between the amount of both transcripts and the G6PDH transcripts (expressed in percentage).
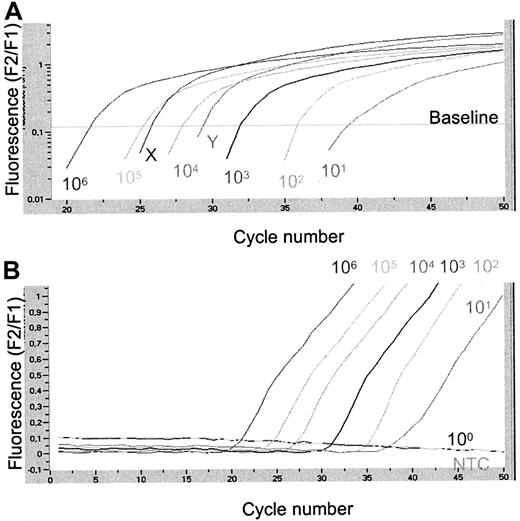
Quantification of IFN-αR2c mRNA levels in CML patient samples at diagnosis using LightCycler technology.
(A) Logarithmic plot (F2/F1) of fluorescence versus cycle number. Six standard dilutions of plasmid pIFN-αR2-2 were compared with 2 patient samples of unknown IFN-αR2c mRNA concentration. Calculated concentrations of 2 patient samples were, respectively, 50 500 (X) and 7,750 (Y) IFN-αR2c transcripts at the start of the reaction. (B) Fluorescence history. Original graph given by the LightCycler. This figure illustrates the possibility of detecting 10 molecules of plasmid pIFN-αR2-2. NTC, no template control.
Quantification of IFN-αR2c mRNA levels in CML patient samples at diagnosis using LightCycler technology.
(A) Logarithmic plot (F2/F1) of fluorescence versus cycle number. Six standard dilutions of plasmid pIFN-αR2-2 were compared with 2 patient samples of unknown IFN-αR2c mRNA concentration. Calculated concentrations of 2 patient samples were, respectively, 50 500 (X) and 7,750 (Y) IFN-αR2c transcripts at the start of the reaction. (B) Fluorescence history. Original graph given by the LightCycler. This figure illustrates the possibility of detecting 10 molecules of plasmid pIFN-αR2-2. NTC, no template control.
Statistical analysis
Quantitative variables were expressed as median and range (minimum to maximum). Comparisons of patients' characteristics between groups were made by the Wilcoxon test for quantitative variables and by the chi-square analysis or the Fisher exact test for qualitative variables. Correlations between 2 continuous variables were studied using the Rho-Spearman test. Cumulative incidences of MCR were estimated by the Kaplan-Meier method and were compared in univariate analysis by the log-rank test,21 and multivariate analysis was conducted using the Cox model.
Results
Sensitivity and accuracy of real-time quantitative polymerase chain reaction
By dilution of plasmids we were able reliably to amplify 10 b3a2 BCR-ABL, 10 IFN-αR2c, and 10 G6PDH molecules per reaction. Figure 1presents an example of real-time q-PCR for the IFN-αR2c plasmid. The sensitivity of the BCR-ABL real-time q-PCR on the cellular level was tested with serial dilutions of K562 cells in HL60 cells. Real-time q-PCR was able to detect one K562 cell in 105 HL60 cells and displayed a linear correlation of the cycle threshold and the log range K562 cell number in the sample over a 5-log range. Intra-assay and inter-assay reproducibility of the quantitative analysis was assessed. Ten identical samples (105 molecules) of plasmid pGD210, pIFN-αR2-2, and pGdBBX were processed and analyzed in one run. The coefficient of variation (CV) for a given concentration and the respective cycle threshold crossing point are presented in Table2 for intra- and inter-assays. Efficiency of real time q-PCR in the various runs was always superior to 92%, and the CV of the calculated value was 6% or less for each target.
Intra-assay and inter-assay comparisons of CV for 105 molecules pGD210, pIFN-αR2-2 and pGdBBX
| Comparison intra-assay (10 tests) . | |||
|---|---|---|---|
| CV . | 105 molecules . | ||
| pGD210 . | pIFN-αR2-2 . | pGdBBX . | |
| of concentration | 0.11 | 0.04 | 0.06 |
| of CT | 0.006 | 0.002 | 0.003 |
| Comparison intra-assay (10 tests) . | |||
|---|---|---|---|
| CV . | 105 molecules . | ||
| pGD210 . | pIFN-αR2-2 . | pGdBBX . | |
| of concentration | 0.11 | 0.04 | 0.06 |
| of CT | 0.006 | 0.002 | 0.003 |
| Comparison inter-assay (> 10 tests) . | |||
|---|---|---|---|
| CV . | 105 molecules . | ||
| pGD210 . | pIFN-αR2-2 . | pGdBBX . | |
| of concentration | 0.18 | 0.14 | 0.17 |
| of CT | 0.029 | 0.079 | 0.035 |
| Comparison inter-assay (> 10 tests) . | |||
|---|---|---|---|
| CV . | 105 molecules . | ||
| pGD210 . | pIFN-αR2-2 . | pGdBBX . | |
| of concentration | 0.18 | 0.14 | 0.17 |
| of CT | 0.029 | 0.079 | 0.035 |
Analysis by real-time q-PCR of BCR-ABL/G6PDH ratio in cell lines and patients with CML
Real-time q-PCR for BCR-ABL mRNA was performed in 2 cell lines, LAMA84 and AR230, and findings were compared to those of their counterparts, which exhibited very high levels of BCR-ABL protein (LAMA84HIGH and AR230HIGH).22 The different levels of BCR-ABL mRNA could be discriminated. Indeed, the ratio BCR-ABL/G6PDH in AR230HIGH and LAMA84HIGHwas increased, respectively 1.65- and 9.2-fold compared to the parental cell lines (data not shown).
Samples from the 74 patients with CML were studied for both BCR-ABL and G6PDH amplification. The median value of the BCR-ABL mRNA level at diagnosis was 2560 (range, 31-28 820). The normalized BCR-ABL mRNA level was calculated as described in “Materials and methods,” by taking into account G6PDH mRNA amplification. Hence, among the 74 patients, the BCR-ABL/G6PDH ratio varied at diagnosis from 0.18% to 41.31%, with a median value of 6.68%.
Association between BCR-ABL/G6PDH ratio and cytogenetic response in CML
The impact of the BCR-ABL/G6PDH ratio at diagnosis and other individual factors given in Table 1 were tested for responses to IFN-α treatment in univariate and multivariate analyses. Individual factors such as sex, age, enlarged spleen, peripheral blood blast cells, myelocytes, peripheral basophils, platelet count, and interval diagnosis-IFN treatment were not significantly associated with the BCR-ABL/G6PDH ratio. In contrast, WBC count (P = .012), hemoglobin level (P = .018), and metamyelocytes (P = .039) were found to be significantly correlated with the BCR-ABL/G6PDH ratio in multivariate analysis. As reported in our previous study,15 Sokal score and achievement of CHR at 3 months were found to be statistically significant for the achievement of MCR for 74 patients with CML. No association between CHR at 3 months and the BCR-ABL/G6PDH ratio or between Sokal score and BCR-ABL/G6PDH ratio was observed.
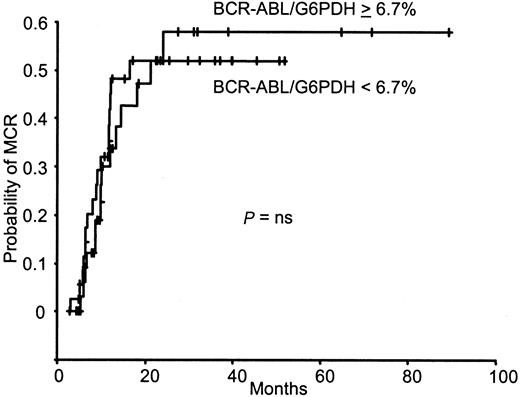
By using the median value (6.68%) of the BCR-ABL/G6PDH ratio as a cutoff to individualize 2 groups, no difference was observed between the latter in the cumulative incidence of MCR (Figure2). The BCR-ABL/G6PDH ratio taken as a continuous variable was not significantly associated with the achievement of MCR. The median value of the BCR-ABL/G6PDH ratio was 6.64% (range, 0.18%-21.53%) for patients who achieved MCR and 6.81% (range, 0.52%-41.31%) for patients who did not (P = .32).
BCR-ABL/G6PDH ratio and MCR achievement.
Kaplan-Meier analysis of the cumulative incidence of MCR achievement according to the BCR-ABL/G6PDH ratio at diagnosis for the 74 patients with CML. ns, not significant.
BCR-ABL/G6PDH ratio and MCR achievement.
Kaplan-Meier analysis of the cumulative incidence of MCR achievement according to the BCR-ABL/G6PDH ratio at diagnosis for the 74 patients with CML. ns, not significant.
Association between IFN-αR2c/G6PDH ratio and cytogenetic response in CML
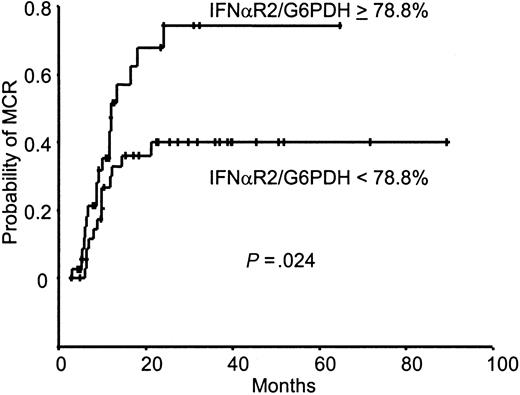
By using the same procedure, the IFN-αR2c mRNA level was quantified at diagnosis, and the IFN-αR2c/G6PDH ratio was calculated. A wide variation of IFN-αR2c/G6PDH ratios was observed (median, 78.83%; range, 5.96%-612.30%). The IFN-αR2c/G6PDH ratio used as a continuous variable was also significantly associated with the cumulative incidence of MCR (P = .0058). The 74 patients were separated into 2 groups with regard to the median value of the IFN-αR2c/G6PDH ratio (Figure 3). For the group of patients with values higher than the median, the probabilities to be in MCR at 12 and 24 months were 52% ± 19% and 75% ± 19%, respectively. In contrast, for the other group, the probabilities to be in MCR at 12 and 24 months were 30% ± 15% and 40% ± 17%, respectively (P = .024). The median value of the IFN-αR2c/G6PDH ratio for patients who achieved MCR was 110.75% (range, 9.47%-612.30%), and it was 64.42% (range, 5.96%-425.40%) for those who did not (P = .037). We also compared for 35 patients the IFN-αR2c/G6PDH ratio and the percentage of Ph− cells 12 and 18 months after IFN-α treatment that were not statistically significant (P = .142 andP = .205, respectively). However, for the group with an IFN-αR2c/G6PDH value higher than the median (78.83%), the median value of Ph− cells after 12 and 18 months was, respectively, 83% and 85% (16 patients; range, 0%-100%). For the group with an IFN-αR2c/G6PDH value lower than 78.83%, the median value of Ph− cells after 12 and 18 months was, respectively, 53% and 48% (19 patients; range, 0%-100%).
IFN-αR2/G6PDH ratio and MCR achievement.
Kaplan-Meier analysis of the cumulative incidence of MCR achievement according to the IFN-αR2/G6PDH ratio at diagnosis for the 74 patients with CML.
IFN-αR2/G6PDH ratio and MCR achievement.
Kaplan-Meier analysis of the cumulative incidence of MCR achievement according to the IFN-αR2/G6PDH ratio at diagnosis for the 74 patients with CML.
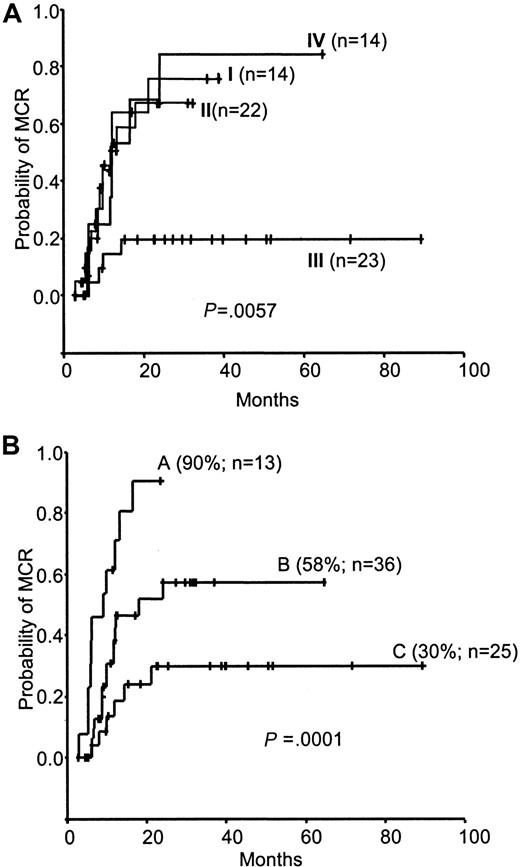
In the 74 patients, we then analyzed by univariate analysis the individual clinical factors described in Table 1. As already reported,15 we analyzed the achievement of CHR at 3 months, which is the most significant factor for predicting MCR (P = .0007). Sokal score (low risk/intermediate and high risk; LR/IR-HR) and the IFN-αR2c/G6PDH ratio were also found to be associated with the cumulative incidence of MCR (P = .012,P = .024). To test the effect of these various factors, different combinations were performed. As shown in Figure4A regarding low-risk Sokal score, the actuarial probability of MCR was very close whatever the IFN-αR2c/G6PDH value (groups I and II) (P = .81). In contrast, regarding the intermediate/high-risk Sokal score (groups III and IV), the probability to achieve MCR was significantly higher in the high IFN-αR2c/G6PDH ratio subgroup (group IV) than in the low subgroup (group III) (P = .0019).
Kaplan-Meier analysis of the cumulative incidence of MCR achievement.
(A) According to combined Sokal score and IFN-αR2c/G6PDH ratio, the 73 patients were separated into 4 groups: group I, low-risk Sokal score (LR) and IFN-αR2c/G6PDH ratio less than 78.83%; group II, low-risk Sokal score and IFN-αR2c/G6PDH ratio greater than 78.83%; group III, intermediate/high-risk Sokal score (IR-HR) and IFN-αR2c/G6PDH ratio less than 78.83%; and group IV, intermediate/high-risk Sokal score and IFN-αR2c/G6PDH ratio greater than 78.83%. The cumulative incidence of MCR achievement was significantly lower for patients with an intermediate/high-risk Sokal score and an IFN-αR2c/G6PDH ratio less than 78.83% (P < .004) than for patients in the 3 other groups, among which there were no significant differences. For groups III, II, I, and IV, the probability of achieving MCR at 12 months was 15% ± 15%, 51% ± 24%, 55% ± 28%, and 54% ± 30%, respectively; at 24 months it was 20% ± 17%, 68% ± 25%, 76% ± 27%, and 85% ± 27%, respectively. (B) According to the combined IFN-αR2/G6PDH ratio at diagnosis and CHR at 3 months for the 74 patients with CML. Group A, patients who had an IFN-αR2/G6PDH ratio of 78.83% or greater and CHR at 3 months. Group B, patients who had an IFN-αR2/G6PDH ratio of 78.83% or greater or CHR at 3 months, but not both. Group C, patients who had neither an IFN-αR2/G6PDH ratio of 78.83% or greater nor CHR at 3 months. For all 3 groups, the cumulative incidence of MCR at 12 months was 72% ± 26%, 43% ± 19%, and 19% ± 17%, respectively. At 24 months, it was 91% ± 17%, 58% ± 20%, and 30% ± 20%, respectively.
Kaplan-Meier analysis of the cumulative incidence of MCR achievement.
(A) According to combined Sokal score and IFN-αR2c/G6PDH ratio, the 73 patients were separated into 4 groups: group I, low-risk Sokal score (LR) and IFN-αR2c/G6PDH ratio less than 78.83%; group II, low-risk Sokal score and IFN-αR2c/G6PDH ratio greater than 78.83%; group III, intermediate/high-risk Sokal score (IR-HR) and IFN-αR2c/G6PDH ratio less than 78.83%; and group IV, intermediate/high-risk Sokal score and IFN-αR2c/G6PDH ratio greater than 78.83%. The cumulative incidence of MCR achievement was significantly lower for patients with an intermediate/high-risk Sokal score and an IFN-αR2c/G6PDH ratio less than 78.83% (P < .004) than for patients in the 3 other groups, among which there were no significant differences. For groups III, II, I, and IV, the probability of achieving MCR at 12 months was 15% ± 15%, 51% ± 24%, 55% ± 28%, and 54% ± 30%, respectively; at 24 months it was 20% ± 17%, 68% ± 25%, 76% ± 27%, and 85% ± 27%, respectively. (B) According to the combined IFN-αR2/G6PDH ratio at diagnosis and CHR at 3 months for the 74 patients with CML. Group A, patients who had an IFN-αR2/G6PDH ratio of 78.83% or greater and CHR at 3 months. Group B, patients who had an IFN-αR2/G6PDH ratio of 78.83% or greater or CHR at 3 months, but not both. Group C, patients who had neither an IFN-αR2/G6PDH ratio of 78.83% or greater nor CHR at 3 months. For all 3 groups, the cumulative incidence of MCR at 12 months was 72% ± 26%, 43% ± 19%, and 19% ± 17%, respectively. At 24 months, it was 91% ± 17%, 58% ± 20%, and 30% ± 20%, respectively.
Regarding the achievement of CHR at 3 months and the IFN-αR2c/G6PDH ratio, 3 populations were studied: one that had both an IFN-αR2c/G6PDH ratio 78.83% or greater and a CHR at 3 months (group A), one that had only one of them (group B), and the other that had none of them (group C). The probability of achieving MCR at 24 months was 91% ± 17% in group A, 58% ± 20% in group B, and 30% ± 20% in group C (P = .0001) (Figure 4B).
When we used multivariate analysis to test the 3 factors in the first model, Sokal score was not significantly associated with the achievement of MCR (P = .1581), in contrast to the achievement of CHR at 3 months (P = .0056) and the IFN-αR2c/G6PDH ratio (P = .0133). Both the achievement of CHR at 3 months and the IFN-αR2c/G6PDH ratio were included in the final model, with P = .0031 and P = .0072, respectively.
Discussion
In the present study, we measured retrospectively the expression level at diagnosis of BCR-ABL and IFN-αR2 genes in patients with CML before starting IFN-α therapy. By using real-time q-PCR, the association between these 2 potential molecular factors and the hematologic and cytogenetic responses to IFN-α treatment were evaluated. Our results show that (1) at diagnosis the expression rate of each gene was heterogeneous among the patients; (2) the amount of BCR-ABL mRNA at diagnosis was not associated with the response to IFN-α treatment; (3) independent of other individual factors, the amount of IFN-αR2c mRNA was associated with the achievement of MCR in CML patients treated with IFN-α.
During recent years, quantification of the BCR-ABL gene and of mRNA has been performed by using several molecular techniques, such as competitive PCR,23 Southern blot analysis,24 capillary electrophoresis,25 and, more recently, real-time q-PCR.26,27 These molecular assays are used to evaluate and monitor minimum residual disease after treatment such as IFN-α therapy or bone marrow transplantation.6,23,26,27 A good correlation was found between the number of Ph+ metaphases in bone marrow and BCR-ABL mRNA levels in the peripheral blood of patients with cytogenetic responses.27 In those reports, variability of BCR-ABL mRNA levels at diagnosis was described in only a few samples. In our study, BCR-ABL mRNA levels were quantified at diagnosis for 74 patients with CML. A reproducible and sensitive test using real-time q-PCR was performed, making it possible to detect a low number of transcripts (10 molecules of plasmid pGD210) and to discriminate variations in high BCR-ABL mRNA levels. Our results confirm the high variability of BCR-ABL gene expression at diagnosis in a larger group of patients.
We and other groups have already demonstrated that the cytogenetic response is significantly associated with survival.7,8,15We tested whether the BCR-ABL mRNA level at diagnosis was a potential prognostic factor for the achievement of MCR, but no statistical association was observed. This is important in light of the results published by Gaiger et al28 showing that BCR-ABL mRNA levels increase with disease progression. In other words, though the increase or decrease in BCR-ABL mRNA levels during IFN-α therapy could be a parameter predicting response to treatment, the level established only at diagnosis does not seems to be a predictive factor for the achievement of MCR.
Although Bcr-Abl protein plays a key role in CML, the prediction of the response to IFN-α therapy depends on other factors. Some studies have already sought the correlation between molecular markers at diagnosis and the response to treatment. For instance, Shepherd et al29 have now established that neither type b3a2 nor type b2a2 BCR-ABL mRNA is correlated with clinical feature, cytogenetic response, duration of chronic phase, or survival. Interestingly, a recent report showed that telomere length measurements could be a good indicator of disease progression.30
Another way to predict the response to IFN-α treatment may be analysis of the factors linked directly with therapy. Genes involved in the IFN-α signaling pathway could be good candidates for IFN-α response prediction, such as interferon regulatory factors (IRF). The IRF1/IRF2 ratio could be associated with cytogenetic response.31 In patients with CML, the expression of interferon consensus sequence binding protein, or ICSBP, was induced with IFN-α treatment but decreased with the progression of CML.32 More recently, the expression of IRF4 in lymphocytes was found to be associated with IFN-α response in patients with CML.33 Here, we focused on the first step of the IFN-α signaling pathway, which is the binding of IFN-α to a specific cell surface receptor. Indeed, Tamura et al34demonstrated, in CML cell lines such as K562, that the response to IFN-α treatment depends on the expression of this receptor. In addition, in hairy cell leukemia, Platanias et al35 showed that lack of expression of IFN-α receptor on the leukemic cells may be associated with resistance to IFN-α therapy. In another model, the determination by RT-PCR of IFN-αR2 and IFN-αR1 mRNA, which code for the IFN-α receptor, was performed in patients with chronic hepatitis C virus (HCV) treated with IFN-α.36 37 The authors showed that expression of IFN-α receptor genes in the liver was a factor for predicting the long-term efficacy of IFN-α therapy in patients with chronic genotype 2a or 2b HCV infection. In the present report, we have specifically quantified the long form of theIFN-αR2 gene. We show for the first time that the expression level of IFN-αR2c mRNA is variable at diagnosis in patients with CML and is statistically associated with MCR achievement after IFN-α treatment. IFN-αR2c/G6PDH ratio and CHR at 3 months are significantly associated with the cumulative incidence of MCR. Both these factors act independently. In our study, Sokal score was not significant when it was included in the initial model of the multivariate analysis of the cumulative incidence of MCR achievement, even though there was a slight trend. By using these 2 independent factors, we were able to identify 3 populations with low, intermediate, and high probability for achieving MCR. Obviously, these preliminary data have to be confirmed in a prospective study in a larger group of patients. Our data suggest that it could also be interesting to study other candidates involved downstream in the IFN-α signaling pathway to predict early the IFN-α response and to propose alternative therapies for nonresponders to IFN-α treatment.
We thank M. Vezon, Director of Etablissement Français du Sang (EFS Aquitaine-Limousin, France) and particularly Mme Hau and M. Comeau for providing access to the LightCycler. We thank G. Daley (Whitehead Institute, Cambridge, MA), G. Uze (IGMM, Montpellier, France), and P. Mason (ICSM, Haematology Department Hammersmith Hospital, London, United Kingdom) for the plasmids pGD210b3a2, BCR-ABL, pIFN-αR2-2IFN-αR2c, and pGdBBXG6PDH. We also thank R. Salmi for his helpful comments regarding statistical analysis and the physicians of the Service des Maladies du Sang: J.M. Boiron, K. Bouabdallah, P. Cony-Makhoul, O. Fitoussi, F. Nicolini, A. Pigneux, and M. Puntous.
C.B. and F.X.M. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Gérald Marit, Laboratoire Universitaire d'Hématologie, Université Victor Segalen Bordeaux 2, Carreire Nord, Bat 1A, 2ème étage, 146, Rue Léo Saignat, 33076 Bordeaux cedex, France; e-mail:gerald.marit@hemato.u-bordeaux2.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal