BCR/ABL fluorescent in situ hybridization study of chronic myeloid leukemia (CML) and Philadelphia+(Ph+) acute lymphoid leukemia (ALL) indicated that approximately 9% of patients exhibited an atypical hybridization pattern consistent with a submicroscopic deletion of the 5′ region ofABL and the 3′ region of the BCR genes on the 9q+ chromosome. The CML patients with deletions had a shorter survival time and a high relapse rate following bone marrow transplant. Since deletions are associated with both Ph+CML and ALL, it seemed probable that other leukemia-associated genomic rearrangements may also have submicroscopic deletions. This hypothesis was confirmed by the detection of deletions of the 3′ regions of theCBFB and the MLL genes in AML M4 patients with inv(16) and in patients with ALL and AML associated withMLL gene translocations, respectively. In contrast, analysis of the AML M3 group of patients and AML M2 showed that similar large deletions were not frequently associated with the t(15;17) or t(8;21) translocations. Analysis of sequence data from each of the breakpoint regions suggested that large submicroscopic deletions occur in regions with a high overall density of Alu sequence repeats. These findings are the first to show that the process of deletion formation is not disease specific in leukemia and also implicate that the presence of repetitive DNA in the vicinity of breakpoint regions may facilitate the generation of submicroscopic deletions. Such deletions could lead to the loss of one or more genes, and the associated haploinsufficiency may result in the observed differences in clinical behavior.
Introduction
Since the discovery of the Philadelphia (Ph) chromosome in patients with chronic myelogenous leukemia (CML), it has become evident that specific chromosomal rearrangements are consistently associated with hematologic malignancies.1Although it is well established that such recurrent chromosome translocations generate several different types of pathognomic fusion oncogenes, the precise details of the molecular processes leading to these rearrangements in leukemias are poorly understood.
The Ph chromosome arises from a reciprocal translocation of the long arms of chromosomes 9 and 22 that transposes the 3′ segment of theABL gene from 9q34 to the 5′ segment of the BCRgene on 22q11. The resulting BCR-ABL gene is transcribed into a chimeric messenger RNA and then translated into fusion proteins of varying size (p190bcr-abl, p210bcr-abl, and p230bcr-abl), depending on the location of the breakpoint of the genes involved. Although these BCR/ABL chimeric fusion proteins play a central role in the pathogenesis of CML, it is unclear whether these fusion oncoproteins alone are sufficient to explain the full range of clinical responses to the disease process.2Recently it was proposed that extensive submicroscopic deletions, 5′ ofABL and 3′ of BCR, on the derivative chromosome 9 could often accompany BCR/ABL rearrangement and that the disease associated with a deletion was more refractile to treatment.3 This investigation mapped the size of the genomic deletions in 16 CML samples and showed that as much as several megabases of DNA flanking the translocation breakpoints was deleted, most likely occurring at the time of the rearrangement. However, the study did not determine whether such deletions were also associated with the p190bcr-abl Ph+ rearrangement or ascertain whether submicroscopic deletions were common in other leukemia-associated translocations.
In this study we identified 23 new examples of deletion from a series of 250 CML patients and confirmed that there was a significant association with poor treatment response. We also performed a retrospective study of the frequency of deletions in Ph+acute lymphoid leukemia (ALL), acute myeloid leukemia (AML) M4Eo, AML M3, AML M2, and ALL or AML associated withMLL translocations to determine the incidence of submicroscopic deletions close to the breakpoints in other common recurrent chromosomal rearrangements of leukemia.
Patients, materials, and methods
Patients, materials, and cytogenetic studies
Informed consent in accordance with the Helsinki protocol was obtained according to our institutional guidelines for all patients. A total of 250 patients with CML, 13 patients with Ph+ ALL, 20 patients with inv(16) AML M4Eo, 30 patients with t(15;17) AML M3, 14 patients with t(8;21) AML M2, and 43 patients with ALL- or AML-associated MLL translocations were analyzed (Table 1). Patients who were enrolled into the study were newly diagnosed between 1987 and 1999. Of the 250 CML patients, 160 were tested by fluorescent in situ hybridization (FISH) alone and 126 by FISH and conventional cytogenetic analysis. All patients with Ph+ ALL, inv(16), t(15;17), t(8;21), andMLL gene translocations were tested by both cytogenetic and FISH methods. Fixed cytogenetic preparations were obtained from cultured (24 and 48 hours) bone marrow samples. Slides for cytogenetic and FISH analyses were prepared directly from methanol/acetic acid-fixed cell pellets of fresh samples or from cell suspension stored in fixative at −20°C. All stored samples exhibited excellent hybridization efficiencies (> 98%), indicating that long-term storage did not affect the quality of the samples. All cytogenetic studies were performed on bone marrow specimens by analyzing up to 20 consecutive G-banded metaphases.
Correlation between the type of leukemia and the presence of the deletions
| Diagnosis . | Total no.of patients . | No. of patients with deletions . | Patients with deletions (%) . |
|---|---|---|---|
| CML | 250 | 23 | 9.2 |
| ALL Ph+ | 13 | 1 | 7.7 |
| AML M4 | 20 | 2 | 10 |
| ALL and AML with MLL gene rearrangements | 43 | 7 | 16.2 |
| AML M3 | 30 | 0 | 0 |
| AML M2 | 14 | 0 | 0 |
| Diagnosis . | Total no.of patients . | No. of patients with deletions . | Patients with deletions (%) . |
|---|---|---|---|
| CML | 250 | 23 | 9.2 |
| ALL Ph+ | 13 | 1 | 7.7 |
| AML M4 | 20 | 2 | 10 |
| ALL and AML with MLL gene rearrangements | 43 | 7 | 16.2 |
| AML M3 | 30 | 0 | 0 |
| AML M2 | 14 | 0 | 0 |
CML indicates chronic myeloid leukemia; ALL, acute lymphoid leukemia; Ph, Philadelphia chromosome; AML, acute myeloid leukemia.
FISH analysis of Ph+ samples was performed, using the commercially available double-fusion signal D-FISH BCR/ABLprobe (Oncor, Gaithersburg, MD). The ABL probe labeled with fluorescein isothiocyanate (green signal) is approximately 600 kilobase (kb), extending from an area well centromeric of the argininosuccinate synthetase (ASS) gene to telomeric of the lastABL exon, spanning the 200-kb breakpoint region ofABL. The BCR probe, directly labeled with Texas Red (red signal), is approximately 500 kb, beginning 5′ of the firstBCR exon and spanning the common breakpoints in both the major and minor bcr and extending well beyond the last exon. FISH analysis for the patients with the inv(16) was carried out, using Vysis (Vysis, Downers Grove, IL) LSI CBFB (Core Binding Factor Beta-subunit) dual-color probe. The Vysis LSI CBFB is a mixture of a 5′ CBFB probe directly labeled with the Spectrum Red fluorophore and a 3′ CBFB probe directly labeled with a Spectrum Green fluorophore. The 5′ CBFB probe is approximately 150 kb and is positioned centromeric to the inv(16) breakpoint region. The 3′ CBFB probe is approximately 170 kb in size and is positioned telomeric to the inv(16) breakpoint, and neither probe extend over the breakpoint. The Vysis LSI MLLprobe consists of a 350-kb portion centromeric of the MLLgene breakpoint region labeled in Spectrum Green and a 190-kb portion that is mostly telomeric of the breakpoint region and is labeled with Spectrum Orange. FISH analysis for the AML M3 patients was carried out with the Vysis LSI PML/RARA translocation probe. The LSIPML probe is approximately 180 kb and hybridizes to chromosome 15q22, and the LSI RARA probe that hybridizes to chromosome 17q12 is approximately 400 kb in size. FISH analysis of the patients with AML M2 was carried out by using the Vysis LSIETO/AML1 dual-fusion DNA probe. The ETO probe (Spectrum Orange) that hybridizes to chromosome 8q22 band is 480 kb, and the AML1 probe (Spectrum Green) that hybridizes to chromosome 22q22 is about 1.3 Mb. Probes were applied and detected according to manufacturer instructions. The slides were coded, and a total of 200 nuclei was analyzed by 2 observers in a blinded manner, using established scoring criteria4 and Vysis scoring criteria for other probes (LSI CBFB, LSI MLL, andAML1/ETO). For each sample found to contain an atypical hybridization pattern that may constitute deletion, a minimum of 10 abnormal metaphase cells was also analyzed by FISH to confirm the presence of deletion. For 7 of these samples, sequential G-banding and D-FISH analyses were performed.
Study design and statistical analysis
For the purpose of statistical analysis, CML patients were divided into 2 groups: patients with deletions, and a control group of patients without an apparent deletion. Duration of chronic phase, relapse rate following bone marrow transplantation (BMT), and survival time in patients with deletions were compared with those of the control group. The following situations resulted in patients from the control group being censored from survival study: if samples were sent from outside hospitals and clinical outcome was not available, if patients presented in blast crisis, and if patients died from unrelated complications of the disease or from transplantation-related complications. We acknowledge that this study is retrospective, covering a 15-year period, with all the inherent problems associated with lengthy studies. Patients included in the study group are derived from 3 institutions with different approaches to treatment, and the type of therapy changed considerably during the study period, as shifts to support transplant or interferon use were made. All statistical calculations were performed, using the S-Plus statistical package Version 5.1 (Mathsoft, Seattle, WA). CML patients with a deletion were compared to those without a deletion (control group) on prognostic parameters at diagnosis (Table 2) and on 3 clinical outcomes: survival time from presentation using the log-rank test, relapse rate after BMT using the Fisher exact test, and chronic-phase duration using the log-rank test.
Patients' clinical data
| Clinical characteristics . | Patients with deletions (n = 23) . | Patients without deletions (n = 163) . | Pvalues . |
|---|---|---|---|
| Sex (M/F) (%) | 60/40 | 61/39 | (Fisher exact test) |
| 1 | |||
| Median age, y (range) | 47 (15-64) | 51 (22-86) | (Wilcoxon rank sum test) |
| .042 | |||
| Peripheral blood prognostic markers* | (Wilcoxon rank sum test) | ||
| Hemoglobin (g/L) | 105.1 ± 26.5 | 116.2 ± 23.7 | .085 |
| White blood cell count (×109) | 181.8 ± 174.1 | 120.3 ± 143.3 | .057 |
| Platelet count (×109) | 768.3 ± 504.0 | 447.4 ± 369.3 | .003 |
| Basophils (%) | 2.4 ± 3.0 | 4.6 ± 3.9 | .039 |
| Blasts (%) | 1.0 ± 1.4 | 2.5 ± 5.2 | .107 |
| No. (%) of patients with poor prognostic markers† | 0 (0%) | 1 (1.9%) | 1.0 |
| Cytogenetics findings (%) | n = 23 | n = 126 | (χ2test) |
| Standard Ph translocation | 74 | 87 | .04 |
| Variant Ph translocation | 17 | 4 | |
| Additional abnormalities | 9 | 9 | |
| Type of treatment (%) | (χ2 test) | ||
| Chemotherapy alone | 18 | 28 | .6 |
| Chemotherapy and interferon | 4 | 8 | |
| Interferon alone | 26 | 22 | |
| BMT‡ | 52 | 42 | |
| Duration of chronic phase2-153 | n = 11 | n = 95 | (log-rank test) |
| 25 mo | 96 mo | .005 | |
| Relapse rate following BMT (%) | n = 12 | n = 58 | (Fisher exact test) |
| 41 | 3.4 | < .001 | |
| Survival time | 36 mo | 84 mo | (log-rank test) |
| .005 |
| Clinical characteristics . | Patients with deletions (n = 23) . | Patients without deletions (n = 163) . | Pvalues . |
|---|---|---|---|
| Sex (M/F) (%) | 60/40 | 61/39 | (Fisher exact test) |
| 1 | |||
| Median age, y (range) | 47 (15-64) | 51 (22-86) | (Wilcoxon rank sum test) |
| .042 | |||
| Peripheral blood prognostic markers* | (Wilcoxon rank sum test) | ||
| Hemoglobin (g/L) | 105.1 ± 26.5 | 116.2 ± 23.7 | .085 |
| White blood cell count (×109) | 181.8 ± 174.1 | 120.3 ± 143.3 | .057 |
| Platelet count (×109) | 768.3 ± 504.0 | 447.4 ± 369.3 | .003 |
| Basophils (%) | 2.4 ± 3.0 | 4.6 ± 3.9 | .039 |
| Blasts (%) | 1.0 ± 1.4 | 2.5 ± 5.2 | .107 |
| No. (%) of patients with poor prognostic markers† | 0 (0%) | 1 (1.9%) | 1.0 |
| Cytogenetics findings (%) | n = 23 | n = 126 | (χ2test) |
| Standard Ph translocation | 74 | 87 | .04 |
| Variant Ph translocation | 17 | 4 | |
| Additional abnormalities | 9 | 9 | |
| Type of treatment (%) | (χ2 test) | ||
| Chemotherapy alone | 18 | 28 | .6 |
| Chemotherapy and interferon | 4 | 8 | |
| Interferon alone | 26 | 22 | |
| BMT‡ | 52 | 42 | |
| Duration of chronic phase2-153 | n = 11 | n = 95 | (log-rank test) |
| 25 mo | 96 mo | .005 | |
| Relapse rate following BMT (%) | n = 12 | n = 58 | (Fisher exact test) |
| 41 | 3.4 | < .001 | |
| Survival time | 36 mo | 84 mo | (log-rank test) |
| .005 |
Ph indicates Philadelphia chromosome; BMT, bone marrow transplant.
Patients who were referred to the center after the treatment was initiated were censored from the peripheral blood prognostic markers study. For the rest of the patients (patients with deletions n = 16, patients without deletions n = 51), laboratory values were obtained at the diagnosis. Values are mean ± SD.
The presence at diagnosis of peripheral blood blasts (15%), peripheral blood basophils (20%), or thrombocytopenia < 100 × 109/L has been identified as features with independent adverse prognostic significance.54 55
One syngeneic transplantation was done in the group of patients with deletions, for the rest of the patients from both groups allogeneic transplantations were done.
Patients who underwent BMT were censored from the duration of chronic-phase study.
Sequence analysis
The analysis was initiated by identification of sequences available for the genes of interest using the National Center fro Biotechnology Information (NCBI) Entrez (http://www.ncbi.nlm.nih.gov/Entrez/),56 the Genome Database (GDB) (http://www.gdb.org/),57 and the Sanger center database search (http://www.sanger.ac.uk/DataSearch/).58Advanced Blast searches were performed against high-throughput genome sequences to identify the clones that encompass the genes of interest and the genomic sequences that flank the breakpoint regions. Sequences obtained from NCBI search and from Blast search hits were subjected to electronic polymerase chain reaction to search dbSTS (http://www.ncbi.nlm.nih.gov/genome/sts/epcr.cgi).59 STSs comparison helped to confirm the hits obtained from the Blast search. For the genes CBFB and MYH11 and the flanking regions, search of the Genome channel database was done as well (http://compbio.ornl.gov).60 For the genes BCR,ABL, MYH11, and CBFB, we were able to obtain sequences for the regions of 1.5-Mb length (5′of ABLand 3′ of BCR, MYH11, and CBFB) that included the genes of interest and the flanking regions. For chromosome 15 and 17, sequence data are limited, but we were able to construct contigs of sufficient length (500-700 Kb) for sequence analysis. DNA sequences were submitted to the repeat identification programs: Censor (http://www.girinst.org/Censor_Server-Data_Entry_Forms.html)61and Repeat Masker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker).62Sequences were also checked against the Blast database and then against the ESTs database and subjected to various gene prediction programs (GENESCAN, http://genes.mit.edu/GENSCAN.html; GRAIL,http://compbio.ornl.gov/Grail-1.3/; FGENES,http://genomic.sanger.ac.uk/gf/gf.shtml) to identify known and novel genes within the deleted regions for the chromosomes 9 and 22.
Results
FISH analysis
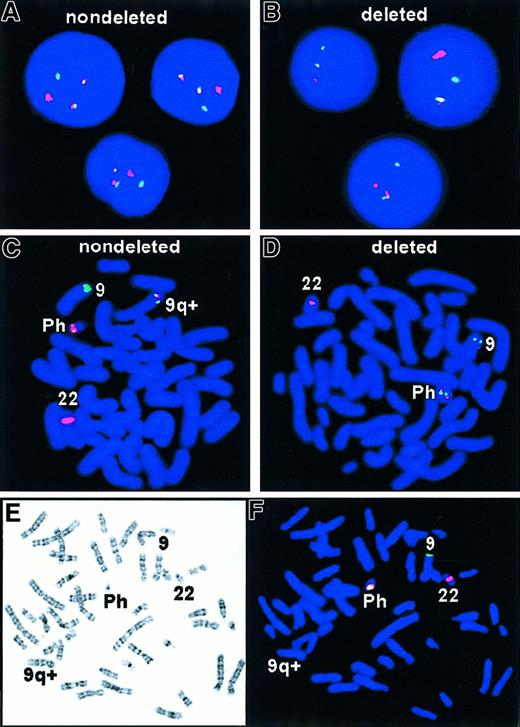
The expected pattern for Ph+ metaphase and interphase cells is shown in Figure 1A,C. The D-FISH probe is designed to produce a 3′ residual BCR signal on chromosome 22 and a 5′ residual ABL signal on chromosome 9. The reciprocal translocation involves the relocation of 3′ABL green probe region next to 5′ BCR red probe region on the Ph chromosome and 3′ BCR red probe to 5′ residual ABL probe on the derivative chromosome 9q+. The resultant FISH signal pattern for this translocation would be 2 yellow fusion signals, one on each of the derivative chromosomes: Ph chromosome and derivative chromosome 9q+, in addition to the single red and green signals expected from the normal chromosomes.
FISH analysis of t(9;22) with deletion.
R = red signal; G = green signal; F = fusion color signal (red/green or yellow). (A) Expected D-FISH signal pattern in Ph+ interphase, 1R1G2F. (B) Atypical D-FISH signal in Ph+ interphase with deletions, 1R1G1F. (C) Expected D-FISH signal pattern in Ph+ metaphase, 1R1G2F. (D) Atypical D-FISH signal in Ph+ metaphase with deletions, 1R1G1F. Colocalization of red and green signals (ABL and BCR) identified the Ph chromosome. A single red signal was seen on the normal chromosome 22 homologue and a single green signal on normal chromosome 9. (E,F) Sequential G-banding and D-FISH analysis of a Ph+ metaphase with deletions of 5′ ABL and 3′ BCR sequences and corresponding loss of the second fusion second signal on the derivative chromosome 9.
FISH analysis of t(9;22) with deletion.
R = red signal; G = green signal; F = fusion color signal (red/green or yellow). (A) Expected D-FISH signal pattern in Ph+ interphase, 1R1G2F. (B) Atypical D-FISH signal in Ph+ interphase with deletions, 1R1G1F. (C) Expected D-FISH signal pattern in Ph+ metaphase, 1R1G2F. (D) Atypical D-FISH signal in Ph+ metaphase with deletions, 1R1G1F. Colocalization of red and green signals (ABL and BCR) identified the Ph chromosome. A single red signal was seen on the normal chromosome 22 homologue and a single green signal on normal chromosome 9. (E,F) Sequential G-banding and D-FISH analysis of a Ph+ metaphase with deletions of 5′ ABL and 3′ BCR sequences and corresponding loss of the second fusion second signal on the derivative chromosome 9.
During routine dual-color BCR/ABL FISH analysis of CML samples, an atypical hybridization pattern was detected in which only one fusion signal was present. (Figure 1B,D). To extend this observation, a large series of 250 CML samples and 13 ALL samples were studied by interphase FISH analysis. Twenty-three of 250 (9.2%) CML samples and 1 of 13 (7.7%) ALL Ph+ samples had one green, one red, and one fusion signal. Sequential G-banding and FISH analysis confirmed that the single fusion signal was derived from the Ph chromosome and that the second fusion signal failed to appear on the derivative chromosome 9 (Figure 1E,F). This analysis indicated that the probe distal to BCR from 22q11 and part of the probe proximal to ABL from 9q34 were unable to hybridize to the 9q+ derivative chromosome, suggesting that submicroscopic deletions extending toward the centromere and spanning at least 600 kb 5′ to ABL gene and also extending about 500 kb 3′ toBCR genes were the cause of the observed atypical hybridization pattern. In all 23 cases with deletions, only 2 populations of cells were observed: cells exhibiting a normal hybridization pattern (2 red and 2 green signals) and cells with an atypical hybridization pattern (one red, one green, and one fused signal) consistent with the deletions of both 5′ ABL and 3′BCR on the derivative chromosome 9. Furthermore, the atypical hybridization pattern was present in all observed metaphases of the diagnostic samples from all 23 patients, suggesting that the deletion was a primary event accompanying the formation of the Philadelphia translocation.
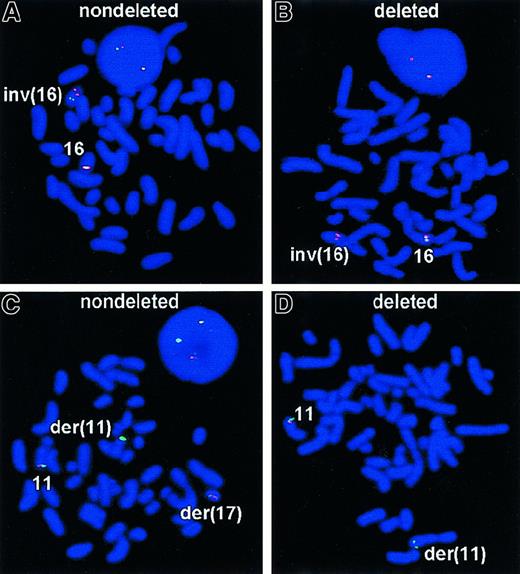
Having determined that submicroscopic deletions often accompany t(9;22), we investigated whether other leukemia-associated chromosomal rearrangements also lead to deletions. Therefore, we tested 20 leukemia preparations with the chromosomal aberration inv(16), which appeared to be balanced by conventional cytogenetic analysis. This inversion is seen in a number of French-American-British subclasses, but it is most commonly associated with acute myelomonocytic leukemia with abnormal eosinophils, M4Eo. It results in the creation of a fusion between the myosin heavy chain gene (MYH11) on the short arm (p13) and the gene for a transcription factor, core binding factor beta (CBFB) on the long arm (q22). The 5′-CBFB/MYH11-3′ fusion gene at 16p13, rather than the reciprocal 5′-MYH11/CBFB-3′ at 16q22, is the critical product for chromosome 16–related leukemogenesis.5 6 When hybridized with the Vysis LSI CBFB dual-color probe, the normal 16q22 region will be seen as 2 adjacent or fused red/green signals, which sometimes may appear yellow. Hybridization with the LSICBFB to a metaphase preparation containing an inv(16) results in red and green signals appearing on opposite arms of the inverted chromosome 16 (Figure 2A). Two of 20 patient samples with an inv(16) had an abnormal hybridization pattern, providing evidence that at least 170 kb of the region that is telomeric to CBFB gene is deleted with this inversion (Figure 2B).
FISH analyses of inv(16) and
MLL deletions. With the LSI CBFBdual-color probe, the normal 16q22 region appears as adjacent red-green or a fused yellow signal. On a metaphase preparation, the inv(16) will have red and green signals on opposite arms of the aberrant chromosome. (A) Expected signal pattern with the LSI CBFB of a metaphase containing an inv(16), with red and green signals appearing on opposite arms of the inverted 16 chromosome. (B) Atypical signal pattern with LSI CBFB of a metaphase containing inv(16) and deletion of 3′ sequences of CBFB gene. Spectrum Green–labeled probe failed to hybridize to the 16q22 region, providing evidence that at least 170 kb of the region 3′ to CBFB gene is deleted with this inversion. (C) Interphase nucleus and metaphase cell showing the results of LSI MLL hybridized to a specimen possessing a t(11;17)(q23;p13). As expected for a rearrangement at theMLL breakpoint, the orange signal has moved to the p arm of chromosome 19, and the green signal has remained on the q arm of chromosome 11. On the normal chromosome 11 the LSI MLL probe green and orange signals (fused yellow) remain unchanged. The interphase nucleus showed one green/orange fused signal representing the normal chromosome 11 and separate green and orange signals representing the translocation chromosomes. (D) Atypical signal pattern with LSI MLL of a metaphase containing t(9;11)(p22;q23) accompanied by the deletion of a 3′ region of MLL gene with the corresponding loss of orange signal. Spectrum Orange–labeled probe failed to hybridize to the 3′ region of MLL gene, providing the evidence that at least 190 kb of the region that is telomeric to the gene was deleted.
FISH analyses of inv(16) and
MLL deletions. With the LSI CBFBdual-color probe, the normal 16q22 region appears as adjacent red-green or a fused yellow signal. On a metaphase preparation, the inv(16) will have red and green signals on opposite arms of the aberrant chromosome. (A) Expected signal pattern with the LSI CBFB of a metaphase containing an inv(16), with red and green signals appearing on opposite arms of the inverted 16 chromosome. (B) Atypical signal pattern with LSI CBFB of a metaphase containing inv(16) and deletion of 3′ sequences of CBFB gene. Spectrum Green–labeled probe failed to hybridize to the 16q22 region, providing evidence that at least 170 kb of the region 3′ to CBFB gene is deleted with this inversion. (C) Interphase nucleus and metaphase cell showing the results of LSI MLL hybridized to a specimen possessing a t(11;17)(q23;p13). As expected for a rearrangement at theMLL breakpoint, the orange signal has moved to the p arm of chromosome 19, and the green signal has remained on the q arm of chromosome 11. On the normal chromosome 11 the LSI MLL probe green and orange signals (fused yellow) remain unchanged. The interphase nucleus showed one green/orange fused signal representing the normal chromosome 11 and separate green and orange signals representing the translocation chromosomes. (D) Atypical signal pattern with LSI MLL of a metaphase containing t(9;11)(p22;q23) accompanied by the deletion of a 3′ region of MLL gene with the corresponding loss of orange signal. Spectrum Orange–labeled probe failed to hybridize to the 3′ region of MLL gene, providing the evidence that at least 190 kb of the region that is telomeric to the gene was deleted.
To determine how frequent submicroscopic deletions were in other recurrent chromosomal rearrangements of leukemia, we studied 43 patients with ALL- or AML-associated MLL translocations, 14 patients with t(8;21), and 30 AML-M3 patients with t(15;17). In each class of rearrangement, the experiments were carried out with dual-color LSI FISH probes. Figure 2C shows the expected signal pattern for the LSI MLL probe. As expected, hybridization with LSIMLL probe to metaphase preparation positive forMLL translocation displays a distinct orange signal that has moved to the translocation partner chromosome and a green signal that remains on the q arm of chromosome 11. On the nonrearranged normal chromosome 11, the LSI MLL probe displays a fused green and orange signal (yellow). Thus positive metaphases exhibit one yellow fused signal, representing the normal chromosome 11, and separate green and orange signals, representing the MLL translocation. In this component of the study, 7 of 43 patients (16%) exhibited an abnormal hybridization pattern in which the Spectrum Orange–labeled probe failed to hybridize to the 3′ region of MLL gene, providing evidence that at least 190 kb of the region that is telomeric to the gene was deleted (Figure 2D). We found no examples of deletions associated with t(15;17) PML/RARA or t(8;21)AML1/ETO rearrangements.
Cytogenetics
Cytogenetic analysis was performed to confirm that deletions were not due to more complex cytogenetic aberrations or gross interstitial deletions adjacent to the point of chromosomal rearrangement. Nineteen of 23 CML patients with the deletions had a standard (9;22) translocation, which appeared to be balanced with no evidence of deletions on the derivative chromosome 9, as determined at the routine banding level (350-400 bands). The remaining 4 patients had a variant Ph translocation with other partner chromosomes being involved in what appeared to be balanced rearrangements. Cytogenetic analysis of metaphase cells derived from the 2 inv(16) samples in which a deletion was present and the 5 11q23 rearrangements in which the MLLgene had undergone translocation demonstrated balanced chromosomal rearrangements without any apparent deletion. This analysis suggested that loss of genomic material is below the level of resolution of classical cancer cytogenetic methods.
Clinical outcome data analysis
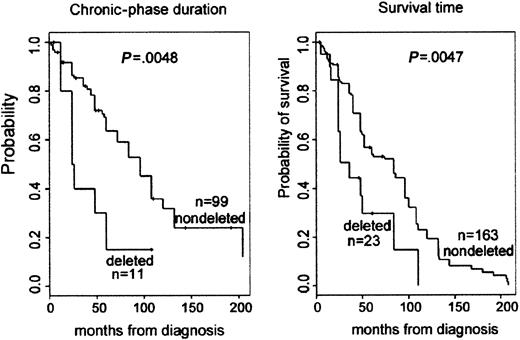
Clinical outcome data of the CML patients was accumulated and analyzed to assess biological and clinical relevance of the deletions close to the ABL and BCR genes associated with the Ph translocation. Clinical outcome data, such as duration of chronic phase, mode of treatment, response to treatment, relapse of disease following BMT, and survival data, were collected for all patients with deletions and for patients from the matched control group. A retrospective assessment of prognosis using the Sokal score was not possible because our institute functions as a major referral center for patients in Eastern Canada. Therefore, many patients are assessed months after diagnosis, and, unfortunately, initial spleen size measurements were usually not noted. Analysis of peripheral blood prognostic markers at diagnosis showed that there was minor difference between the percentages of platelets when the deleted group was compared to controls. However, analysis of subsequent clinical outcome revealed a statistically significant association between the presence of the deletion and a poor prognosis (Table 2). The difference in duration of chronic phase was significant: log-rank test provided aP = .005, with a median duration (by Kaplan-Meier estimator) of 96 months for patients with no deletion and 25 months for patients with a deletion. The relapse rate following BMT was 2 of 58 (3.4%) for the control group and 5 of 12 (41%) for the patients with deletion (P < .001, Fisher exact test). For survival time, the median survival time (by Kaplan-Meier estimator) for the patients with no apparent deletion was 84 months, whereas those patients with a deletion had a median survival time of only 36 months. This difference was significant; the log-rank test provided aP value of .005 (Figure 3).
Kaplan-Meier plot demonstrating reduced survival and duration of chronic phase for patients with deletions.
Kaplan-Meier plot demonstrating reduced survival and duration of chronic phase for patients with deletions.
AML associated with the inversion chromosome 16, inv(16) (p13q22), has a favorable prognosis and is known to be chemosensitive. In keeping with this observation, all patients in our study without a 3′CBFB deletion achieved remission within a short period of time and are off treatment at the moment. Two patients with a deletion had unusually aggressive disease. One patient did not respond to treatment and died 5 months after diagnosis. The second patient had a transient remission and relapsed 1 month later. Because of the small sample size of this study group, a statistical analysis is not possible.
Sequence analysis
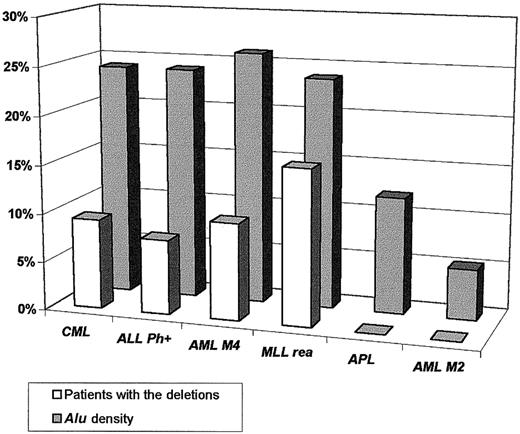
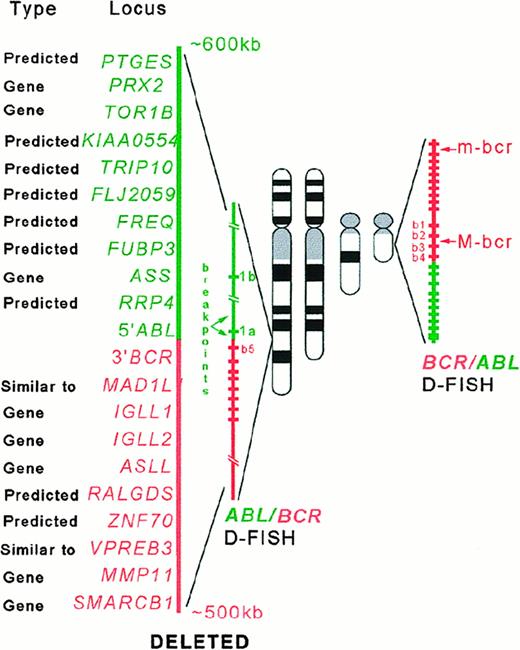
Sequence data analysis suggested a significant association between the presence of the deletions and the high density of theAlu repeats in the regions containing the genes involved in the chromosomal rearrangements. Alu sequences are overrepresented in the DNA sequences of the genes involved in the t(9;22) (ABL and BCR), inv(16) (CBFBand MYH11), MLL gene, and their flanking regions compared with their density in the genes involved in the t(15;17) (PML and RARA), t(18;21) (ETO andAML), and overall incidence throughout the genome (5%-10%)7 (Figure 4). Preliminary DNA sequence analysis of the minimally deleted region (as defined by D-FISH probe on chromosomes 9 and 22) allowed us to identify either known or predicted genes (with similarity to known genes) that map to the deleted region (Figure 5). Some of these genes could in some circumstances function as a tumor suppressor gene and possibly modify aspects of the disease phenotype of CML. For example, deletion of the gene similar to MAD1L1gene on chromosome 22 could possibly lead to abrogation of the mitotic cell cycle checkpoint.8 Homozygous deletions ofSMARCB1 (SWI/SNF-related gene) were identified in rhabdoid tumors, choroids plexus carcinomas, and loss of heterozygosity in breast cancers, Wilms tumors, gliomas, sarcomas, and other tumor types.9-11 Evolutionary conserved, the SWI/SNF complexes, which have been identified from yeast to humans, play an important role in the remodeling of chromatin structure.12-14 Altered chromatin organization at specific DNA sites may be crucial in the process of oncogenesis. The prostaglandin E synthase (PTGES) encoding gene that maps to the chromosome 9–deleted region was first identified as a p53-induced gene.15PTGES encodes redox-controlling protein, which is a potent inducer of apoptosis.15 Altered apoptosis may play a critical role in the disease progression in CML.16-18 Similarly, it was proposed that the thyroid hormone receptor interactor 10 (TRIP 10) gene that maps 5′ to ABL gene is a downstream target of activated GTP-bound CDC42 and may act as a link between CDC42 signaling and regulation of the actin cytoskeleton19—one of the components ofBCR/ABL signaling pathway.20-22
Correlation between the presence of the deletions associated with chromosomal rearrangements and the density of the
Alu repeats in the genes involved in the rearrangements and their flanking regions.
Correlation between the presence of the deletions associated with chromosomal rearrangements and the density of the
Alu repeats in the genes involved in the rearrangements and their flanking regions.
Schematic representation of the
ABL/BCR and BCR/ABLrearrangements with D-FISH configuration and the genes (known and predicted) that map to the deleted regions.
Schematic representation of the
ABL/BCR and BCR/ABLrearrangements with D-FISH configuration and the genes (known and predicted) that map to the deleted regions.
Discussion
In this study we have found that submicroscopic deletions are more commonly associated with recurrent chromosomal rearrangements of leukemia than previously suspected. Our findings confirm and extend those reported previously for CML,3 23 which showed that the deletions probably involve several hundred kilobases of DNA and many different genes. We have identified 23 (9.2%) new cases of CML with deletions from a study group of 250 and have shown that the deletions affect 2 regions: proximal to the rearranged ABLand distal to BCR gene on the 9q+ derivative chromosome. We found that patients with deletions (who did not undergo BMT) had a shorter chronic phase and overall survival time in comparison to a similar group of patients without apparent deletions. For those patients who underwent BMT, a statistically significant difference was observed in the relapse rate following transplantation. Patients with deletions had a significantly higher frequency of early relapses within the first 12 months; patients without deletions had lower relapse rates and no first-year relapses. Although patients with the deletions have poorer prognosis when compared to the CML patients without the deletion, these 2 groups of patients exhibited only minor differences indistinguishable using conventional clinical prognosticators at diagnosis. Thus, it is important that the laboratory work-up of all new CML patients should involve a careful FISH analysis to identify those patients with a deletion who may subsequently have an elevated risk of poor response to treatment. In light of potential clinical implications of these findings, it is also important that diagnostic laboratories engaged in routine FISH analysis are cognizant of the presence of a relatively high frequency of submicroscopic deletion associated with these recurrent chromosomal rearrangements. We have studied 13 cases of Ph+ ALL and detected one example of a deletion, suggesting that the mechanism of submicroscopic deletion is not disease specific but more probably associated with the translocation process itself.
To determine the frequency of cryptic deletions, other leukemia-associated recurrent chromosomal rearrangements were studied. Of these leukemias, the AML M4Eo samples with inv(16) also had submicroscopic deletions not previously reported. It is well established that there are often deletions 3′ from MYH11, involving a region of 160-350 kb centromeric to the 16p13 short-arm inversion breakpoint cluster region.5,6,24 However, we found 2 examples of deletions telomeric to the CBFBbreakpoint cluster region at band 16q22. For both of these deletion patients, it is noteworthy that each had an unusually unfavorable disease course. In our study we also detected deletions 3′ to theMLL gene associated with translocations t(9;11), t(4;11), and t(11;17) as reported previously.25 26 The reason why deletions are so commonly associated with inv(16) and MLLgene rearrangements is poorly understood but may be mechanistically similar to the deletions observed for ABL/BCR. In our small study group we found no example of deletions associated withPML/RARA or AML1/ETO rearrangements, suggesting that large deletions may not be such a frequent feature of all chromosomal rearrangements of leukemias.
All Ph deletions in this study were detected in all the abnormal metaphases of diagnostic patient samples, suggesting that the deletion most likely arose simultaneously with the initial translocation or inversion event. In keeping with other studies we found that the prevalence of deletions proximal to the rearranged ABL and distal to BCR was significantly higher in patients with variant Ph chromosomes in comparison to the deletion rate in standard Ph translocations (P = .004).3,23,27 This observation would support a mechanism whereby the complex molecular rearrangements that occur during the formation of variant Ph translocations are associated with an increased probability of deletion. This higher incidence of concomitant deletion in variant Ph translocations may explain some of the variable results obtained in the assessment of the clinical effect of variant translocations in CML.28
Our findings are the first to implicate a process of deletion that is not disease specific per se but more likely is determined by factors that influence the somatic process of chromosomal rearrangement itself. Several potential molecular mechanisms have been proposed to explain nonrandom leukemia-associated chromosomal rearrangements,1including illegitimate V(D)J recombination,29 homologous recombination mediated via Alu elements,30,31translin activity,32,33 cleavage at sites of Z-DNA structure,34 topoisomerase II subunit exchange,35 repair of DNA breaks with nonhomologous chromosome, and presence of fragile sites.36 37 These mechanisms are not mutually exclusive, so any molecular event leading to chromosomal rearrangement may in fact involve more than one of these mechanisms.
Since deletions are associated with different chromosomal rearrangements, it is possible that the mechanism for deletion formation may be dependent on the nature of sequences that flank translocation breakpoint regions. We, therefore, examined the distribution of Alu repeats in the vicinity of the genes subject to rearrangement since Alu sequences are known to facilitate recombination processes. It has previously been proposed that reciprocal rearrangement such as those underlying the formation of complex BCR/ABL genes in cells of CML patients might be mediated by mutual attraction of Alu sequences located in heterologous chromosomes.30 Jeffs et al30showed that the 3′ part of M-Bcr recombined within, or immediately adjacent to, Alu elements at the additional sites of the variant chromosome partners in all 5 complex BCR-ABLrearrangements examined. This finding suggests that Alusequences may facilitate the BCR/ABL recombination process in variant translocations and provides intriguing evidence that these elements may play a direct role in the recombination process inBCR-ABL translocations. Sequence analysis ofBCR-ABL and/or ABL-BCR recombination products from 21 standard t(9;22) rearrangements of CML38,39analysis of M-Bcr40 as well as minor breakpoint41,42 has provided indications thatAlu repeat elements may play a role in the recombination process, since Alu elements were present at or adjacent to the breakpoints on the chromosome 22. Members of the Alurepetitive DNA family are often found at illegitimate junctions such as deletion or translocation breakpoints.31,43 FlankingAlu elements may undergo homologous base pairing so that recombination is more likely to occur in their vicinity. A requirement of the double-strand recombination model44 is that small deletions (up to several bases) can be generated by translocations. The precise mechanism, which underlies creation of the more extensive deletions observed in this and other studies,3,23 remains to be defined. An attractive hypothesis is that Alu elements mediate homologous recombination and may also be responsible for generating more extensive deletions. In this model the deletions that accompany translocation formation may arise as a result of base pairing of fortuitously located direct repeats, which may be several hundred kilobases away. When the double-strand DNA broken ends are ligated, the intervening DNA fragment between the direct repeats is deleted. Our analysis of the density of repetitive sequences in the regions subject to deletion supports the notion that an increased density ofAlu elements widely distributed either side of the breakpoint regions could facilitate homologous recombination with deletion (Kolomietz et al, manuscript in progress). The observation by Sinclair et al3 that the deletions have variable breakpoints also supports the hypothesis, since Alu-mediated base pairing need not always be exact. Because of the wide distribution of Alu elements in this region, pairing could occur at some distance from the ABL/BCR region. Thus, it would be expected that diverse Alu-rich sites in the regions flanking theABL and BCR genes could facilitate recombination and lead to variable segments of intervening DNA being deleted.
The differences in the clinical behavior of CML patients with deletions observed in this study and by others3 suggest that the concurrent haploinsufficiency of one or more genes in the 2 flanking regions may be responsible. In keeping with this idea, it is noteworthy that deletion and loss of heterozygosity of these regions have previously been observed in several different types of tumors45-53 (see also Breakpoint Map of Recurrent Chromosome Aberrations at CGAP, the Cancer Genome Anatomy Project:http://www.ncbi.nlm.nih.gov/CCAP/mitelsum.cgi).63 In common with strategies pursued for other deletion syndromes, further extensive molecular studies are necessary to delineate the minimally deleted region and to localize the genes within the region that may be causative. More detailed characterization of the genes that map to the minimally deleted regions of 9q34, 22q11, 16q22, and 11q23 will help to understand how the disease course can be modified by chromosomal losses in these regions.
We are indebted to Dr Mark Minden, Dr Suzanne Kamel-Reid, and Jane Bayani for critically reviewing this manuscript and to Ben Beheshti and Lada Vorobyova for technical assistance as well as all members of the Cancer Cytogenetics program at the Banting Institute, Toronto.
Supported by the Canadian Cancer Society and National Cancer Institute of Canada.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jeremy A. Squire, Division of Cellular and Molecular Biology, Ontario Cancer Institute, Princess Margaret Hospital, 610 University Ave, Rm 9-721, Toronto, Ontario, M5G 2M9 Canada; e-mail: jeremy.squire@utoronto.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal