The BCR-ABL chimeric gene results from a reciprocal translocation, t(9;22)(q34;q11). The breakpoint on chromosome 9 is in the majority of cases 5′ to ABL exon 2, whereas the breakpoints on chromosome 22 can occur in various regions within theBCR gene, giving rise to different BCR-ABLjunction types. Breakpoints within the so-called major BCR (M-BCR) lead to a BCR-ABL messenger RNA with e13a2 or e14a2 junction in 99% of patients with chronic myeloid leukemia (CML). Two other breakpoint types outside the M-BCR have been described: the m-BCR (e1a2 junction), mainly detected in Ph+ acute lymphoblastic leukemia, and the μ-BCR(e19a2 junction) associated with neutrophilic-CML.1-6
We report a novel BCR-ABL fusion transcript with e15a2 junction detected in 2 patients with a presumptive diagnosis of atypical chronic myeloproliferative disorder.7 Patient 1, a 65-year-old male with an 8-year history of persistent unexplained neutrophilic leukocytosis (15 × 109/L) was referred to us in December 1998 with a 5-month history of recurrent fever, weight loss, fatigue, and anorexia, with no apparent evidence of infectious diseases. Blood film examination showed 74% neutrophils with no shift to the left in the granulocytic lineage, 24% lymphocytes, and 2% monocytes with enlarged platelets. The patient received no treatment and currently remains asymptomatic with persistent moderate leukocytosis. Patient 2, a 59-year-old male with a history of diabetes, hypertension, and alcoholism, was referred to our hospital in September 1998 because of leukocytosis (20 × 109/L) detected in a routine blood testing. Blood film showed megaloblastic anemia and neutrophilic leukocytosis with no shift to the left. Serum tests showed low iron levels and increased ferritin (1000 ng/mL). Bone marrow smear and biopsy disclosed dysplastic features and maturation defects of the erythropoietic lineage and a hypogranular granulocytic series. Neutrophilic leukocytosis persisted with no other relevant associated symptoms until last control (September 1999) after which the patient was lost to follow-up. More detailed clinical and laboratory features of the 2 patients are summarized in Table1.
Patient clinical and laboratory features
| . | Patient 1 . | Patient 2 . |
|---|---|---|
| Age | 65 | 59 |
| Karyotype | 46, XY (20/20) | 46, XY (20/20) |
| Hemoglobin (g/dL) | ND | 10.7 |
| White cell count (/L) | 15 × 109 | 20 × 106 |
| Platelet count (/L) | 400 × 109 | 527 × 109 |
| Bone marrow | ND | Dysplastic features |
| NAP score | 70 | 39 |
| Splenomegaly | No | No |
| . | Patient 1 . | Patient 2 . |
|---|---|---|
| Age | 65 | 59 |
| Karyotype | 46, XY (20/20) | 46, XY (20/20) |
| Hemoglobin (g/dL) | ND | 10.7 |
| White cell count (/L) | 15 × 109 | 20 × 106 |
| Platelet count (/L) | 400 × 109 | 527 × 109 |
| Bone marrow | ND | Dysplastic features |
| NAP score | 70 | 39 |
| Splenomegaly | No | No |
ND indicates not determined; NAP, neutrophil alkaline phosphatase.
Cytogenetic analyses were performed in bone marrow cells and showed in both cases a normal karyotype (46,XY), with no evidence of the Philadelphia chromosome. Molecular analyses were performed on total cellular RNA from peripheral mononuclear blood. RNA isolation, the reverse transcription–polymerase chain reaction (RT-PCR) assay for detection of BCR-ABL transcripts and semiquantitativeG3PDH expression (to ensure the efficiency of the reverse-transcription step and to confirm successful amplification of intact complementary DNA) were performed according to previously reported protocols.8-10
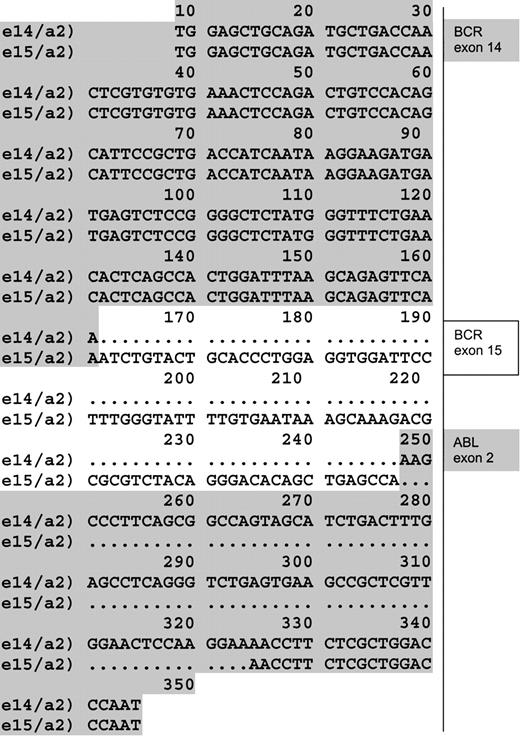
In both patients, no PCR products were detected after the first PCR round. False-negative results were ruled out, due to successful amplification of the control gene.9 10 But an unpredicted product distinct from the already described BCR-ABL fusion transcripts was detected after nested PCR (with a sensitivity of 10-5 to 10-6 μg of RNA). Direct sequencing of this fragment revealed a previously unreported BCR-ABLhybrid originated from an e15a2 junction. In fact, 86 base pairs of BCR exon 15 were joined in frame with ABL exon 2 at nucleotide 78 (GenBank accession number GI7406986). The difference between total base pairs gained and lost was 9, or 3 amino acids (Figure 1). Thus translation of such an e15a2 transcript would result in a protein that is only 3 amino acids longer than the classical P210 BCR-ABL, though quite different in the composition of amino acid residues (Figure2).
e15a2 sequence.
Alignment of a sequence obtained from a junction e14/a2 found in the cell line K562 (first line) and the atypical e15/a2 sequence obtained from patients (second line). (GenBank acession number GI7406986.)
e15a2 sequence.
Alignment of a sequence obtained from a junction e14/a2 found in the cell line K562 (first line) and the atypical e15/a2 sequence obtained from patients (second line). (GenBank acession number GI7406986.)
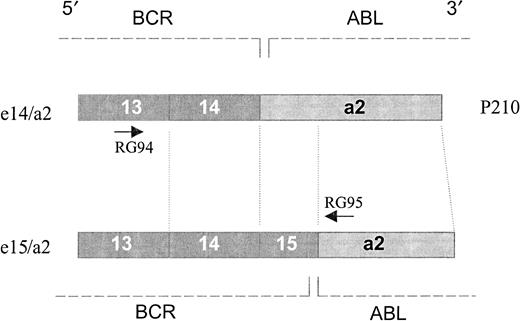
Protein scheme.
The scheme represents a structural view of the translated products from e14/a2 and e15/a2 junctions. Translation of e15/a2 transcript would result in a protein that is only 3 amino acids longer than the classical P210 BCR/ABL, although quite different in the composition of amino acid residues. The primers used are indicated by arrows.
Protein scheme.
The scheme represents a structural view of the translated products from e14/a2 and e15/a2 junctions. Translation of e15/a2 transcript would result in a protein that is only 3 amino acids longer than the classical P210 BCR/ABL, although quite different in the composition of amino acid residues. The primers used are indicated by arrows.
Some rare BCR-ABL fusion types have occasionally been reported in the literature.4 The 2 cases hereby described were identified among 225 patients with myeloproliferative syndromes studied in our laboratory. To the best of our knowledge, these are the first reported cases with a variant BCR-ABLfusion transcript involving BCR exon 15. Interestingly, such molecular aberration was only detected in a minor clone after nested PCR and was associated with mild clinical symptoms (moderate leukocytosis that did not increase with time) and an indolent clinical course.
As to the reasons underlying the detection of very low levels of e15a2 transcript in our patients, we believe that the successful amplification of the endogenous G3PDH may rule out the occurrence of technical problems. Two other explanations that may account for this features include (a) the presence of a small subpopulation of BCR-ABL–expressing cells and(b) a low level of BCR-ABL transcript expressed by the entire cell population. In light of the absence of the Philadelphia chromosome after conventional karyotypic analysis, we are more prone to favor the former hypothesis.
Recently, the presence of BCR-ABL transcripts only detectable at the RT-PCR level has been associated with some cases of Ph− essential thrombocytemia,11 though these findings were not confirmed by others.12 13
In conclusion, the novel BCR-ABL fusion type hereby described and/or its low levels detected might be associated with a phenotype of mild leukocytosis. But more cases showing these molecular and clinical features should be described before such a hypothesis is confirmed. The identification of new transcripts confirms the heterogeneity of breakpoints in BCR-ABL rearrangements. The roles of different BCR-ABL fusion proteins and their relationships to distinct leukemic or indolent phenotypes still deserve further investigation.
We thank Drs Francesco Lococo and Gustavo Folle for valuable discussions and critical reading of the manuscript. In addition, we are grateful to Dr Cristina Mogdasy, head of Clinical Laboratory, Asociación Española Primera de Socorros Mutuos, for her support.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal