Severe iron overload usually develops in patients with hereditary hemochromatosis (HHC), but variability in the phenotypic expression of the disease has been reported. This study assessed whether tumor necrosis factor α (TNF-α) plays a role in phenotypic expression of HHC. Sixty-four patients with HHC and 172 healthy volunteers (controls) were studied. Release of TNF-α from stimulated peripheral blood monocytes was measured by enzyme-linked immunosorbent assay, and 308 and 238 TNF-α polymorphisms were detected with polymerase chain reaction and restriction fragment-length polymorphism analysis. The relation between TNF-α polymorphisms and clinical expression of HHC was evaluated. Patients with HHC released less TNF-α than controls, but the difference was significant only in homozygotes for the C282Y mutation. The prevalence of the 308 TNF-α polymorphism was similar in patients and controls, whereas the prevalence of the 238 polymorphic allele was significantly lower in patients (3% versus 16%;P = .002). A lower prevalence of cirrhosis was observed in patients with TNF-α polymorphism than in those without it (4 of 15 [27%] versus 28 of 49 [57%]), but the difference was not significant (P = .07). In nonhomozygotes for the C282Y mutation, severe liver siderosis was less prevalent in patients with the 308 polymorphism than in those without it (P = .05). Alanine aminotransferase (ALT) values were significantly lower in patients with TNF-α polymorphism (P = .006), even when patients with other hepatotoxic factors were excluded. Multivariate analysis showed that TNF-α polymorphism was independently associated with ALT values (P = .0008 and P = .045, respectively, in homozygotes and nonhomozygotes for the C282Y mutation) and siderosis in nonhomozygotes (P = .047). Thus, TNF-α appears to play a role in HHC by modulating the severity of liver damage.
Introduction
Hereditary hemochromatosis (HHC) is characterized by progressive iron overload in parenchymal tissue that may lead to hepatic cirrhosis. The disease is associated with a high risk of development of hepatocellular carcinoma, cardiomyopathy, diabetes, hypogonadotropic hypogonadism, skin hyperpigmentation, and arthritis. In most patients (more than 80% of patients of Northern European descent and 64% of patients in Italy), HHC is due to homozygosity for the point mutation C282Y in the HFE gene. The role of the second mutation, H63D, in the pathogenesis of the disease is still uncertain.1,2 Half of the subjects homozygous for the C282Y mutation identified in epidemiological studies did not have clinical features of HHC, and about one third did not have evidence of iron overload.3 This suggests the existence of various acquired and genetic factors that can modify the phenotype of HHC. Involvement of genetic factors was also suggested by the greater concordance between clinical manifestations and biochemical markers of iron within families than between families.4
Tumor necrosis factor α (TNF-α), a cytokine with multiple functions that is mapped on 6p21.3 in the major histocompatibility complex, contributes to the regulation of iron metabolism by stimulating synthesis of ferritin and the transferrin receptor in different cell types,5,6 inhibiting iron release from peritoneal macrophages,7 and decreasing plasma iron uptake in erythroid precursors.8 Two polymorphisms have been identified in the TNF-α promoter—one at position 308 (the TNF2 allele)9 and the other at position 238 (the TNFA allele).10 Studies with the TNF-α promoter showed that the TNF2 allele leads to increased constitutive and inducible expression compared with the wild type (TNF1),11,12whereas data on the TNFA allele were conflicting.13-15 The presence of TNF-α polymorphisms appears to influence the prognosis of hepatic diseases with various causes (autoimmune and alcoholic).14 16-22
Macrophages from patients with HHC have repeatedly been reported to have a defect in iron metabolism, since a low iron content is usually found in circulating monocytes, their precursors, and Kupffer cells. In addition, monocytes from patients with HHC release an increased amount of ferritin after erythrophagocytosis because they are unable to store iron.23-25 A possible link between TNF-α and HHC has been hypothesized on the basis of the observation that TNF-α release from lipopolysaccharide (LPS)-stimulated monocytes from patients with HHC was lower than in monocytes from controls, and it was suggested that this could account for the low iron content in monocyte-macrophage cells from patients with HHC.26-28
The aim of this study was to establish whether TNF-α plays a role in the phenotypic expression of HHC. To achieve this aim, we studied TNF-α release from macrophages from patients with HHC typed for HFE gene mutations, the prevalence of TNF-α polymorphism at positions 238 and 308, and the relation between TNF-α polymorphism and clinical expression of HHC.
Patients, materials, and methods
Patients
Of patients with HHC in a larger series, we studied 64 unselected, unrelated subjects for whom DNA and liver biopsy results were available and geographical origin was ascertained up to the third generation. Forty-eight (75%) were men and 16 (25%) were women; their mean (± SD) age was 53.8 ± 11.2 years. The diagnosis of HHC was based on standard criteria: (1) exclusion of other known causes of iron overload; (2) hepatocellular hemosiderin deposits of grade III or IV according to the definition of Scheuer et al,29 with evidence of a gradient of siderosis from periportal hepatocytes to centrolobular veins; and (3) hepatic iron index (HII) greater than 1.9 or amount of iron removed by weekly phlebotomy to reach depletion more than 5 g in men and more than 3 g in women. Forty-six patients were from Northern Italy and 18 were from Southern Italy.
All patients were tested for HFE gene mutations; examined for liver and spleen enlargement with use of abdominal ultrasonography; and assessed for skin hyperpigmentation, diabetes (World Health Organization criteria), cardiomyopathy (electrocardiographic and echocardiographic examinations), hypogonadism (clinical and hormonal evaluations), and arthropathy (clinical and radiographic evaluations). All patients were screened for hepatitis B virus (HBV) surface antigen, anti–hepatitis C virus (HCV) antibodies, and HCV RNA, and assessed for alcohol abuse, which was defined as a daily intake of more than 60 g in men and more than 40 g in women.
Liver biopsy specimens, which were obtained from all patients, were processed routinely. Tissue sections were stained with hematoxylin and eosin, impregnated with silver to examine the reticulin framework, and stained with periodic acid–Schiff for glycogen, periodic Schiff diastase for nonglycogen proteins, Perls stain for iron, and trichrome for collagen. Liver iron concentration (LIC) was determined by atomic absorption spectrophotometry as described previously,30and the HII, ratio of LIC (μM/g dry weight) to age (years), was calculated. The amount of iron removed was determined as reported previously.31
Controls
The control group, which consisted of 172 healthy subjects with the same geographical origin as the patients, was formed by enrolling volunteers from among hospital staff, medical students, and nonblood relatives of the patients. HFE gene mutations in most of these subjects were reported previously.2
Isolation of peripheral blood monocytes
Peripheral blood monocytes were obtained from patients with iron overload and controls by using the Lymphoprep-Percoll method (Nycomed Amersham, Little Chalfont, United Kingdom)32; a few patients were also tested after iron depletion. Briefly, 30 mL venous blood (plus EDTA) was obtained from each patient and control and diluted with Hanks balanced salts. Blood was then stratified in Lymphoprep solution (ratio of blood to Lymphoprep, 2:1) and centrifuged at 1680 revolutions per minute (rpm) for 30 minutes. The mononuclear cell layer was aspirated and washed 3 times in saline solution. Monocytes were isolated by another separation in Percoll solution by using centrifugation at 2100 rpm for 30 minutes at 4°C and then washed 3 times. Finally, cells were plated (500 000 cells/mL) in 30-mm wells in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% β-mercaptoethanol, and 1% streptomycin in the presence or absence of LPS (5 μg/mL; Sigma Chemical, St Louis, MO). After 24 hours, the supernatants were collected, centrifuged at 2000 rpm for 10 minutes, and assayed for TNF-α concentration.
To assess the effect of cellular iron content on TNF-α cellular release, cells were preincubated for 24 hours with ferric ammonium citrate (100 μM; Sigma Chemical) or desferrioxamine (40 μM; Sigma Chemical) and then treated with LPS (5 μg/mL) for 24 hours. Control cells were cultured for 24 hours without addition of ferric ammonium citrate or desferrioxamine and then treated with LPS. Supernatants were collected and assayed for TNF-α concentration.
Assessment of TNF-α concentration
TNF-α release from monocyte-macrophage cells from 50 patients and 34 controls was measured by using a commercially available enzyme-linked immunosorbent assay (Nycomed Amersham).
Analysis of genomic TNF-α locus polymorphism
Genomic DNA was extracted from whole blood preserved in EDTA by using the phenol-chloroform method. TNF2 and TNFA alleles were detected by using mutagenic primers containing a single base-pair mismatch adjacent to the polymorphic site to introduce a restriction site into the wild-type nucleotide sequences after amplification.14We used the following 4 primers (M-medical, Florence, Italy; mismatch positions underlined): F308 (5′-GGG ACA CAC AAG CAT CAA GG-3′), R308 (5′-AAT AGG TTT TGA GGG CCA TG-3′), F238 (5′-ATC TGG AGG AAG CGG TAG TG-3′), and R238 (5′-AGA AGA CCC CCC TCG GAACC-3′). The fragment containing the TNF2 polymorphism was amplified by using F308 and R308 primers; TNFA was amplified with F238 and R238.
DNA samples were amplified in 50 μL ammonia reaction buffer (Bioline, London, United Kingdom) containing 200 μM deoxynucleoside triphosphate, 10 μM each primer, 2 μL DNA sample, and 2 U Taq polymerase (Bioline) for one cycle at 94°C for 4 minutes, 59°C for 1 minute, and 70°C for 45 seconds, followed by 33 cycles at 94°C for 1 minute, 59°C for 1 minute, and 70°C for 45 seconds, followed by one cycle at 70°C for 10 minutes. The polymerase chain reaction (PCR) products were digested at 37°C with NcoI to detect the TNF2 allele and with MspI to detect the TNFA allele and then subjected to 4% agarose-gel electrophoresis. Each PCR batch included a “blank” to which no DNA had been added to ensure that no contamination of samples had occurred. None of the blank reactions yielded any visible product after gel electrophoresis.
HFE gene mutations
C282Y and H63D HFE mutations were sought in genomic DNA extracted from peripheral leukocytes as described previously.1
Statistical analysis
Results were expressed as mean (± SD) and considered significant when the P value was less than .05 (2-tailed test). Mean values were compared by using t tests for unequal variances. Frequencies were compared by performing χ2 tests. Genotype distributions were compared with the Fisher exact test. Multivariate analysis was done to assess the association between TNF-α polymorphism and several variables of phenotype expression in HHC (age at diagnosis, amount of iron removed, liver siderosis, HCV and HBV infection, HFE genotype, alanine aminotransferase [ALT] values, and presence of liver cirrhosis).
Results
Demographic information and HFE gene mutations in patients with HHC
Thirty-eight patients (59.4%) were homozygous for the C282Y mutation (C282Y +/+) and 26 (40.6%) were nonhomozygous (non-C282Y +/+). Two were heterozygous for the C282Y mutation, 2 were homozygous and 7 heterozygous for the H63D mutation, and 15 had neither; none had compound heterozygosity. Three non-C282Y +/+ patients had a family history of HHC. Patients homozygous for the C282Y mutation were younger and had more severe iron overload (Table1). The prevalence of exogenous hepatotoxic factors (alcohol abuse and HBV and HCV infection) was higher in non-C282Y +/+ patients than C282Y +/+ patients, as was reported previously.2 Alcohol abuse was present in 6 of 38 C282Y +/+ patients (16%) and 9 of 26 non-C282Y +/+ patients (35%), HCV infection in 4 of 38 C282Y +/+ patients (11%) and 9 of 26 non-C282Y +/+ patients (35%; P = .03), and HBV infection in 1 of 38 C282Y +/+ patients (2%) and 4 of 26 non-C282Y +/+ patients (15%).
Characteristics of 48 men and 16 women with hereditary hemochromatosis according to homozygosity for the C282Y mutation
| Variable . | C282Y +/+ (n = 38) . | Non-C282Y +/+ (n = 26) . | P value . |
|---|---|---|---|
| Age, y | 46 ± 13 (20-69) | 52 ± 7 (36-60) | .04 |
| Transferrin saturation, % | 86 ± 14 (54-100) | 75 ± 17 (46-100) | .006 |
| Ferritin, μg/L | 2103 ± 1801 (422-8000) | 1558 ± 1281 (380-5800) | NS |
| Iron removed, g | 11.6 ± 7 (3.5-30) | 7.8 ± 5.5 (3-13.5) | .02 |
| No. (%) patients with grade IV siderosis* | 20 (53%) | 14 (54%) | NS |
| Variable . | C282Y +/+ (n = 38) . | Non-C282Y +/+ (n = 26) . | P value . |
|---|---|---|---|
| Age, y | 46 ± 13 (20-69) | 52 ± 7 (36-60) | .04 |
| Transferrin saturation, % | 86 ± 14 (54-100) | 75 ± 17 (46-100) | .006 |
| Ferritin, μg/L | 2103 ± 1801 (422-8000) | 1558 ± 1281 (380-5800) | NS |
| Iron removed, g | 11.6 ± 7 (3.5-30) | 7.8 ± 5.5 (3-13.5) | .02 |
| No. (%) patients with grade IV siderosis* | 20 (53%) | 14 (54%) | NS |
Values are mean ± SD (range) unless otherwise indicated.
NS indicates not significant.
On Perls staining.29
TNF-α release from stimulated monocytes
Monocyte stimulation with LPS (5 μg/mL) was followed by an increase in TNF-α release in both patients and controls. TNF-α release from monocytes from patients with HHC who were nonhomozygous for the C282Y mutation was similar to that from monocytes from controls (424.01 ± 244.6 ng/L versus 478.75 ± 150.7 ng/L), whereas levels of the cytokine were significantly lower in C282Y homozygous patients (363.4 ± 157.9 ng/L; P = .01; Table2). A few patients were retested after reaching iron depletion, and there was no evidence of any significant difference in TNF-α release.
Release of tumor necrosis factor α from monocytes cultured for 24 hours with lipopolysaccharide (5 μg/mL), according to subject group
| Subject group . | TNF-α (ng/L) . |
|---|---|
| Controls (n = 34)* | 478.5 ± 150.7 |
| Patients with HHC (n = 50) | 385.2 ± 193.4 |
| C282Y +/+ patients (n = 32) | 363.4 ± 157.9† |
| Non-C282Y +/+ patients (n = 18) | 424.0 ± 244.6 |
| Subject group . | TNF-α (ng/L) . |
|---|---|
| Controls (n = 34)* | 478.5 ± 150.7 |
| Patients with HHC (n = 50) | 385.2 ± 193.4 |
| C282Y +/+ patients (n = 32) | 363.4 ± 157.9† |
| Non-C282Y +/+ patients (n = 18) | 424.0 ± 244.6 |
Values are mean ± SD.
TNF-α indicates tumor necrosis factor α; HHC, hereditary hemochromatosis.
Two controls were C282Y +/−, and none were C282Y +/+ or had compound heterozygosity.
P = .01 compared with controls.
Of the 34 healthy subjects in whom TNF-α release was measured, 2 were heterozygous for C282Y (C282Y +/−) and none were C282Y +/+ or had compound heterozygosity. Monocytes from the 2 C282Y +/− controls released an amount of TNF-α similar to the quantity released by monocytes from subjects with wild-type HFE.
TNF-α release in iron-depleted or enriched cells
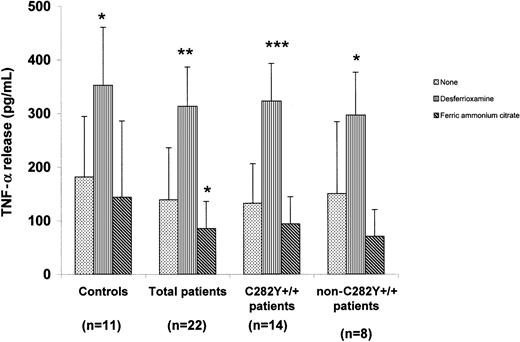
Results of measurement of TNF-α release from iron-depleted or enriched monocytes are shown in Figure 1. In all subject groups, iron depletion led to a significant increase in TNF-α release compared with the amount released from control cells. Iron addition led to a decrease in TNF-α release from monocytes from all groups. TNF-α release from monocytes from patients with HHC was lower than that from cells from controls cultured under the same experimental conditions.
TNF-α release from iron-manipulated monocytes from patients with HHC and controls.
Shown is TNF-α release from monocytes incubated for 24 hours with desferrioxamine (40 μM), ferric ammonium citrate (100 μM), or nothing and then treated with LPS (5 μg/mL) for 24 hours. Values are mean ± SD. Cells preincubated for 24 hours before LPS stimulation released less TNF-α than cells stimulated with LPS immediately. Preincubation in the presence of desferrioxamine caused a significant increase in TNF-α release, whereas preincubation in the presence of ferric ammonium citrate induced a decrease in TNF-α release. *P < .05, **P < .01, and ***P < .0001 for cells grown in the same experimental conditions and not preincubated with iron or desferrioxamine.
TNF-α release from iron-manipulated monocytes from patients with HHC and controls.
Shown is TNF-α release from monocytes incubated for 24 hours with desferrioxamine (40 μM), ferric ammonium citrate (100 μM), or nothing and then treated with LPS (5 μg/mL) for 24 hours. Values are mean ± SD. Cells preincubated for 24 hours before LPS stimulation released less TNF-α than cells stimulated with LPS immediately. Preincubation in the presence of desferrioxamine caused a significant increase in TNF-α release, whereas preincubation in the presence of ferric ammonium citrate induced a decrease in TNF-α release. *P < .05, **P < .01, and ***P < .0001 for cells grown in the same experimental conditions and not preincubated with iron or desferrioxamine.
TNF-α gene polymorphisms
The frequency distribution of the 308 TNF-α polymorphism genotype in healthy controls and patients with HHC is shown in Table3. In patients with HHC, the prevalence of the TNF1/TNF2 genotype was 23%, independent of HFE genotype. This frequency was identical to that in controls. The allele frequency for TNF1 was 0.84 in HHC patients homozygous for C282Y, 0.92 in HHC patients with other HFE genotypes, and 0.88 in healthy controls.
Prevalence of 308 tumor necrosis factor α polymorphism (TNF1, G at position 308; and TNF2, A at position 308), according to subject group
| Subject group . | TNF1/TNF1 . | TNF1/TNF2 . | TNF2/TNF2 . |
|---|---|---|---|
| Controls (n = 172) | 134 (77) | 38 (22) | 2 (1) |
| Patients with HHC (n = 64) | 49 (77) | 15 (23) | 0 |
| C282Y +/+ patients (n = 38) | 29 (76) | 9 (24) | 0 |
| Non-C282Y +/+ patients (n = 26) | 20 (77) | 6 (23) | 0 |
| Subject group . | TNF1/TNF1 . | TNF1/TNF2 . | TNF2/TNF2 . |
|---|---|---|---|
| Controls (n = 172) | 134 (77) | 38 (22) | 2 (1) |
| Patients with HHC (n = 64) | 49 (77) | 15 (23) | 0 |
| C282Y +/+ patients (n = 38) | 29 (76) | 9 (24) | 0 |
| Non-C282Y +/+ patients (n = 26) | 20 (77) | 6 (23) | 0 |
Results regarding the 238 TNF-α gene polymorphism are shown in Table4. Sixty-three of the 64 HHC patients (98.4%) were homozygous for TNFG; the other (1.6%) had TNFG/TNFA heterozygosity. The TNFG/TNFA genotype was observed significantly more often in controls (13%) than in patients with HHC (1.6%;P = .0022). The allele frequency for TNFG was 0.99 in HHC patients homozygous for C282Y, 1.0 in HHC patients with other HFE genotypes, and 0.92 in controls.
Prevalence of 238 tumor necrosis factor α polymorphism (TNFG, G at position 238; and TNFA, A at position 238), according to subject group
| Subject group . | TNFG/TNFG . | TNFG/TNFA . | TNFA/TNFA . |
|---|---|---|---|
| Controls (n = 172) | 146 (85) | 23 (13) | 3 (2) |
| Patients with HHC (n = 64) | 63 (98)4-150 | 1 (2) | 0 |
| C282Y +/+ patients (n = 38) | 37 (97)4-151 | 1 (3) | 0 |
| Non-C282Y +/+ patients (n = 26) | 26 (100)‡ | 0 | 0 |
| Subject group . | TNFG/TNFG . | TNFG/TNFA . | TNFA/TNFA . |
|---|---|---|---|
| Controls (n = 172) | 146 (85) | 23 (13) | 3 (2) |
| Patients with HHC (n = 64) | 63 (98)4-150 | 1 (2) | 0 |
| C282Y +/+ patients (n = 38) | 37 (97)4-151 | 1 (3) | 0 |
| Non-C282Y +/+ patients (n = 26) | 26 (100)‡ | 0 | 0 |
Values are number (%) of subjects in each group.
P = .0022 compared with controls.
P = .0344 compared with controls.
P = .029 compared with controls.
Relation between TNF-α polymorphism and clinical expression of HHC
None of the demographic or clinical characteristics assessed (sex, age, alcohol abuse, chronic viral hepatitis, hepatomegaly, splenomegaly, diabetes, cardiomyopathy, hyperpigmentation, hypogonadism, arthropathy, portal hypertension, and liver cirrhosis) were found to be significantly associated with TNF-α polymorphism. A trend toward a lower prevalence of cirrhosis in patients with TNF-α polymorphism was observed, although the difference was not significant (4 of 15 patients [26.7%] versus 28 of 49 patients [57.1%];P = .07). The trend was independent of HFE genotype; in fact, cirrhosis was present in 2 of 9 C282Y +/+ patients (22%) and 2 of 6 non-C232Y +/+ patients (33%) with TNF-α polymorphism and in 15 of 29 C282Y +/+ patients (52%) and 13 of 20 non-C232Y +/+ patients (65%) without TNF-α polymorphism.
Severe liver siderosis (grade IV on Perls staining), which was not related to TNF-α polymorphism in the larger series of patients, was significantly less frequent in non-C282Y homozygous patients with TNF-α polymorphism than in those homozygous for the wild-type TNF-α allele (1 of 6 [16.7%] versus 14 of 20 [70%];P = .05). The HII, determined in 27 patients, was lower in patients with TNF-α polymorphism than in those without, independent of HFE genotype (3.8 ± 1.1 versus 6.3 ± 2.9;P = .05). A separate evaluation in patients divided according to HFE genotype was not possible because of the limited number of patients in whom LIC was determined. Transferrin saturation and the amount of iron removed to reach depletion were not related to TNF-α polymorphism, with no difference between C282Y +/+ and non-C282Y +/+ patients. A trend toward lower serum ferritin levels was observed in patients with TNF-α polymorphism (1284 ± 1024 μg/L versus 2031 ± 1706 μg/L), both in C282Y +/+ patients (1728 ± 1377 μg/L versus 2300 ± 1940 μg/L) and in non-C282Y +/+ patients (906 ± 454 μg/L versus 1535 ± 1236 μg/L), but the differences were not significant.
Serum ALT levels were significantly lower in patients with polymorphism than in those without it (37.5 ± 25 U/L versus 77.2 ± 53 U/L;P = .006). When the 23 patients with coexistent hepatotoxic factors (HBV and HCV infections and alcohol abuse) were not included in the statistical analysis, this difference increased (P = .004). Logistic regression analysis indicated that TNF-α polymorphism was independently associated with liver siderosis in non-C282Y +/+ patients (P = .047) and with ALT values in both C282Y +/+ patients and non-C282Y +/+ patients (P = .0008 and P = .045, respectively).
Relation between TNF-α polymorphism and TNF-α release
Ten patients and 6 controls in whom TNF-α release was measured had the 308 polymorphism. TNF-α release in those with TNF-α polymorphism did not differ from that in subjects without it (patients, 422 ± 232 ng/L and 379 ± 224 ng/L; and controls, 420 ± 145 ng/L and 500 ± 115 ng/L, respectively). The difference between patients and controls without TNF-α polymorphism was significant (P = .01).
Discussion
In this study, we found that, compared with controls, patients with HHC had a lower prevalence of the polymorphism at position 238 of the TNF-α promoter and that their monocytes released a lower amount of TNF-α, with the second finding being more pronounced in patients who were homozygous for the C282Y HFE mutation. Patients with the 308 polymorphism (we did not identify any patient with the 238 polymorphism alone) had milder liver damage. We therefore suggest that TNF-α plays a role in the phenotypic expression of HHC by influencing the severity of liver disease and that it is at least partly responsible for the scanty iron deposits found in monocyte-macrophage cells from these patients.
Our study was prompted by studies showing that TNF-α is involved in the regulation of iron metabolism. This cytokine modulates expression of H and L ferritin and of the transferrin receptor in different cell types, including monocytes,33hepatocytes,34,35 and duodenocytes,36 as well as of HFE protein.36 A relation between TNF-α and HHC was first suggested after it was observed that stimulated monocytes from patients with HHC released a lower amount of TNF-α than did monocytes from controls.28 However, a subsequent study by Recalcati et al32 did not confirm these results. In the current study, we reproduced the findings of Gordeuk et al28 and demonstrated a relation between TNF-α release and HFE genotype, since only monocytes from patients homozygous for the C282Y mutation released an amount of TNF-α that was significantly lower than that released by monocytes from controls. Because of the high prevalence of C282Y homozygosity in patients with HHC in the United States (> 95%), it is likely that all the subjects studied by Gordeuk et al28 were homozygous for the C282Y mutation. As previously suggested,28 the reduction in TNF-α release could account for the reported defects in iron metabolism in monocytes, because of the decrease in TNF-α–induced ferritin synthesis in these cells.
Alternatively, the decreased TNF-α release could be a result of the iron-deficient phenotype of reticuloendothelial cells in C282Y +/+ patients, an observation confirmed by Montosi et al37 by means of wild-type HFE transfection of monocytes from C282Y +/+ patients. However, this hypothesis contrasts with our current evidence of an increased TNF-α release from desferrioxamine iron-depleted monocytes from both patients with HHC and controls and, conversely, a reduced release from iron-enriched monocytes. Moreover, even after iron manipulation, TNF-α release from cells from patients was lower than that from cells from controls, suggesting an intrinsic defect of C282Y+/+ cells.
A possible consequence of the reduced TNF-α release is increased iron absorption.38,39 It has been shown that TNF-α can decrease intestinal iron absorption, thereby inducing H ferritin synthesis in enterocytes and promoting their differentiation, apoptosis, and shedding into the lumen. A role for TNF-α in modulating iron absorption is also suggested by the finding that both β2-microglobulin and γ-δ T-cell-receptor knockout mice, in which enteral administration of iron is not followed by the expected increase in TNF-α release from intraepithelial lymphocytes, develop parenchymal iron overload similar to that occurring in HHC.39
We analyzed the prevalence of 308 and 238 polymorphisms in patients with HHC and found that the 238 polymorphism, but not the 308 polymorphism, was significantly less prevalent in patients than in healthy controls. We did not find any relation between TNF-α release from monocytes and polymorphic alleles, as previously reported.40 However, we were able to test only one patient with the TNFG allele and 10 patients with the TNF2 allele. It is also possible that the experimental conditions used to assess TNF-α release were not suitable for detection of minor changes in release of the cytokine. The low prevalence of the 238 TNF-α polymorphism in patients with HHC may have been due to the fact that this polymorphism occurs more readily in chromosomes that do not carry HFE gene mutations (both genes are mapped on chromosome 6, at a distance of 5 centimorgans). However, in our study, TNFG allele prevalence was not related to HFE gene mutations.
An alternative hypothesis is that the polymorphism makes the phenotype of patients with HHC milder, possibly as a consequence of the increased TNF-α release, and thereby leads to an underdiagnosis of the disease. This hypothesis is supported by the finding that the patients with the 308 TNF-α polymorphism had milder liver involvement, indicated by significantly lower ALT values; a trend toward a lower prevalence of cirrhosis; and a significantly lower prevalence of severe liver siderosis in those with non-C282Y homozygosity. The lack of relation between TNF-α polymorphisms and liver siderosis in homozygotes for the C282Y mutation could have been due to the more severe liver siderosis in these patients2 and the poor sensitivity of Perls staining in discriminating different degrees of severe siderosis. A multivariate analysis confirmed that there was a significant association between TNF-α polymorphisms and liver siderosis in non-C282Y +/+ patients and transaminase levels in both C282Y +/+ patients and non-C282Y +/+ patients. It may be that TNF-α polymorphism does interfere with liver iron deposits but that it interferes to a lesser extent, if at all, with extrahepatic iron accumulation, given the lack of differences in the total iron burden as determined by the amount of iron removed to reach depletion and in extrahepatic involvement in patients with the 308 TNF-α polymorphism compared with those without it. Interestingly, the effect of the 308 TNF-α polymorphism on transcription of this cytokine has been shown to be tissue specific.41
The milder liver involvement we observed in patients with TNF-α polymorphism, which was associated with an increased expression of TNF-α in most previous studies,11,12 may reflect the hepatoprotective activity of this cytokine.42Alternatively, the reduction in liver necrosis may have been due to a lower accumulation of iron in hepatocytes, perhaps in favor of Kupffer cells and macrophages, although we have no clear evidence indicating that patients with TNF-α polymorphism had more severe siderosis in this cellular compartment.
In conclusion, our data suggest that TNF-α may play a role in HHC, ie, it may affect cellular iron distribution, modulate the severity of liver iron overload, and influence the phenotypic expression of the disease. Additional studies to clarify the relation between TNF-α and HHC are needed to explore possible differences in patients with different geographic origins.
Supported by MURST 2000, Ricerca Finalizzata IRCCS 1998, and MURST 1999.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Silvia Fargion, Dipartimento di Medicina Interna, Ospedale Maggiore IRCCS, Pad Granelli, Via F Sforza 35, 20122 Milan, Italy; e-mail: silvia.fargion@unimi.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal