X-linked lymphoproliferative disease (XLP) is a rare immune disorder commonly triggered by infection with Epstein-Barr virus. Major disease manifestations include fatal acute infectious mononucleosis, B-cell lymphoma, and progressive dys-gammaglobulinemia. SAP/SH2D1A, the product of the gene mutated in XLP, is a small protein that comprises a single SH2 domain and a short tail of 26 amino acids. SAP binds to a specific motif in the cytoplasmic tails of the cell surface receptors SLAM and 2B4, where it blocks recruitment of the phosphatase SHP-2. Here it is reported that Ly-9 and CD84, 2 related glycoproteins differentially expressed on hematopoietic cells, also recruit SAP. Interactions between SAP and Ly-9 or CD84 were analyzed using a novel yeast 2-hybrid system, by COS cell transfections and in lymphoid cells. Recruitment of SAP is most efficient when the specific tyrosine residues in the cytoplasmic tails of Ly-9 or CD84 are phosphorylated. It is concluded that in activated T cells, the SAP protein binds to and regulates signal transduction events initiated through the engagement of SLAM, 2B4, CD84, and Ly-9. This suggests that combinations of dysfunctional signaling pathways initiated by these 4 cell surface receptors may cause the complex phenotypes of XLP.
Introduction
After infection with Epstein-Barr virus (EBV), patients with X-linked lymphoproliferative disease (XLP) mount a vigorous, uncontrolled polyclonal expansion of both T and B cells. The primary cause of death is hepatic necrosis and bone marrow failure, which appears to stem from uncontrolled T-cell responses. However, 2 other major manifestations of the XLP syndrome, B-cell lymphomas of the gastrointestinal tract and dys-gammaglobulinemia, can develop in the absence of EBV infection.1Collectively, genetic and functional studies of patients with XLP suggest that a mutation in SAP causes an intrinsic T or natural killer (NK) cell defect that becomes particularly life threatening with EBV infection.
The XLP gene SAP (or SH2D1A)2-4encodes a 15-kd single SH2 domain that can function as a natural inhibitor of signal transduction events initiated by the cell surface receptors SLAM (CD150) and 2B4.1,2,5-10 In fact, on phosphorylation, both receptors recruit the tyrosine phosphatase SHP-2, which is blocked by SAP. SAP binds to the cytoplasmic tail of SLAM in the absence of phosphorylation in yeast, COS cells, and T lymphocytes.2
SLAM (CD150), a member of the immunoglobulin superfamily, is a glycosylated transmembrane protein expressed on all activated T, B, and dendritic cells.11-16 Engagement of SLAM by specific monoclonal antibodies induces interferon γ (IFN-γ) production and redirects Th2 responses of antigen-specific T-cell clones to a Th1 or a Th0 phenotype.14 In B cells, SLAM triggering with anti-CD40 antibodies or by the soluble SLAM ectodomain leads to B-cell proliferation and production of IgM, IgG, and IgA.15Because SLAM appears to be a self-ligand, it is likely that it signals bidirectionally.15 16 However, because SLAM associates with the natural inhibitor SAP in T cells and not in B cells, a SLAM-induced signal transduction pathway could be different in T and B cells.
SAP also associates with 2B4 (CD244), a membrane protein that has sequence homologies with SLAM in its ectodomain and its cytoplasmic tail and that is expressed constitutively on the surfaces of NK cells.17 In addition, 2B4 is expressed on the surfaces of a subset of human and mouse CD8+ T cells and human monocytes. Engagement of 2B4 induces cytokine secretion (IFN-γ) and enhances non–major histocompatibility complex–restricted killing by NK cells.17-21 The mechanism by which SAP controls 2B4 function in NK cells may be different than in CD8+ T cells, because regulation of expression of the SAP gene is different in NK cells than in T lymphocytes.8 SAP mRNA levels fall rapidly on triggering of the T-cell receptor for antigen. By contrast, the level of SAP expression in very low in resting mouse NK cells but increases after virus infection.8Interestingly, recent studies show that 2B4 inhibits cytolysis induced by its ligand CD48 or by anti-CD3 in NK cells from patients with XLP. It appears, therefore, that SAP blocks a negative signal induced by the engagement of 2B4.7-9 This is of particular interest for the dissection of the complex phenotypes of XLP because the 2B4 ligand, CD48, is expressed on the surfaces of all EBV-induced B-cell blasts.
To identify other proteins that interact with SAP, a novel yeast 2-hybrid system was used in which tyrosine residues of the bait-protein could be phosphorylated. This altered yeast 2-hybrid system uses a mutated form of the src-family kinase c-fyn to phosphorylate proteins in the yeast cell. The mutations are designed to eliminate toxicity for the yeast cell. Using this method, 2 cell surface proteins, Ly-9 (CD299) and CD84, were found to interact with phosphorylated SAP. Further analyses indicate that the binding properties of SAP and the cytoplasmic tails of Ly-9, CD84, and 2B4 are slightly different from those of SAP and SLAM. Nevertheless, the current study shows that SAP can be recruited to the cytoplasmic tail of at least 4 cell surface molecules on the surfaces of hematopoietic cells.
Materials and methods
Plasmid construction
For expression in yeast, a sequence encoding human SAP was amplified by polymerase chain reaction (PCR) from the original yeast-expression plasmid pGAD4242 and cloned into theEcoRI/BamHI sites in the multiple cloning site of a vector termed pBRIDGE. The SAP 5′ sense primer was 5′-ATCGAATTCATGGACGCAGTGGCTGTGTAT-3′, and the SAP 3′ antisense primer was 5′ ATCGGATCCTCATGGGGCTTTCAGGCAGAC 3′.
DNA sequences encoding mutants of human c-fyn—either Fyn 531 Y-F, Fyn420, 531 Y-F or Fyn420, 531 Y-F, 176 R-Q—were subcloned in the second multiple cloning site of pBRIDGE using the BglII site. Mutant Fyn 531 Y-F was amplified by PCR fromc-fyn in plasmid pMES (Fyn 5′ sense primer, 5′ CCGAGATCTATGGGCTGTGTGCAATGTAAG 3′; Fyn 3′ antisense primer, 5′ CCGAGATCTTTACAGGTTTTCACCAGGTTGGAA 3′). For the construction of mutant Fyn 420 531 Y-F, 2 c-fyn cDNA fragments were amplified using Fyn 531 Y-F as a template, then annealed at overlapping ends containing 420 Y-F substitution, filled in, and further amplified to produce the double mutant. (First fragment was generated using Fyn 5′ sense primer and primer Fyn 420 F 3′ antisense, 5′ TTGTCTTGCTGTGAACTCATTGTCTTCTATCAA 3′; and the second cDNA fragment using primers Fyn 420 F 5′ sense, 5′ GAAGACAATGAGTTCACAGCAAGACAAGGTGCA 3′ and Fyn 3′ antisense primer.) For the construction of mutant Fyn 420, 531 Y-F, 176 R-Q, 2c-fyn cDNA fragments were amplified using Fyn420 531 Y-F as a template, then annealed at overlapping ends containing 176 R-Q substitution, filled in, and further amplified to produce the triple mutant. (First fragment was generated using Fyn 5′ sense primer and primer Fyn 176 R 3′ antisense, 5′ GGTTTCACTCTCCTGGATAAGAAAGGTACCTCT 3′; and the second cDNA fragment using primers Fyn176R 5′ sense, 5′ ACCTTTCTTATCCAGGAGAGTGAAACCACCAAA 3′ and Fyn 3′ antisense primer.)
SLAM-, 2B4-, and CD84-GAL4 DNA activation domain chimeric proteins were obtained by cloning cDNA encoding for the cytoplasmic tail of these proteins in the vector pGAD424. SLAM cDNA fragment was obtained from the pGBT9/SLAM construct2 by cutting with EcoRI and BamHI and cloned in theEcoRI/BamHI sites. 2B4 was amplified by PCR using as a template m2B4 in plasmid pC1-Neo and cloned in theEcoRI/BglII sites (2B4 5′ sense primer, 5′-CCGGAATTCAAGAAGAGGAAGCAGTTACAG TTC-3′; and 2B4 3′ antisense primer 5′-GGAAGATCTCTAGGAGTAGACATCAAAGTTCTC-3′). CD84 was amplified by PCR using as a template hCD84 in plasmid pC1-Neo22 and cloned in the EcoRI/BamH1 sites. (CD84 5′ sense primer, 5′-ATCGAATTCTTCCGTTTGTTCAAGAGAAGA-3′; and CD84 3′ antisense primer, 5′-ATCGGATCCCTAGATCACAATTTCATAGCT-3′.)
Three mutated DNA sequences encoding for the last 107 amino acids of human Ly-9 cytoplasmic domain (mutants Ly-9 558 Y-F, Ly-9581 Y-F, and Ly-9558, 581 Y-F) were subcloned in the GAL4 DNA activation domain vector pGAD424 using the EcoRI/BamH1 sites. For the construction of mutant Ly-9 558 Y-F, 2 Ly-9 cDNA fragments were amplified using human pGAD424/Ly-9 (clone 4-1) as a template, then annealed at overlapping ends containing the 558 Y-F substitution, filled in, and further amplified to produce the mutant. (First fragment was generated using the Ly-9 5′ sense primer, 5′ GATGATGAAGATACCCCACCAAA 3′; and primer Ly-9 558 F 3′ antisense, 5′ CACTTGTGCAAACACGGTGTTCTCTCCAAC 3′; and the second cDNA fragment using primers Ly-9 558 F 5′ sense, 5′ GAGAACACCGTGTTTGCACAAGTGTTCAAC 3′ and Ly-9 3′ antisense primer, 5′ ATCGGATCCCTGAGGTGCTTCTGTCCTGCGAGC.) Mutant Ly-9581 Y-F was generated in a similar way using the primers Ly-9 581 F 5′ sense, 5′ TCAGCCACAATCTTCTGCTCCATACGGAAACCT 3′; and Ly-9 581 F 3′ antisense, 5′ CCGTATGGAGCAGAAGATTGTGGCTGAGCTCTC 3′. Mutant Ly-9558, 581 Y-F was obtained in the same way as Ly-9 581 F but using Ly-9 558 F as a template for the PCR reactions.
An altered yeast 2-hybrid system
Human SAP cloned in pBRIDGE and transformed in the yeast strain CG1945 was used as bait to screen a human T-cell cDNA library in pGAD424 (KT3).2 The principle of the altered 2-hybrid system was that SAP and mutated c-fyn were inserted in one bi-cistronic vector pBRIDGE as described above. In mutantc-fyn, the regulatory tyrosines 420 and 531 were substituted by phenylalanine (Fyn 531 Y-F, Fyn420, 531 Y-F). Conditional expression of the resultant fusion proteins was driven by the MET25 promoter in response to methionine levels in the medium. In the presence of 1 mM methionine, the expression of the protein was repressed, whereas the absence of methionine in the medium induced protein expression. Mutated c-fyn still phosphorylated endogenous yeast proteins, and its capability to become autophosphorylated was maintained (data not shown).
For the 2-hybrid screen in the presence of Fyn420, 531 Y-F, the yeast strain CG1945 was cotransformed with the vector (pBRIDGE), containing both SAP and the c-fyn mutant. These transformants were selected on SD media lacking tryptophan for 3 days. Next, these transformants were transformed for a second time with 1 mg cDNA library derived from the human T-cell line KT3 in pGAD424. Double transformants were then plated in SD media lacking Trp, Leu, His, and Met in the presence of 5 mM 3-amino triazole.
Yeast clones that grew under these restrictive conditions were then tested by the β-galactosidase assay. Plasmid DNA was extracted from clones that were positive by both criteria. These plasmid DNAs were then expanded after transformation of HB101 bacteria and selection in M9 medium media lacking Leu to isolate the GAL4 activation domain plasmid pGAD424. Purified plasmids were analyzed by restriction analysis and sequenced.
The β-galactosidase colony-lift filter assay and liquid culture assay using ONPG as a substrate were carried out as described in the Clontech (Palo Alto, CA) yeast protocols handbook. The vector pBRIDGE containing SAP in the absence of mutated c-fyn was used as an alternative to repressing c-fyn expression with methionine because in the high-density cultures necessary for transformation, methionine can be depleted, thus activating expression of c-fyn.
Cells and antibodies
EL-4/SLAM4,2 Jurkat cells, Jurkat stably transfected with human CD84 cells, and Raji cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. COS cell transfections were carried out as previously described.2 8
An antihuman 2B4 monoclonal antibody (C1.7) was purchased from Immunotech, and monoclonal antimouse Ly-9 (clone 30C7) hybridoma from ATTC (Manassas, VA).23 Antihuman Ly-9 (clone HLy-9.1.84) and CD84 (clone CD84.1.21 and CD84.1.7) were produced by immunizing BALB/c mice with 300.19 murine cells stably transfected with full-length cDNA. The antihuman SLAM antibody (A12) was a gift from DNA-X (Palo Alto, CA).2
Monoclonal antihuman SAP was obtained by standard procedures by immunizing BALB/c mice with the synthetic peptide CQGTTGIREDPDV coupled to KLH (Pierce, Rockford, IL). Phosphotyrosine monoclonal antibody cocktail horseradish peroxidase–conjugated (PY-7E1, PY-1B2, PY20) and horseradish peroxidase–conjugated streptavidin were from Zymed (San Francisco, CA).
Cell activation, immunoprecipitation, and immunoblotting
Mouse thymocytes (BALB/c), Jurkat, Jurkat/CD84, Raji, and EL-4/SLAM4 cells (50 × 106/mL) were activated with 1 mM pervanadate for 20 minutes at 37°C. Lysis was carried out with 2% Triton X-100 as described before.2 Cell lysates were centrifuged at 14 000g for 15 minutes at 4°C, and the crude lysate was precleared for 1 hour with 50 μL protein G–agarose beads (Gibco BRL, Rockville, MD) and 5 μL normal mouse serum. Immunoprecipitations used 1μg indicated antibody and 30 μL protein G–agarose beads for 3 hours at 4°C. Beads were then washed as described.2 Crude lysates and immunoprecipitates were subjected to SDS-PAGE and transferred onto nitrocellulose filters (Millipore, Bedford, MA). Filters were blocked for 1 hour with 5% skim milk (or 3% bovine serum albumin) and then probed with the indicated antibodies. Bound antibody was detected using horseradish peroxide–conjugated secondary antibodies and enhanced chemiluminescence (Supersignal; Pierce).
Immunofluorescence microscopy
COS-7 cells were transfected with human Ly-9 or human CD84 cDNA using the lipofectamine method (Roche, Pleasanton, CA). After 48 hours, cells were labeled with biotinylated antihuman mAb Ly-9 or (HLy-9.1.84), biotinylated antihuman mAb CD84 (CD84.1.7), or biotinylated anti-SLAM (A12) (5 μg/mL) at 4°C for 30 minutes. After 2 washes with ice-cold PBS, cells were incubated with streptavidin–fluorescein isothiocyanate (FITC) (Dako, Carpinteria, CA) or streptavidin-Cy3 (Dako). Cells were then washed twice with ice-cold PBS, immobilized in poly-L-lysine–treated coverslips at 4°C for 15 minutes, and fixed in −20°C methanol for 15 minutes. After 2 washes, cells were incubated for 30 minutes at room temperature with blocking buffer (PBS containing 0.2% skim milk, 2% fetal bovine serum, 1% bovine serum albumin, 0.1 mM Gly) and then with Cy3-conjugated anti-SAP antibody (10C4.2) or FITC-conjugated anti-SAP antibody for 30 minutes at room temperature. Controls used an isotypic IgG1 conjugated with Cy3 or FITC. Cells were washed twice with PBS and mounted in Fluoromount-G (Southern Biotechnology, Birmingham, AL). Fluorescence images were obtained using a confocal microscope (TCS NT; Leica, Heidelberg, Germany).
Jurkat cells were stained with biotinylated antihuman CD84 (CD84.1.7), biotinylated antihuman Ly9 (HLy-9.1.84), or mouse IgG1 isotype control (5 μg/mL) at 4°C for 30 minutes as described above.
Results
A novel yeast 2-hybrid system for binding studies between SAP and other proteins
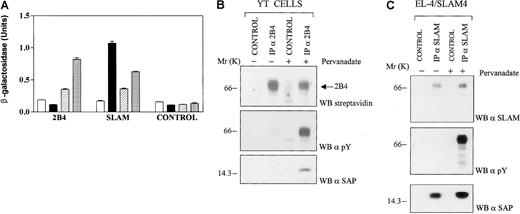
To study binding of the SAP SH2 domain to phosphorylated and nonphosphorylated proteins, a novel yeast 2-hybrid system was set up using c-fyn. Efficiency of the method was first tested using the cytoplasmic tail of 2B4 and full-length SAP (see “Materials and methods”). Only when a yeast cell coexpressed SAP, 2B4, and Fyn420, 531 Y-F was an interaction detected in the β-galactosidase assay (Figure 1A, hatched bars). Cotransfection of SAP with 2B4 in the presence of Fyn420, 531 Y-F, 176 R-Q resulted in higher values in the β-galactosidase assay (Figure 1A, dotted bars), implying a stronger interaction between SAP and 2B4 than that detected in the presence of Fyn420, 531 Y-F, which had an intact SH2 domain. This demonstrated that 2B4 and SAP interact in yeast, but only when 2B4 is phosphorylated. The experiment suggests that the c-fyn SH2 domain might interfere with the binding of SAP to the cytoplasmic tail of 2B4 (see below).
The cell surface molecule 2B4 recruits the
XLP gene product SAP after phosphorylation of the tyrosine residues in its cytoplasmic tail. (A) Comparison of the interactions between 2B4/SAP and SLAM/SAP in yeast cells. To detect binding between the cytoplasmic tail of 2B4 and SAP, a yeast 2-hybrid system was adapted to measure interactions with phospho-proteins. To this end, mutations of c-fyn were cotransfected with SAP into the yeast cell. Binding between 2 proteins in yeast cell extracts was detected by a β-galactosidase assay, as described in “Materials and methods.” For each construct, at least 3 independent colonies were tested in the β-galactosidase assay. Open bars: cells transfected with empty pBRIDGE vector and pGAD424 encoding the cytoplasmic tail of 2B4 or SLAM; solid bars: cells transfected with pBRIDGE-SAP and pGAD424 encoding the cytoplasmic tail of 2B4 or SLAM; hatched bars: cells transfected with pBRIDGE-SAP and Fyn420, 531 Y-F and with pGAD424 encoding the cytoplasmic tail of 2B4 or SLAM; dotted bars: cells transfected with pBRIDGE-SAP and Fyn 420, 531 Y-F, 176 R-Q and with pGAD424 encoding the cytoplasmic tail of 2B4 or SLAM. Control, empty pGAD424 with pBRIDGE encoding the indicated DNA sequences. (B) SAP binds to phosphorylated 2B4 in NK cell line. Interactions between SAP and 2B4 in the YT cell line were studied by immunoprecipitation of 2B4 followed by Western blot analysis with anti-SAP. YT cells (50 × 106 cells/mL) were biotinylated and then incubated in the presence or absence of 1 mM pervanadate. Cells were lysed in detergent, and 2B4 was immunoprecipitated with the 2B4 specific monoclonal antibody C1.7 (α-2B4). After SDS-PAGE, proteins were transferred to a PVDF membrane and were identified with streptavidin, antiphosphotyrosine (α-PY), or antihuman SAP 10C4.2 (α-SAP). CONTROL, immunoprecipitation with an irrelevant monoclonal antibody. (C) SAP binds to nonphosphorylated and phosphorylated SLAM in a T-cell line. Interactions between SAP and SLAM were studied in the T-cell transfectant cell line EL-4/SLAM4 by immunoprecipitation of SLAM, followed by Western blot analysis with anti-SAP. EL-4/SLAM4 cells (20 × 106 cells/mL) expressing human SLAM were treated with pervanadate, as described in panel B, and SLAM was immunoprecipitated by an antihuman SLAM monoclonal antibody (α-SLAM).2 After SDS-PAGE, proteins were transferred to a PVDF membrane and identified by Western blot analysis with a rabbit antihuman SLAM antibody, antiphosphotyrosine (α-PY), or antihuman SAP (α-SAP) 10C4.2 CONTROL, immunoprecipitation with an irrelevant monoclonal antibody.
The cell surface molecule 2B4 recruits the
XLP gene product SAP after phosphorylation of the tyrosine residues in its cytoplasmic tail. (A) Comparison of the interactions between 2B4/SAP and SLAM/SAP in yeast cells. To detect binding between the cytoplasmic tail of 2B4 and SAP, a yeast 2-hybrid system was adapted to measure interactions with phospho-proteins. To this end, mutations of c-fyn were cotransfected with SAP into the yeast cell. Binding between 2 proteins in yeast cell extracts was detected by a β-galactosidase assay, as described in “Materials and methods.” For each construct, at least 3 independent colonies were tested in the β-galactosidase assay. Open bars: cells transfected with empty pBRIDGE vector and pGAD424 encoding the cytoplasmic tail of 2B4 or SLAM; solid bars: cells transfected with pBRIDGE-SAP and pGAD424 encoding the cytoplasmic tail of 2B4 or SLAM; hatched bars: cells transfected with pBRIDGE-SAP and Fyn420, 531 Y-F and with pGAD424 encoding the cytoplasmic tail of 2B4 or SLAM; dotted bars: cells transfected with pBRIDGE-SAP and Fyn 420, 531 Y-F, 176 R-Q and with pGAD424 encoding the cytoplasmic tail of 2B4 or SLAM. Control, empty pGAD424 with pBRIDGE encoding the indicated DNA sequences. (B) SAP binds to phosphorylated 2B4 in NK cell line. Interactions between SAP and 2B4 in the YT cell line were studied by immunoprecipitation of 2B4 followed by Western blot analysis with anti-SAP. YT cells (50 × 106 cells/mL) were biotinylated and then incubated in the presence or absence of 1 mM pervanadate. Cells were lysed in detergent, and 2B4 was immunoprecipitated with the 2B4 specific monoclonal antibody C1.7 (α-2B4). After SDS-PAGE, proteins were transferred to a PVDF membrane and were identified with streptavidin, antiphosphotyrosine (α-PY), or antihuman SAP 10C4.2 (α-SAP). CONTROL, immunoprecipitation with an irrelevant monoclonal antibody. (C) SAP binds to nonphosphorylated and phosphorylated SLAM in a T-cell line. Interactions between SAP and SLAM were studied in the T-cell transfectant cell line EL-4/SLAM4 by immunoprecipitation of SLAM, followed by Western blot analysis with anti-SAP. EL-4/SLAM4 cells (20 × 106 cells/mL) expressing human SLAM were treated with pervanadate, as described in panel B, and SLAM was immunoprecipitated by an antihuman SLAM monoclonal antibody (α-SLAM).2 After SDS-PAGE, proteins were transferred to a PVDF membrane and identified by Western blot analysis with a rabbit antihuman SLAM antibody, antiphosphotyrosine (α-PY), or antihuman SAP (α-SAP) 10C4.2 CONTROL, immunoprecipitation with an irrelevant monoclonal antibody.
In contrast to 2B4, SLAM interacted with SAP in a phosphotyrosine-independent fashion. β-Galactosidase activity was detected when SLAM was coexpressed with either SAP alone or with SAP and Fyn420, 531 Y-F (Figure 1A). In yeast cells cotransformed with SLAM, SAP, and Fyn420, 531 Y-F (Figure 1A, hatched bars), β-galactosidase activity was lower than in yeast cells that express SAP and SLAM (Figure 1A, solid bars). One possible reason for this was that c-fyn's own SH2 domain would bind to a SLAM phosphotyrosine, thus competing with SAP for the same docking site. To test this possibility, arginine 176 in the c-fyn sequence was substituted by a glutamine Fyn420, 531 Y-F. This mutation disabled the binding properties of the SH2 domain of c-fyn. Indeed, the β-galactosidase assay showed a stronger response when the triple mutant of c-fyn, Fyn420, 531 Y-F, 176 R-Q, was used (Figure 1A, dotted bars). This suggests that c-fynbinds to phosphorylated SLAM through its SH2 domain. Taken together, the data show that in yeast, SAP interacts with phosphorylated 2B4 only and with both nonphosphorylated and phosphorylated SLAM.
To confirm that SAP bound to phospho-2B4 preferentially, cells of the NK line YT were used for coimmunoprecipitation of SAP and 2B4. Only when 2B4 was phosphorylated by pervanadate treatment of the cells did this surface receptor bind to SAP (Figure 1B). By contrast, SAP binds to both phospho- and nonphospho-SLAM in the T-cell line EL4 (Figure1C). Thus, the binding experiments in yeast truthfully represented interactions found in hematopoietic cells.
SAP binds specifically to the cytoplasmic tail of the hematopoietic cell surface receptor Ly-9
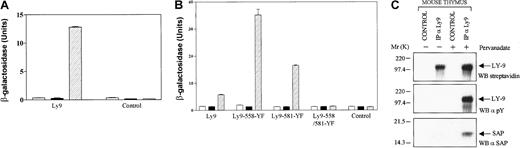
When 1 × 106 clones of a yeast T-cell library (KT3) were screened using SAP as bait in the presence of Fyn 420, 531 Y-F, 5 clones were isolated. These were all positive, as judged by their ability to grow in media lacking histidine and by their β-galactosidase activity. Each clone encoded a fragment with the exact nucleotide sequence of the cytoplasmic tail of human Ly-9. Ly-9 is a glycoprotein whose extracellular domain belongs to the same subfamily of the immunoglobulin superfamily of proteins as SLAM and 2B4. When Ly-9 was subsequently cotransfected in yeast with SAP alone, no β-galactosidase activity was detected. However, in the presence of Fyn420, 531 Y-F, the interaction between phospho-Ly-9 and SAP resulted in detectable β-galactosidase activity (Figure 2A).
SAP interacts with the phosphorylated cytoplasmic tail of Ly-9.
A human cDNA library made from poly A+ RNA of the human T-cell line KT3 in pGAD424 was screened with the altered yeast 2-hybrid system. Thus, 5 cDNA clones encoding the cytoplasmic tail of Ly-9 were isolated; an example is shown in panel A. To map the binding sites in the cytoplasmic tail of human Ly-9, 3 mutations of Ly-9 were analyzed in panel B. SAP binding to phospho-Ly-9 was shown in murine thymocytes in panel C. (A) Interactions of SAP and the cytoplasmic tail of Ly-9 in yeast are dependent on the presence of Fyn 420, 531 Y-F. The interaction of SAP with the cytoplasmic tail of Ly-9 in the presence or absence of Fyn 420, 531 Y-F took place in the yeast cell and was measured in a β-galactosidase assay. For each construct, at least 3 independent colonies were tested in the galactosidase assay. Open bars: cells transfected with empty pBRIDGE vector and pGAD424 encoding the cytoplasmic tail of Ly-9; solid bars: cells transfected with pBRIDGE-SAP and pGAD424 encoding the cytoplasmic tail of Ly-9; hatched bars: cells transfected with pBRIDGE-SAP and Fyn420, 531 Y-F and with pGAD424 encoding the cytoplasmic tail of Ly-9. Control, empty pGAD424 with pBRIDGE encoding the indicated DNA sequences. (B) SAP interacts with 2 phosphotyrosine motifs in the cytoplasmic tail of Ly-9. The interaction of SAP with 3 Ly-9 cytoplasmic tail mutations in the presence or absence of Fyn420, 531 Y-F took place in the yeast cell and was measured in a β-galactosidase assay. For each construct, at least 3 independent colonies were tested in the galactosidase assay. The cytoplasmic tail of Ly-9 was mutated in tyrosine residue 558 (558-YF), tyrosine 581 (581-YF), or in both tyrosine 558 and tyrosine 581 (558-581-YF). Open bars: cells transfected with empty pBRIDGE vector and pGAD424 encoding the cytoplasmic tail of Ly-9. Solid bars: cells transfected with pBRIDGE-SAP and pGAD424 encoding the cytoplasmic tail of Ly-9 or one of the Ly-9 mutants. Hatched bars: cells transfected with pBRIDGE-SAP and Fyn420, 531 Y-F and with pGAD424 encoding the cytoplasmic tail of Ly-9 or one of the Ly-9 mutants. CONTROL, empty pGAD424 with pBRIDGE encoding the indicated DNA sequences. (C) Association of SAP and Ly-9 in mouse thymocytes. Mouse thymocytes (50 × 106 cells/mL) were biotinylated and then incubated in the presence or absence of 1 mM pervanadate. Cells were lysed, and Ly-9 was immunoprecipitated with 1 μg antimouse Ly-9 monoclonal antibody (IP aLy9). Proteins were transferred to PVDF, and Western blot analysis (WB) identified specific proteins with streptavidin, antiphosphotyrosine (WB α-PY) and a rabbit antimouse SAP antibody (WB α-SAP). CONTROL, immunoprecipitation with a monoclonal hamster antibody.
SAP interacts with the phosphorylated cytoplasmic tail of Ly-9.
A human cDNA library made from poly A+ RNA of the human T-cell line KT3 in pGAD424 was screened with the altered yeast 2-hybrid system. Thus, 5 cDNA clones encoding the cytoplasmic tail of Ly-9 were isolated; an example is shown in panel A. To map the binding sites in the cytoplasmic tail of human Ly-9, 3 mutations of Ly-9 were analyzed in panel B. SAP binding to phospho-Ly-9 was shown in murine thymocytes in panel C. (A) Interactions of SAP and the cytoplasmic tail of Ly-9 in yeast are dependent on the presence of Fyn 420, 531 Y-F. The interaction of SAP with the cytoplasmic tail of Ly-9 in the presence or absence of Fyn 420, 531 Y-F took place in the yeast cell and was measured in a β-galactosidase assay. For each construct, at least 3 independent colonies were tested in the galactosidase assay. Open bars: cells transfected with empty pBRIDGE vector and pGAD424 encoding the cytoplasmic tail of Ly-9; solid bars: cells transfected with pBRIDGE-SAP and pGAD424 encoding the cytoplasmic tail of Ly-9; hatched bars: cells transfected with pBRIDGE-SAP and Fyn420, 531 Y-F and with pGAD424 encoding the cytoplasmic tail of Ly-9. Control, empty pGAD424 with pBRIDGE encoding the indicated DNA sequences. (B) SAP interacts with 2 phosphotyrosine motifs in the cytoplasmic tail of Ly-9. The interaction of SAP with 3 Ly-9 cytoplasmic tail mutations in the presence or absence of Fyn420, 531 Y-F took place in the yeast cell and was measured in a β-galactosidase assay. For each construct, at least 3 independent colonies were tested in the galactosidase assay. The cytoplasmic tail of Ly-9 was mutated in tyrosine residue 558 (558-YF), tyrosine 581 (581-YF), or in both tyrosine 558 and tyrosine 581 (558-581-YF). Open bars: cells transfected with empty pBRIDGE vector and pGAD424 encoding the cytoplasmic tail of Ly-9. Solid bars: cells transfected with pBRIDGE-SAP and pGAD424 encoding the cytoplasmic tail of Ly-9 or one of the Ly-9 mutants. Hatched bars: cells transfected with pBRIDGE-SAP and Fyn420, 531 Y-F and with pGAD424 encoding the cytoplasmic tail of Ly-9 or one of the Ly-9 mutants. CONTROL, empty pGAD424 with pBRIDGE encoding the indicated DNA sequences. (C) Association of SAP and Ly-9 in mouse thymocytes. Mouse thymocytes (50 × 106 cells/mL) were biotinylated and then incubated in the presence or absence of 1 mM pervanadate. Cells were lysed, and Ly-9 was immunoprecipitated with 1 μg antimouse Ly-9 monoclonal antibody (IP aLy9). Proteins were transferred to PVDF, and Western blot analysis (WB) identified specific proteins with streptavidin, antiphosphotyrosine (WB α-PY) and a rabbit antimouse SAP antibody (WB α-SAP). CONTROL, immunoprecipitation with a monoclonal hamster antibody.
To refine the specificity of the interaction and to determine which of the 3 phosphotyrosines in the cytoplasmic tail of Ly-9 were involved in SAP binding, Ly-9 mutants were tested in the yeast 2-hybrid system. Because the 2 (T V/I pY x x V/I)-motifs (see Figure 4), in the cytoplasmic tail of Ly-9 were the most logical candidates for SAP binding, 3 mutants were made. Tyrosine 558 (Ly9-558-YF), tyrosine 581 (Ly9-581-YF), or both tyrosine residues (Ly9-558581-YF) were substituted by phenylalanine. The Ly-9 mutants were then subcloned in the GAL4-binding domain of pGAD424, and these constructs were cotransfected in yeast with SAP alone or with SAP and Fyn 420, 531 Y-F. As shown in Figure 2B, the phosphorylated form of each single mutant (Ly9-558-YF or Ly9-581-YF) binds SAP, in fact better than the wt Ly9 segment. However, no interaction was detected when both tyrosine residues were absent in the cytoplasmic tail of Ly-9 (Ly9-558581-YF). We conclude that each of the 2 phosphotyrosine motifs binds SAP specifically and that no other binding sites are involved.
To verify the interaction between SAP and Ly-9, mouse thymocytes, which express large amounts of both Ly9 and SAP, were activated with pervanadate and subjected to immunoprecipitation using an anti-Ly-9 antibody. As predicted by the yeast data, SAP interacted with Ly-9, but only after the cells were treated with pervanadate and Ly-9 became tyrosine-phosphorylated (Figure 2C). No interaction was detected in untreated cells. We conclude that Ly-9, like 2B4 and SLAM, recruits SAP but that, unlike SLAM, this binding appears to be dependent on the phosphorylation status of the tyrosine motif in the cytoplasmic tail of Ly-9.
SAP binds to the cytoplasmic tail of the cell surface receptor CD84
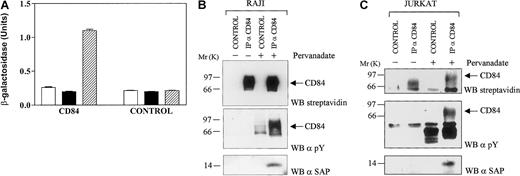
CD84, a member of the SLAM family expressed on the surface of B and T lymphocytes and monocytes, contains 3 potential SAP-binding motifs in its cytoplasmic tail (Figure 4). That prompted us to examine the interaction of CD84 with SAP in yeast and in lymphocytes. CD84 was transfected into yeast together with SAP or with SAP and Fyn420531 Y-F. As judged by the β-galactosidase assay, CD84 interacted with SAP (Figure 3A). Again this interaction could be detected only when c-fyn was cotransfected and no binding was detected in the absence of the tyrosine kinase.
Interactions of SAP with the phosphorylated cytoplasmic tail of CD84.
(A) CD84 and SAP in yeast. The interaction of SAP with the cytoplasmic tail of human CD84 in the presence or absence of Fyn 420, 531 Y-F took place in the yeast cell and was measured in a β-galactosidase assay. For each construct, at least 3 independent colonies were tested in the galactosidase assay. Open bars: cells transfected with empty pBRIDGE vector and pGAD424 encoding the cytoplasmic tail of CD84; solid bars: cells transfected with pBRIDGE-SAP and pGAD424 encoding the cytoplasmic tail of CD84; hatched bars: cells transfected with pBRIDGE-SAP and Fyn420, 531 Y-F and with pGAD424 encoding the cytoplasmic tail of CD84. Control, empty pGAD424 with pBRIDGE encoding the indicated DNA sequences. (B) SAP coprecipitates with phosphorylated CD84 in a B-cell line. Cells of a variant of the Burkitt lymphoma cell line Raji, which was known to express SAP in its cytoplasm,2 were biotinylated and incubated in the presence or absence of 1 mM pervanadate. After lysis in detergent, CD84 was immunoprecipitated with 1 μg antihuman CD84 monoclonal antibody (IP α-CD84) (CD84.1.21). Proteins were transferred to PVDF membranes and Western blotted (WB) with streptavidin (WB streptavidin), antiphosphotyrosine (α-pY), and monoclonal antihuman SAP (α-SAP)10C4.2 CONTROL, immunoprecipitation with mouse immunoglobulin. (C) SAP coprecipitates with phosphorylated CD84 in Jurkat. Stable CD84 transfectants of the T- cell line Jurkat were biotinylated and then incubated in the presence or absence of 1 mM pervanadate. After lysis in detergent, CD84 was immunoprecipitated with 1 μg antihuman CD84 monoclonal antibody (IP α-CD84) (CD84.1.21). Proteins were transferred to PVDF membranes and Western blotted (WB) with streptavidin (WB streptavidin), antiphosphotyrosine (α-pY) and monoclonal antihuman SAP (α-SAP) (10C4.2). CONTROL, immunoprecipitation with mouse immunoglobulin.
Interactions of SAP with the phosphorylated cytoplasmic tail of CD84.
(A) CD84 and SAP in yeast. The interaction of SAP with the cytoplasmic tail of human CD84 in the presence or absence of Fyn 420, 531 Y-F took place in the yeast cell and was measured in a β-galactosidase assay. For each construct, at least 3 independent colonies were tested in the galactosidase assay. Open bars: cells transfected with empty pBRIDGE vector and pGAD424 encoding the cytoplasmic tail of CD84; solid bars: cells transfected with pBRIDGE-SAP and pGAD424 encoding the cytoplasmic tail of CD84; hatched bars: cells transfected with pBRIDGE-SAP and Fyn420, 531 Y-F and with pGAD424 encoding the cytoplasmic tail of CD84. Control, empty pGAD424 with pBRIDGE encoding the indicated DNA sequences. (B) SAP coprecipitates with phosphorylated CD84 in a B-cell line. Cells of a variant of the Burkitt lymphoma cell line Raji, which was known to express SAP in its cytoplasm,2 were biotinylated and incubated in the presence or absence of 1 mM pervanadate. After lysis in detergent, CD84 was immunoprecipitated with 1 μg antihuman CD84 monoclonal antibody (IP α-CD84) (CD84.1.21). Proteins were transferred to PVDF membranes and Western blotted (WB) with streptavidin (WB streptavidin), antiphosphotyrosine (α-pY), and monoclonal antihuman SAP (α-SAP)10C4.2 CONTROL, immunoprecipitation with mouse immunoglobulin. (C) SAP coprecipitates with phosphorylated CD84 in Jurkat. Stable CD84 transfectants of the T- cell line Jurkat were biotinylated and then incubated in the presence or absence of 1 mM pervanadate. After lysis in detergent, CD84 was immunoprecipitated with 1 μg antihuman CD84 monoclonal antibody (IP α-CD84) (CD84.1.21). Proteins were transferred to PVDF membranes and Western blotted (WB) with streptavidin (WB streptavidin), antiphosphotyrosine (α-pY) and monoclonal antihuman SAP (α-SAP) (10C4.2). CONTROL, immunoprecipitation with mouse immunoglobulin.
SAP and the cytoplasmic tail of CD84 interact in a variant of the human B-cell line Raji, but only when the cells were treated with pervanadate (Figure 3B). Pervanadate treatment results in a strong phosphorylation of CD84, which permits association with SAP. No SAP binding was observed on untreated cells. Because SAP is primarily a T-cell protein, the CD84-SAP interaction was also examined in JURKAT cells, which had been stably transfected with human CD84. Once again, SAP coprecipitated with the phosphorylated form of CD84 (Figure 3C).
Taken together, the observations demonstrate that the cell surface receptor CD84 interacts with SAP. The results also indicate that the mode of interaction between SAP and SLAM differs from interactions between SAP and Ly-9 or CD84. This suggests that the apparent affinity between the SH2 domain of SAP and its recognition sites in the cytoplasmic tail of Ly-9 or CD84 must be lower. Because the minimal binding motifs (Figure 4), however, are similar if not identical, interactions with other segments might play a role.
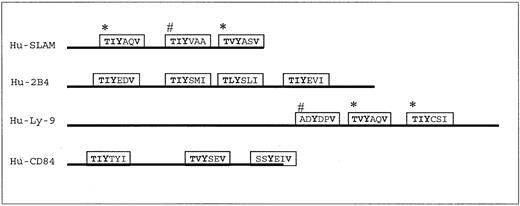
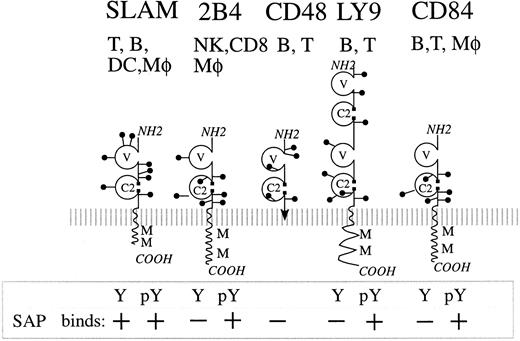
Schematic representation of the cytoplasmic domains of 4 SLAM family members.
SAP has been shown to bind preferentially to a consensus binding motif: T (I/V)Y x x (V/I), where x represents any amino acid. The cytoplasmic domains of the SLAM family members SLAM, 2B4, Ly9, and CD84 contain multiple SAP-binding motifs. Lines indicate the cytoplasmic domain of the SLAM family members. Blocks represent real (*) or putative SAP-binding motifs. Blocks labeled with # are putative motifs proven to be unable to bind SAP, whereas those without marks have not been tested for SAP binding.
Schematic representation of the cytoplasmic domains of 4 SLAM family members.
SAP has been shown to bind preferentially to a consensus binding motif: T (I/V)Y x x (V/I), where x represents any amino acid. The cytoplasmic domains of the SLAM family members SLAM, 2B4, Ly9, and CD84 contain multiple SAP-binding motifs. Lines indicate the cytoplasmic domain of the SLAM family members. Blocks represent real (*) or putative SAP-binding motifs. Blocks labeled with # are putative motifs proven to be unable to bind SAP, whereas those without marks have not been tested for SAP binding.
SAP inhibits the association of SHP-2 with phosphorylated Ly-9 and CD84
The cytoplasmic tail of SLAM contains 2 binding sites for SAP—one is in a peptide segment that includes Y281, and one includes Y327. Optimal binding of SAP occurs when each site is phosphorylated, but SAP also binds strongly to the Y281 site in the absence of phosphorylation. For activation of the tyrosine phosphatase SHP-2, both of its SH2-domains are required to bind to their docking sites. In the cytoplasmic tail of SLAM, the SHP-2 docking sites are the same as the SAP docking sites (the pY281 and the pY327 motif) (D.H., unpublished data, February 2000). As expected, SHP-2 does not bind to nonphosphorylated SLAM.2 10
As reported previously, SAP blocks recruitment of SHP-2 to the phosphorylated cytoplasmic tail of SLAM.2 The structure of SAP is consistent with this role as a natural inhibitor, and the affinity of SAP for a phosphorylated tyrosine pY281-peptide (binding constant, approximately 120 nM) is higher than the affinity of other SH2 domains for their phosphotyrosine-binding motifs.10
Because Ly-9 and CD84 each contain 2 SAP-binding motifs (Figure 4), the ability of SAP to block recruitment of SHP-2 to these receptors was tested in COS cells.2 As expected, SHP-2 binds to phosphorylated Ly-9 only (Figure 5A), and SAP interferes with that binding. Similarly, SHP-2 binds to CD84, ifc-fyn is cotransfected into the same COS cells (Figure 5B), and this binding is blocked by the presence of SAP.
SAP blocks binding of SHP-2 to phosphorylated Ly-9 and CD84.
To detect interactions between Ly-9 or CD84 and SHP-2, COS-7 cells were cotransfected with cDNAs encoding one of the receptors, humanc-fyn, in the presence or absence of human SAP. On immunoprecipitation with monoclonal antibody directed at either Ly-9 or CD84, the receptor-associated protein complexes were analyzed by Western blotting. (A) SHP-2 binds to phosphorylated Ly-9 in the absence of SAP. COS-7 cells were transfected with the indicated constructs, and 48 hours later cell surface proteins were biotinylated, cells were lysed, and Ly-9 was immunoprecipitated as described in “Materials and methods.” Immunoprecipitates and whole-cell lysates were run on SDS-PAGE and transferred to PVDF. Ly-9 was detected using streptavidin conjugated to horseradish peroxidase, and phosphotyrosine, SAP, and SHP-2 were detected using the antibodies described in “Materials and methods.” Whole-cell lysates were Western blotted for SAP and SHP-2 to control for gel loading. (B) SHP-2 binds to phosphorylated CD84 in the absence of SAP. COS-7 cells were transfected with the indicated constructs, and 48 hours later cell surface proteins were biotinylated, cells were lysed, and CD84 immunoprecipitated as described in “Materials and methods.” Immunoprecipitates and whole-cell lysates were run on SDS-PAGE and transferred to PVDF. CD84 was detected using streptavidin conjugated to horseradish peroxidase, and phosphotyrosine, SAP, and SHP-2 were detected using the antibodies described in “Materials and methods.” Whole-cell lysates were Western blotted for SAP and SHP-2 to control for gel loading.
SAP blocks binding of SHP-2 to phosphorylated Ly-9 and CD84.
To detect interactions between Ly-9 or CD84 and SHP-2, COS-7 cells were cotransfected with cDNAs encoding one of the receptors, humanc-fyn, in the presence or absence of human SAP. On immunoprecipitation with monoclonal antibody directed at either Ly-9 or CD84, the receptor-associated protein complexes were analyzed by Western blotting. (A) SHP-2 binds to phosphorylated Ly-9 in the absence of SAP. COS-7 cells were transfected with the indicated constructs, and 48 hours later cell surface proteins were biotinylated, cells were lysed, and Ly-9 was immunoprecipitated as described in “Materials and methods.” Immunoprecipitates and whole-cell lysates were run on SDS-PAGE and transferred to PVDF. Ly-9 was detected using streptavidin conjugated to horseradish peroxidase, and phosphotyrosine, SAP, and SHP-2 were detected using the antibodies described in “Materials and methods.” Whole-cell lysates were Western blotted for SAP and SHP-2 to control for gel loading. (B) SHP-2 binds to phosphorylated CD84 in the absence of SAP. COS-7 cells were transfected with the indicated constructs, and 48 hours later cell surface proteins were biotinylated, cells were lysed, and CD84 immunoprecipitated as described in “Materials and methods.” Immunoprecipitates and whole-cell lysates were run on SDS-PAGE and transferred to PVDF. CD84 was detected using streptavidin conjugated to horseradish peroxidase, and phosphotyrosine, SAP, and SHP-2 were detected using the antibodies described in “Materials and methods.” Whole-cell lysates were Western blotted for SAP and SHP-2 to control for gel loading.
In the COS cell experiments, SAP was found to bind frequently to Ly-9 or CD84 after transfection with SAP but in the absence of transfection with c-fyn. This could be explained by the small percentage of Ly-9 or CD84 molecules phosphorylated in COS cells in the presence of SAP, probably by an endogenous COS cell tyrosine kinase; SAP may bind to those phosphorylated forms exclusively. Induction of phosphorylation of SLAM in COS cells transfected with SAP had been observed previously.2 In addition, the high levels of SAP and receptor in the COS cell expression system could be favorable to an interaction between SAP and nonphospho Ly9 or CD84. Collectively, these results support a model in which SAP acts as a natural inhibitor of the docking sites for SH2-containing enzymes and adapters in the cytoplasmic tail of Ly-9 or CD84.
SAP is recruited to the cell surface on phosphorylation of Ly-9 and CD84
To examine whether the associations between Ly-9 or CD84 and SAP took place on the cell surfaces, COS cell transfectants were analyzed by immunofluorescence techniques. As shown in Figure6, SAP is evenly distributed throughout the cytoplasm of COS cells. By contrast, most Ly-9 and CD84 molecules are expressed in the plasma membrane. In double transfectants, a large proportion of the immunofluorescence developed with α-SAP colocalizes with α-Ly-9 or α-CD84 staining (Figure 6). The selected pictures are representative of observations made in 3 independent COS cell experiments.
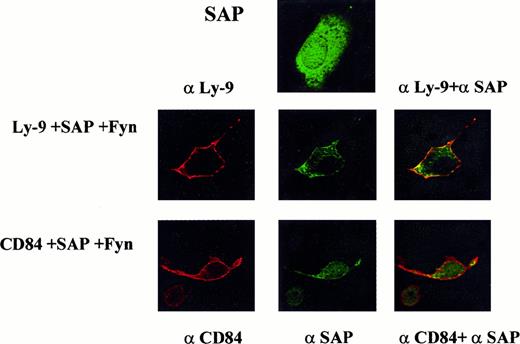
Phosphorylated CD84 and Ly-9 associate with SAP at the plasma membrane.
To verify the results obtained with the yeast 2-hybrid system and immunoprecipitation in COS-7 cells, a fluorescence microscopy approach was used. COS-7 cells were transfected with SAP and Fyn along with CD84 or Ly-9. The cells were then stained for SAP and Ly-9 or CD84. Colocalization of SAP and Ly-9 or CD84 at the cell membrane can be visualized by yellow fluorescence where the 2 fluorochromes colocalize.
Phosphorylated CD84 and Ly-9 associate with SAP at the plasma membrane.
To verify the results obtained with the yeast 2-hybrid system and immunoprecipitation in COS-7 cells, a fluorescence microscopy approach was used. COS-7 cells were transfected with SAP and Fyn along with CD84 or Ly-9. The cells were then stained for SAP and Ly-9 or CD84. Colocalization of SAP and Ly-9 or CD84 at the cell membrane can be visualized by yellow fluorescence where the 2 fluorochromes colocalize.
To test whether SAP was recruited to the plasma membrane in T cells, cocapping experiments were done in Jurkat cells with α-SAP and α-CD84 or α-Ly-9 antibodies. First, Jurkat cells were stained with α-CD84 for 10 minutes at 4°C. Then SAP was visualized with a directly labeled monoclonal antibody using standard cytoplasmic staining techniques. CD84 was homogeneously distributed on the membrane of nonactivated Jurkat cells, whereas SAP was mostly expressed in the cytosolic compartment (Figure 7A-B). When cells were treated with biotinylated anti-CD84 antibody followed by streptavidin-FITC for 30 minutes at 4°C and were incubated for 10 minutes at 37°C, the CD84 molecules capped (Figure 7C). No capping was detected when cells were incubated under the same conditions with an isotypic control (IgG1) instead of anti-CD84 (data not shown). Jurkat cells that had been treated with anti-CD84 under capping condition were then stained with an CY3 conjugated anti-SAP antibody (10C4).2 Under those conditions, the distribution of SAP and CD84 on the plasma membrane of capped cells overlapped (Figure 7D). No cocapping was detected in CD84 capped cells stained with CY3-conjugated mouse IgG1 isotypic control (data not shown).
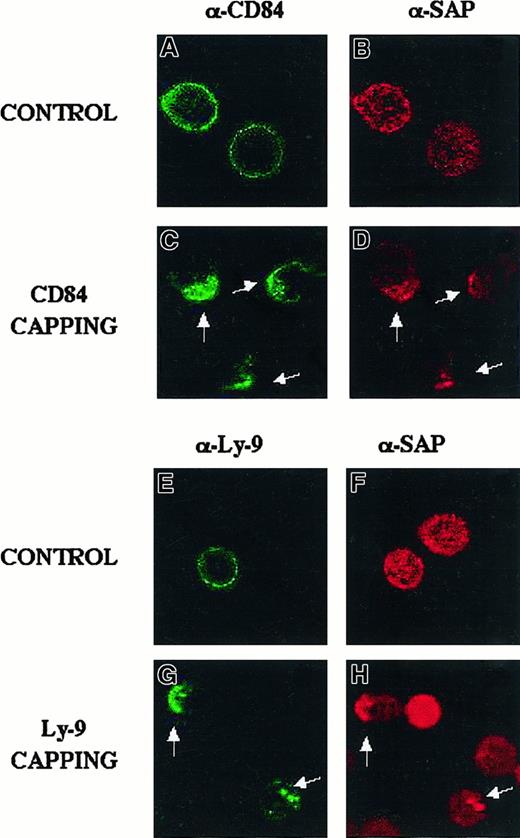
CD84 and Ly-9 colocalize with SAP in Jurkat after activation of the receptors with monoclonal antibody.
A fluorescence microscopy approach was used in the human T-cell line Jurkat to confirm the SAP/Ly-9 and SAP/CD84 interaction in T cells. Jurkat cells were incubated with 5 μg/mL biotinylated anti-CD84 or anti-Ly9 antibody for 30 minutes at 4°C, as detailed in “Materials and methods.” Cells were then incubated with streptavidin-FITC and incubated at 4°C (control) or 10 minutes at 37°C (capping). Cells were then stained with Cy3-SAP, as indicated. Green fluorescence shows CD84 (A, C) or Ly 9 (E, G). Staining with red fluorescence indicates SAP (B, D, F, H). White arrows show colocalization.
CD84 and Ly-9 colocalize with SAP in Jurkat after activation of the receptors with monoclonal antibody.
A fluorescence microscopy approach was used in the human T-cell line Jurkat to confirm the SAP/Ly-9 and SAP/CD84 interaction in T cells. Jurkat cells were incubated with 5 μg/mL biotinylated anti-CD84 or anti-Ly9 antibody for 30 minutes at 4°C, as detailed in “Materials and methods.” Cells were then incubated with streptavidin-FITC and incubated at 4°C (control) or 10 minutes at 37°C (capping). Cells were then stained with Cy3-SAP, as indicated. Green fluorescence shows CD84 (A, C) or Ly 9 (E, G). Staining with red fluorescence indicates SAP (B, D, F, H). White arrows show colocalization.
Identical results were obtained when Jurkat cells were treated with an anti–Ly-9 monoclonal antibody in a similar set of experiments. Under capping conditions, SAP colocalized with Ly-9 (Figure 7E-H). We conclude from the immunofluorescence studies that on triggering of Ly-9 and CD84 with their respective monoclonal antibodies, SAP colocalizes with the receptors at the plasma membrane.
Discussion
Ly-9 and CD84 are both members of the immunoglobulin superfamily, and their ectodomains are related to the other 2 SAP-binding proteins SLAM and 2B4 (Figure 8).2,5-10,23,24 All 4 molecules comprise 2 immunoglobulin-like domains (V and C2 domains) with the exception of Ly-9, which contains a tandem repeat of V-C2 domains (Figure 8). By contrast, CD48 does not have a cytoplasmic tail and has been shown to be attached to the plasma membrane by a conventional phosphoinositol lipid tail.25CD48 serves as the ligand for 2B4,19 and a weak but measurable homophilic interaction between SLAM has been reported.16
Summary of the SAP-binding proteins.
Binding of SAP to the cytoplasmic tail of the cell surface receptors is indicated by a plus sign, and absence of binding to SAP is indicated by a minus sign. SAP binds to the phosphorylated (pY) cytoplasmic tails of SLAM, 2B4, Ly-9, and CD84 and only to the nonphosphorylated (Y) tail of SLAM. CD48 is the ligand for 2B4 (see “Discussion”). B, B lymphocyte; T, T lymphocyte; CD8, CD8+ lymphocyte; COOH, carboxy-terminus; DC, dendritic cell; Mφ, macrophage; NH2, amino-terminus; M, SAP-binding motif; –, N-linked carbohydrate side chain.
Summary of the SAP-binding proteins.
Binding of SAP to the cytoplasmic tail of the cell surface receptors is indicated by a plus sign, and absence of binding to SAP is indicated by a minus sign. SAP binds to the phosphorylated (pY) cytoplasmic tails of SLAM, 2B4, Ly-9, and CD84 and only to the nonphosphorylated (Y) tail of SLAM. CD48 is the ligand for 2B4 (see “Discussion”). B, B lymphocyte; T, T lymphocyte; CD8, CD8+ lymphocyte; COOH, carboxy-terminus; DC, dendritic cell; Mφ, macrophage; NH2, amino-terminus; M, SAP-binding motif; –, N-linked carbohydrate side chain.
The interaction between SAP and the Y281 motif of the cytoplasmic tail of SLAM appears to be unique in that it does not require tyrosine phosphorylation. Understanding the physicochemical parameters of the interactions between SAP and SLAM has been facilitated by the 3-dimensional structure of SAP associated with a peptide that included the binding motif in the cytoplasmic tail of SLAM.10 This peptide binds to SAP regardless of whether its essential tyrosine residue Y281 is phosphorylated. However, the apparent binding constant, as determined by fluorescence polarization, was different with the phosphophorylated peptide (120 nM) than with the nonphosphorylated peptide (830 nM),10 indicating a higher affinity for the phosphorylated peptide. Thus, the structure of SAP and physicochemical studies support the notion that SAP can act as a natural inhibitor.
The physicochemical parameters, which determine the differences in affinity of binding between SAP and the 4 members of the SLAM family, are unknown. However, it is likely that segments of their cytoplasmic tails located outside the binding motif areas could affect the binding affinity. It is an attractive speculation that the observed dependence on phosphorylation of the interaction of SAP and 2B4, Ly9, or CD84 may indicate differences in the way SAP governs signal transduction pathways initiated by these receptors.
Although the function of Ly-9 and CD84 is unknown, it has been suggested that they may participate in adhesion between T lymphocytes and antigen-presenting cells by homophilic interactions.24CD84 is expressed on macrophages and T and B lymphocytes, whereas Ly-9 expression is restricted to T and B cells.22 Thus, 3 known SAP-binding cell surface structures—namely, SLAM, Ly-9, and CD84—are found on the surfaces of activated T cells, whereas 2B4 is detected on the surfaces of CD8 cells and NK cells. Because the SAP gene is expressed in activated NK cells, SAP controls the signal transduction pathways initiated by 2B4 in this cell type.5-9 Given that SLAM expression was induced on the surfaces of mouse NK cells after infection with LCMV, but not after MCMV infection,8 SLAM-SLAM interactions might play a role in NK cells as well.
As expansion of CD8 and CD4 cells has been observed in XLP patients infected with EBV and because NK cell functions are impaired in a subset of XLP patients, the 4 SAP-binding cell surface molecules are likely to have a cumulative effect on the pathogenesis of the disease.
Based on their tissue distribution, we speculate that all 4 receptors are engaged on the surface of a T cell (CD8 cell in the case of 2B4) that recognizes an EBV-infected B cell. Moreover, in the absence of SAP, the CD48/2B4 pair induces abnormal signaling in NK cells. This might happen in concordance with aberrant SLAM signaling in NK cells.
Taken together, our observations are consistent with a model in which T and NK cells, through a group of signaling pathways generated by different receptors, control EBV-transformed B-cell proliferation. The absence of SAP in XLP patients produces a functional impairment of these pathways and results in a deficient or unbalanced production of cytokines (IFN-γ) and in the ineffective NK cell and CD8+T-cell response in sustaining elimination of EBV-infected B cells. However, XLP manifests itself as at least 3 disease states in addition to fatal infectious mononucleosis. Because 3 different disease phenotypes occur within one family, we speculate that the balance of signals initiated through the 4 cell surface receptors may vary from patient to patient. Precise dissection of the pathogenesis of XLP will therefore require an understanding of the signal transduction pathways induced by SLAM, Ly-9, CD84, and 2B4.
We thank Charles Gullo, Ype de Jong, and Kareem Clarke for a critical review of the manuscript, and Martin Sickler for editorial assistance.
Supported by National Institutes of Health grant PO1-AI-35714 (C.T.), the National Foundation March of Dimes (C.T.), and Comision Interministerial de Ciencia y Tecnologia grants SAF97-0136 and SAF00-37 (P.E.). M.M. and M.S. are supported by a Fellowship from Ministerio de Educacion y Cultura, and D.H. is supported by a Postdoctoral Fellowship from the Leukemia and Lymphoma Society.
J.S. and M.M. contributed equally to this project.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Cox Terhorst, Division of Immunology, RE-204, Beth Israel Deaconess Medical Center, Harvard Medical School, Brookline Ave, Boston, MA 02215; e-mail: terhorst@caregroup.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal