Efflux of Hoechst 33342 from normal hematopoietic cells identifies a “side population” (SP+) of negatively staining cells that, in the mouse, are largely CD34− and are enriched for primitive progenitors. To further characterize human SP+cells, blood or bone marrow from 16 patients with acute myeloid leukemia (AML) was analyzed for their presence, immunophenotype, and cytogenetic and functional properties, and for the relation between SP phenotype and multidrug resistance-1 (MDR-1) expression. The mean percentages of SP+ and MDR+ cells was 8.1% (range, 0.5%-29.9%) and 12.8% (range, 0%-54.8%), respectively, with no correlation between the 2 values. The percentages of SP+ cells that were CD34+CD38−, CD34+CD38+, or CD34− were 12% (range, 0.4%-50%), 25% (range, 0.5%-96%), and 63% (range, 4%-99%). Cytogenetically abnormal cells were always detected in the SP−CD34+CD38− and SP+CD34− fractions, and abnormal colonies (CFC), long-term culture-initiating cells (LTC-IC), and nonobese diabetic-severe combined immunodeficiency (NOD/SCID) mouse leukemia–IC were detected in the former fraction. No progenitors were detected among SP+CD34− cells in any of these assays from 9 of 10 samples. In contrast, exclusively normal cells were detected in the SP+CD34+CD38−fraction from 9 of 15 samples, and CFC, LTC-IC, and multilineage engraftment in NOD/SCID mice from this subpopulation were also cytogenetically normal in 6 of 8, 6 of 7, and 2 of 2 cases studied, respectively. In contrast to murine studies, primitive progenitors are enriched among SP+CD34+CD38− cells from patients with AML. The molecular basis for Hoechst dye efflux is uncertain because it does not appear to be related to MDR-1 expression.
Introduction
Acute myeloid leukemia (AML) arises from the clonal expansion of a malignant transformed progenitor cell.1-3Leukemic cells that initiate long-term hematopoiesis in stromal co-cultures (AML LTC-IC) and suspension cultures and engraft in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (NOD/SCID leukemia-initiating cells) are rare progenitors that express a cell surface phenotype similar to that seen on normal multipotential long-term culture-initiating cells (LTC-IC) and lympho-myeloid NOD/SCID mouse competitive repopulating units (CRU).2,4-9 Thus, these assays identify candidate leukemic “stem cells” that may be responsible for maintaining the malignant clone in patients with AML. The immunophenotypic profile of primitive AML progenitor cells is usually characterized by the expression of CD34 but the lack of CD38, Thy-1, CD71, HLA-DR, and c-kitexpression.1,2,10-15 However, in some reports, CD34− leukemic progenitors cells were isolated from patients with AML with blasts that were also largely CD34−.11 16
Several reports demonstrate normal hematopoietic stem cells that lack the expression of CD34, CD38, and lineage-associated markers in murine systems and, using the sheep in utero transplantation model, in primitive human hematopoiesis.17-19 In the NOD/SCID mouse model CD34− lineage-negative (Lin−) human cells with minimum clonogenic and LTC-IC activity had in vivo repopulating activity comparable to that previously described for CD34+ SCID-repopulating cells (SRCs).6,20 In contrast to CD34+ SRCs, the CD34− SRCs did not express Thy-1 or HLA-DR antigens. Furthermore, these CD34−SRCs could be expanded in suspension culture whereas the CD34+ SRCs lost their repopulating capability if grown under the same conditions.20
Goodell et al18 21 have identified murine bone marrow cells with stem cell characteristics by their rapid efflux of the fluorescent vital DNA binding dye, Hoechst 33342. The simultaneous display of the fluorescent staining of cells exposed to Hoechst 33342 at 2 different wavelengths identifies a small and distinct subpopulation termed the side population (SP). These SP+cells are largely CD34− and directly nonclonogenic, and they are enriched for LTC-IC and cells that initiate multilineage marrow reconstitution in mice. SP+ cells have also been demonstrated in other species, including humans. However, functional characterization of the human SP+ cells remains incomplete.
These interesting findings suggest that CD34 may not be as reliable a marker for the identification of hematopoietic stem cells as was previously thought. The experiments described in this report were designed to further investigate Hoechst 33342 dye efflux as a marker for normal and malignant human progenitors. Our goals were, first, to identify and quantitate SP+ cells in AML samples and, second, to demonstrate the functional properties of these cells. Third, we investigated whether SP+ cells in AML samples were part of the leukemic clone. We determined that SP+CD34+CD38− cells from most patients newly diagnosed with AML were highly enriched for normal progenitor cells, including those that initiate long-term cultures and multilineage engraftment in NOD/SCID mice. In contrast, cells with the SP+34−38− phenotype consistently showed the leukemia-specific clonal chromosomal abnormality and no stem cell characteristics in the assay systems we used (LTC-IC growth and NOD/SCID engraftment). These latter data contrast with the original data on SP cells derived from the murine system in which SP+ cells capable of initiating long-term, multilineage hematopoiesis failed to express CD34 or lineage markers.18 21
Materials and methods
Patient samples
Peripheral blood (n = 6) or bone marrow (n = 10) cells were obtained from 16 patients with newly diagnosed AML after informed consent and with the approval of the Clinical Research Ethics Board of the University of British Columbia. Diagnosis and classification of AML were based on the criteria of the French-American-British (FAB) group.22 Cytogenetic analysis was performed on the bone marrow at initial diagnosis. Mononuclear cells were isolated by Ficoll Hypaque density gradient centrifugation (Pharmacia, Uppsala Sweden) and cryopreserved in Iscoves modified Dulbecco medium (IMDM) with 50% fetal calf serum (FCS) (both from Stemcell Technology, Vancouver, Canada) and 10% dimethyl sulfoxide.
Analysis of acute myeloid leukemia cells
Frozen mononuclear peripheral blood or bone marrow cells from patients with AML were rapidly thawed, washed twice in IMDM with 20% FCS, resuspended in serum-free media containing 10−4 M β-mercaptoethanol, 2 mM glutamine, 20 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL G-CSF, 20 ng/mL GM-CSF, and 50 ng/mL Steel factor (SF) in IMDM with 20% BIT (Stemcell) and were incubated overnight at 4°C. Immediately before Hoechst staining, cells were counted and resuspended at 106 cells/mL in DMEM–2% FCS–1 mM HEPES warmed to 37°C as described previously18 and incubated with 5 μg/mL Hoechst 33342 for 120 minutes at 37°C. An aliquot of the Hoechst-stained cells was removed immediately, additionally stained with 50 μM verapamil (Sigma-Aldrich Canada, Oakville, ON, Canada) to block Hoechst dye efflux, and also incubated at 37°C for 120 minutes. After Hoechst staining, cells were centrifuged and resuspended at 107 cells/mL in ice-cold Hanks balanced salt solution (Stemcell) containing 2% FCS for antibody staining. For the sorting experiments, cells that were stained with the Hoechst dye were also stained with antihuman CD34 fluorescein isothiocyanate (FITC) (HPCA-2) and anti–CD38 PE (HB-7) (Becton Dickinson, San Jose, CA). For evaluation of lineage-specific antigen expression, aliquots of cells that were stained with the Hoechst dye were also stained with the following lineage markers: anti–CD15 PE (MMA), anti–CD19 PE (4G7), anti–CD33 PE (P67.6), anti–HLADR PE (all from Becton Dickinson), anti–Thy 1 PE (5E10), anti–c-kit (CD117) PE (YB5.B8), anti–glycophorin A PE (10F7), anti–CD61 PE (V1-PL2) (all Pharmingen, Mississauga, ON, Canada) for 20 minutes at 4°C and were washed with HFN. The monoclonal antibody used for P-glycoprotein (Pgp) staining was UIC2-PE Ab (Pharmingen), which reacts with membrane surface domains of Pgp. After antibody staining, cells were resuspended in HFN including 2 μg/mL propidium iodide (PI) to stain dead cells. Control staining with IgG FITC (Becton Dickinson) and IgG PE (Pharmingen) was used to evaluate nonspecific immunofluorescence. Analysis and sorting experiments were performed on a dual-laser fluorescence-activated cell sorter (FACStar Plus) from Becton Dickinson. The percentage of positively stained cells was determined after setting gates that excluded 99.9% of cells labeled with the isotype control and nonviable cells.
Fluorescence-activated cell sorting
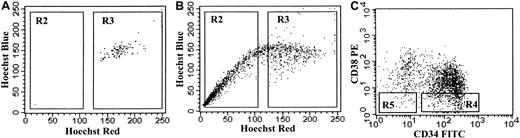
Hoechst dye was excited at 350 nm, and its fluorescence was measured at 2 wavelengths using a 424 bandpass (BP) 44 and a 660 BP 20 optical filter (Omega Optical, Battleboro, VT). A 640-nm long-pass filter was used to separate the emission wavelengths. PI fluorescence was measured through a 715-nm long-pass filter. Hoechst blue represents the 424 BP filter, the standard analysis wavelength for DNA content. Both Hoechst blue and Hoechst red fluorescence are shown on a linear scale. PI staining was used to exclude dead cells. SP+ was defined using control cells stained with both Hoechst and verapamil to establish the SP gate (Figure 1). The sort gates for CD34+/− and CD38+/− cells among SP+ and SP− cells were established using an isotype control. The following subpopulations were sorted: SP+CD34+CD38−, SP+CD34−CD38−, and SP−CD34+CD38−. Cells were sorted into Eppendorf tubes containing serum-free media or directly onto 10-well glass slides.
Sorting strategy for AML cells.
(A) Control staining of AML cells with Hoechst 33342 and verapamil. (B) The FACS profile of AML cells stained with Hoechst 33342 alone, with gate R2 defined as SP+ cells. (C) FACS profile of AML cells stained with CD34-FITC and CD38-PE. Gates were set using the verapamil and IgG FITC/PE controls. The following subpopulations were sorted: R4 and R5 within R2 and R4 within R3.
Sorting strategy for AML cells.
(A) Control staining of AML cells with Hoechst 33342 and verapamil. (B) The FACS profile of AML cells stained with Hoechst 33342 alone, with gate R2 defined as SP+ cells. (C) FACS profile of AML cells stained with CD34-FITC and CD38-PE. Gates were set using the verapamil and IgG FITC/PE controls. The following subpopulations were sorted: R4 and R5 within R2 and R4 within R3.
Cultures of acute myeloid leukemia cells
Assays for AML CFCs were performed by plating sorted cells at 150 to 10 000 cells/mL in methylcellulose medium (0.92% methylcellulose, 30% FCS, 2 mM l-glutamine, 10−4 M β-mercaptoethanol, 1% BSA in IMDM; Stemcell) with 3 U/mL human erythropoietin (EPO; Stemcell), 10 ng/mL GM-CSF (Norvartis, Basel, Switzerland), 10 ng/mL IL-3, (Norvartis) 50 ng/mL SF (Terry Fox Laboratory) and 50 ng/mL Flt-3 ligand (Immunex, Seattle, WA). Cultures were scored after 14 days for the presence of clusters (4-20 cells) and colonies (more than 20 cells). In patients with a leukemia-specific clonal chromosomal abnormality, colonies were plucked onto 10-well glass slides for fluorescence in situ hybridization (FISH) analysis. The relative number of colonies from the sorted subpopulations that would be contained in 1 × 106unsorted cells was calculated by multiplying the percentage of the SP+ (or SP−) sorted subpopulation in the total cell population by the number of colonies per 106 plated cells from that sorted subpopulation.
LTCs of AML cells were established as previously described.23 AML cells at a concentration of 85 to 7500 cells/mL in Myelocult LTC media (Stemcell) with 10−6 M solucortef (Sigma-Aldrich Canada) were cocultured with irradiated (80 Gy) Sl/Sl-J-IL3 feeders, consisting of Sl/Sl fibroblasts, originally obtained from Sl/Sl mouse embryos, and engineered to produce human IL-3 at a concentration of 16.5 ng/mL.23 LTCs were incubated at 37°C in 5% CO2 and received weekly half-media changes and twice-weekly additions of SF to give a final concentration of 50 ng/mL. After 6 weeks, the dishes were harvested by trypsinization, and all cells were placed into methylcellulose assay for the detection of AML-CFC as described above. The relative number of LTC-IC derived colonies from the sorted subpopulations that would be contained in 1 × 106 unsorted cells was calculated by multiplying the percentage of the SP+ (or SP−) sorted subpopulation in the total cell population by the number of colonies per 106 plated cells from that sorted subpopulation.
NOD/SCID mice
NOD/LtSz-scid/scid (NOD/SCID) mice24 were bred and maintained under sterile conditions in the British Columbia Cancer Research Center Joint Animal Facility according to protocols approved by the Animal Care Committee of the University of British Columbia. Eight- to 10-week-old mice were irradiated with 350 cGy from a cesium Cs 137 source 24 hours before injection of AML cells. Sorted AML cells were kept in α-MEM (Stemcell) with 50% FCS and injected into mice through the tail veins. Because of concerns that the seeding efficiency of the AML cells would be reduced when very small numbers of cells were injected, 2 × 106 irradiated (15 Gy) normal human bone marrow cells were co-injected with the sorted cells. After the injection of AML cells into mice, bone marrow aspiration from the femur was performed every 4 weeks after anesthesia with 11 μL/g Avertin (2,2,2-Tribromoethanol; Aldrich, Milwaukee, WI) per mouse injected intraperitoneally. Cells were stored in α-MEM with 50% FCS and were used for FACS staining. Eight to 16 weeks after injection, the mice were killed by CO2 inhalation. Blood cells were harvested by cardiac puncture. Bone marrow was obtained from the 4 long bones by flushing the bones with α-MEM with 50% FCS. The spleen was removed, and a single cell solution was prepared by flushing the cells with α-MEM with 50% FCS. Mice were checked frequently and euthanized earlier if apparent disease or distress developed.
Analysis of mice
To prepare cells from mouse tissue for FACS analysis, samples were incubated with 7% ammonium chloride (Stemcell) to lyse red blood cells, pelleted, and resuspended in Hanks balanced salt solution (Stemcell) with 2% FCS (HFN) and 5% human serum to block human Fc receptors. Cells were then incubated for 10 minutes on ice with an antimouse IgG Fc receptor monoclonal antibody (2.4G2, provided by SyStemix, Palo Alto, CA) for blocking of nonspecific binding to mouse Fc receptors. Then 105 cells were incubated for 30 minutes on ice with a mouse IgG1 isotype control (Becton Dickinson Immunocytometry Systems, San Jose, CA) to evaluate nonspecific immunofluorescence or with fluorescein anti-CD45, a human-specific pan-leukocyte marker (prepared in our center from clone HB10508, American Type Culture Collection, Rockville, MD) to detect human cells. Separate aliquots of cells were incubated for 30 minutes. at 4°C with the following combinations of antibodies against human antigens for the detection of lymphoid and myeloid engraftment: (1) antihuman CD34-FITC (8G12) plus anti–CD19-PE and anti–CD20-PE (both from Becton Dickinson) and (2) anti–CD71-PE (OKT-9) plus anti–CD45-PE, anti–CD15-FITC, anti–CD66b-FITC (all from Pharmingen). Cells were washed twice with HFN, the second wash containing 1 μg/mL PI (Sigma Chemical, St Louis, MO). FACS analysis was performed on a FACScan or a FACSort flow cytometer (Becton Dickinson). Positive cells were defined as those exhibiting a level of fluorescence that exceeded 99.98% of that obtained with isotype-matched control antibodies labeled with the same fluorochromes. Mice were considered negative if there were fewer than 5 CD34+CD19/20+ human cells or fewer than 5 CD45/71+CD15/66b+ human cells per 2 × 104 viable cells analyzed.25
To allow morphologic confirmation of the presence of leukemic cells, smears of mouse bone marrow or cytospins of flow-sorted CD45+ cells were prepared on glass slides. After staining with Wright Giemsa, slides were examined by bright-field microscopy, and cells were scored to determine percentages of AML blasts.
Fluorescence in situ hybridization
Cytospin preparations were obtained from the AML samples after FACS sorting. Colonies from methylcellulose assay of 5-week-old LTCs were plucked onto 10-well slides and fixed in methanol–glacial acetic acid 3:1 for 10 minutes. The centromeric repeat probe used to detect the +8 abnormality (plasmid D8Z2 from American Type Culture Collection), which is specific for human chromosome 8, was labeled with digoxigenin (DIG; Boehringer Mannheim, Mannheim, Germany) by nick translation. For the inv(16) rearrangement, a yeast artificial chromosome clone containing 550 kb human DNA encompassing the breakpoint on 16p13 (CEPHy904E02 854; Max Planck Institut, Berlin, Germany) was labeled by nick translation with digoxigenin. To detect the +13 abnormality, the Quint-Essential (Oncor, Gaithersburg, MD) 13-specific DNA probe labeled with DIG was purchased and used as specified by the manufacturer. The 11q23 MLL probe labeled with DIG was purchased from Oncor. Chromosomes 8 and 16 probes and all slides were treated as previously described.23 Probes were denatured, applied to denatured cells on slides, and hybridized overnight at 37°C. For probe detection slides were incubated with a sheep anti–DIG-FITC antibody (Boehringer Mannheim) at 37°C for 1 hour in the dark, washed, and further incubated for 1 hour with rabbit-antisheep FITC (Vector Labs, Burlingame, CA) and counterstained with PI as previously described.23
Analysis of the slides was performed on a Zeiss (Oberkochen, Germany) Axioplan fluorescence microscope equipped with a double bandpass filter to allow simultaneous visualization of the FITC signal and the PI counterstain. Each colony was scored as either normal or abnormal only if a minimum of 5 cells showed a clear signal and if at least 80% of the cells showed either the normal or the abnormal signal.
Results
Patient characteristics
Leukemic blast samples from 16 patients (12 men, 4 women) with newly diagnosed AML were studied. In 10 patients the bone marrow (BM) was analyzed, and in 6 patients the peripheral blood (PB) was analyzed. The mean age was 54.6 years (range, 25-77 years). The FAB subtype for 2 patients was M1, M2 for 1, M4 for 2, M4Eo for 6, and M5 (a or b) for 4 patients. Refractory anemia with excess of blasts in transformation (RAEB-IT) was diagnosed in 1 patient. Mean white blood cell count was 99 × 109/L (range, 11-370 × 109/L) with a mean blast percentage of 53.3% (range, 5-91) (Table1).
Patient characteristics
| Patient . | Age . | Sex . | Material . | FAB . | WBC (% blasts) (× 109/L) . | Marrow cytogenetics (% abnormal) . |
|---|---|---|---|---|---|---|
| 1 | 48 | M | BM | M4Eo | 101 (71) | 46 XY inv(16)(p13q22)(100) |
| 2 | 77 | M | BM | M2 | 56 (53) | 47 XY, +8, t(12;22); del 20q 8(100) |
| 3 | 36 | M | BM | M4Eo | 85 (90) | 47 XY, inv(16)(p13q22) +22 (100) |
| 4 | 59 | M | PB | M5b | 42 (45) | 48 XY +21 +8 (+1q)(100) |
| 5 | 62 | M | BM | M4 | 14.2 (20) | 48 XY, +8, +20 (12) |
| 6 | 68 | M | BM | RAEB-IT | 63.2 (5) | 48 XY, +8+8 (12) |
| 7 | 58 | F | PB | M4 | 370 (47) | 47 XX, +13 (38) |
| 8 | 46 | M | BM | M5a | 41.3 (5) | 48 XY, +8, +13 (100) |
| 9 | 71 | M | BM | M1 | 10.7 (75) | 47 XY, +8 (100) |
| 10 | 59 | M | BM | M4Eo | 76.1 (91) | 46 XY, inv(16)(p13q22)(93) |
| 11 | 25 | F | PB | M4Eo | 19.6 (9.8) | 46 XX, inv(16)(p13q22)(80) |
| 12 | 68 | M | PB | M4Eo | 27.4 (24) | 45 X-Y, inv(16)(p13q22)(79) |
| 46X-Y, +8, inv(16)(p13q22)(14) | ||||||
| 13 | 74 | F | BM | M 5 | 260 (90) | 46 XX del(11)(q14q25)(100) |
| 14 | 72 | F | BM | M5b | 51 (65) | 47 XX +8 (74) |
| 15 | 30 | M | PB | M4Eo | 47.2 (72) | 46 XY inv(16)(p13q22)(83) |
| 16 | 21 | M | PB | M1 | 320 (90) | 46 XY, add (6) (p23), t(6;11)(q27;q23)(100) |
| Patient . | Age . | Sex . | Material . | FAB . | WBC (% blasts) (× 109/L) . | Marrow cytogenetics (% abnormal) . |
|---|---|---|---|---|---|---|
| 1 | 48 | M | BM | M4Eo | 101 (71) | 46 XY inv(16)(p13q22)(100) |
| 2 | 77 | M | BM | M2 | 56 (53) | 47 XY, +8, t(12;22); del 20q 8(100) |
| 3 | 36 | M | BM | M4Eo | 85 (90) | 47 XY, inv(16)(p13q22) +22 (100) |
| 4 | 59 | M | PB | M5b | 42 (45) | 48 XY +21 +8 (+1q)(100) |
| 5 | 62 | M | BM | M4 | 14.2 (20) | 48 XY, +8, +20 (12) |
| 6 | 68 | M | BM | RAEB-IT | 63.2 (5) | 48 XY, +8+8 (12) |
| 7 | 58 | F | PB | M4 | 370 (47) | 47 XX, +13 (38) |
| 8 | 46 | M | BM | M5a | 41.3 (5) | 48 XY, +8, +13 (100) |
| 9 | 71 | M | BM | M1 | 10.7 (75) | 47 XY, +8 (100) |
| 10 | 59 | M | BM | M4Eo | 76.1 (91) | 46 XY, inv(16)(p13q22)(93) |
| 11 | 25 | F | PB | M4Eo | 19.6 (9.8) | 46 XX, inv(16)(p13q22)(80) |
| 12 | 68 | M | PB | M4Eo | 27.4 (24) | 45 X-Y, inv(16)(p13q22)(79) |
| 46X-Y, +8, inv(16)(p13q22)(14) | ||||||
| 13 | 74 | F | BM | M 5 | 260 (90) | 46 XX del(11)(q14q25)(100) |
| 14 | 72 | F | BM | M5b | 51 (65) | 47 XX +8 (74) |
| 15 | 30 | M | PB | M4Eo | 47.2 (72) | 46 XY inv(16)(p13q22)(83) |
| 16 | 21 | M | PB | M1 | 320 (90) | 46 XY, add (6) (p23), t(6;11)(q27;q23)(100) |
BM indicates bone marrow; PB, peripheral blood; FAB, French-American-British group; WBC, white blood cell; RAEB-IT, refractory anemia with excess of blasts in transformation.
Frequency of total side population cells and immunophenotypic profiles
Figure 1 shows a typical FACS profile of AML cells stained with Hoechst 33342 with and without verapamil to identify SP+cells, followed by an analysis of gated cells for the expression of CD34 and CD38. This strategy was used to analyze the 16 AML patient samples as shown on Table 2. Mean frequency of SP+ nucleated cells in the BM or PB was 8% (range, 0.53%-29.9%). Mean frequency of the CD34+CD38−, CD34+CD38+, CD34−CD38+, and CD34−CD38− subpopulations among the SP+ cells was 12.3% (range, 0.4%- 49.5%), 24.9% (range, 0.5%-96%), 23.9% (range, 0%-90%), and 31.8% (range, 1.4%-76.3%), respectively, with no significant difference between PB and BM. Staining for the expression of lineage markers on SP+ cells revealed a mean percentage of 0.6% (range, 0%-1.7%) Thy 1+, 0.6% (range, 0%-3.5%) c-kit+, 44% (range, 16.5%-68%) CD33+, 66% (range, 1.7%-93.5%) HLA-DR+, 50% (range, 11%-83%) CD15+, 1.6% (range, 0.6%-2.5%) GlyA+, 20% (range, 7.2%-39.3%) CD61+, and 24% (range, 2.1%-68.4) CD19+ cells. Within the SP, it was also possible to detect cells with the aberrant co-expression of lineage markers often seen on AML blasts—a mean percentage of 23.2% (range, 0%-71.4%) cells co-expressed CD34 and CD15, 5.5% (range, 0%-16.4%) co-expressed CD34 and CD33, and 30.8% (range, 0%-61.3%) co-expressed CD34 and HLA-DR. On average, the expression of Thy 1 was found on 0.2% (range, 0%-0.9%) and c-kit on 0.1% (range, 0%-0.6%) of the SP+CD34+ cells.
Frequency of the side population, MDR (UIC2), and CD34/CD38 expression in acute myeloid leukemia blood and marrow
| Patient . | SP+ (%) . | MDR+(%) . | SP+ cells in different subpopulations (%) . | ||||
|---|---|---|---|---|---|---|---|
| MDR+ . | CD34+CD38− . | CD34+CD38+ . | CD34−CD38+ . | CD34−CD38− . | |||
| 1 | 9.1 | 8.1 | 2.3 | 24 | 8.1 | 39.5 | 28.4 |
| 2 | 4.8 | 17.3 | 0 | 20.5 | 35.9 | 3.7 | 39.9 |
| 3 | 5.7 | 4.9 | 1.0 | 34.3 | 11.9 | 25.7 | 28.1 |
| 4 | 18.1 | 10.2 | 5.8 | 0.4 | 1.3 | 22 | 76.3 |
| 5 | 2.8 | 22 | 14.2 | 1.9 | 0 | 58.1 | 40 |
| 6 | 4.9 | 0.1 | 0 | 49.5 | 2 | 16.7 | 31.9 |
| 7 | 13.3 | 9.4 | 1.1 | 1.1 | 18.2 | 74.7 | 6.0 |
| 8 | 1.3 | 54.8 | 0.3 | 0.6 | 0.4 | 45.0 | 54.0 |
| 9 | 2.8 | 0 | 0 | 19.3 | 54 | 6.9 | 19.8 |
| 10 | 29.9 | 13 | 16.6 | 5.7 | 2.7 | 90.3 | 1.4 |
| 11 | 3.8 | 0 | 0 | 13.8 | 51.1 | 22.2 | 12.9 |
| 12 | 19.7 | 11.5 | 11.4 | 4.7 | 47.2 | 6.5 | 41.7 |
| 13 | 0.5 | 36.1 | 36.4 | 12.2 | 17.1 | 34.2 | 36.6 |
| 14 | 1.2 | 5.1 | 4.1 | 1.5 | 5.2 | 45.6 | 47.8 |
| 15 | 6.4 | 0 | 0 | 5.9 | 48.5 | 5.0 | 40.6 |
| 16 | 5.6 | ND | ND | 1.8 | 94.3 | 0 | 3.9 |
| Patient . | SP+ (%) . | MDR+(%) . | SP+ cells in different subpopulations (%) . | ||||
|---|---|---|---|---|---|---|---|
| MDR+ . | CD34+CD38− . | CD34+CD38+ . | CD34−CD38+ . | CD34−CD38− . | |||
| 1 | 9.1 | 8.1 | 2.3 | 24 | 8.1 | 39.5 | 28.4 |
| 2 | 4.8 | 17.3 | 0 | 20.5 | 35.9 | 3.7 | 39.9 |
| 3 | 5.7 | 4.9 | 1.0 | 34.3 | 11.9 | 25.7 | 28.1 |
| 4 | 18.1 | 10.2 | 5.8 | 0.4 | 1.3 | 22 | 76.3 |
| 5 | 2.8 | 22 | 14.2 | 1.9 | 0 | 58.1 | 40 |
| 6 | 4.9 | 0.1 | 0 | 49.5 | 2 | 16.7 | 31.9 |
| 7 | 13.3 | 9.4 | 1.1 | 1.1 | 18.2 | 74.7 | 6.0 |
| 8 | 1.3 | 54.8 | 0.3 | 0.6 | 0.4 | 45.0 | 54.0 |
| 9 | 2.8 | 0 | 0 | 19.3 | 54 | 6.9 | 19.8 |
| 10 | 29.9 | 13 | 16.6 | 5.7 | 2.7 | 90.3 | 1.4 |
| 11 | 3.8 | 0 | 0 | 13.8 | 51.1 | 22.2 | 12.9 |
| 12 | 19.7 | 11.5 | 11.4 | 4.7 | 47.2 | 6.5 | 41.7 |
| 13 | 0.5 | 36.1 | 36.4 | 12.2 | 17.1 | 34.2 | 36.6 |
| 14 | 1.2 | 5.1 | 4.1 | 1.5 | 5.2 | 45.6 | 47.8 |
| 15 | 6.4 | 0 | 0 | 5.9 | 48.5 | 5.0 | 40.6 |
| 16 | 5.6 | ND | ND | 1.8 | 94.3 | 0 | 3.9 |
ND indicates not done; SP, side population; MDR, multidrug resistance-1.
Multidrug resistance-1 expression on SP cells
To investigate one possible reason for the relatively high proportion of SP+ cells in AML samples, multidrug resistance-1 (MDR-1) expression was measured by FACS using the UIC2-PE antibody. The mean frequency of MDR+ cells in the samples was 12.8% (range, 0%-54.8%). Within the side population, a mean of 8.3% of cells was positive for UIC2 (range, 0%-43.2%). There was no correlation between the frequency of SP+ cells and the frequency of MDR+ cells. In fact, 3 samples (numbers 9, 11, 15) in which no cells stained with the UIC2 antibody had easily detectable SP+ cells, and 1 sample (number 2) with a large fraction of UIC2+ cells in the total population showed no expression of MDR among SP+ cells (Table 2).
Fluorescence in situ hybridization of sorted subpopulations
Initial analysis of unsorted AML cells confirmed that the FISH probes selected could detect the abnormality diagnosed by conventional bone marrow cytogenetics in all patient samples. The proportion of abnormal cells detected by FISH varied from 6% to 100% among the unsorted samples and, in most cases, was similar to the proportion of metaphase cells found abnormal by conventional cytogenetics (Tables 1,3). AML cells were sorted using Hoechst 33342 dye efflux and expression of CD34 and CD38 to isolate 3 subpopulations of interest. SP+CD34−CD38− cells were studied because, in the mouse, SP+ cells with long-term hematopoietic reconstituting ability lack the expression of CD34 and lineage markers.21 Hoechst dye efflux was used to further characterize CD34+CD38− cells into SP+CD34+CD38− and SP−CD34+CD38− fractions given that the CD34+CD38− phenotype is known to identify normal and AML cells that initiate long-term cultures and growth in NOD/SCID mice. FISH analysis of the 3 sorted subpopulations was successful with 15 patient samples (Table 3). In 14 of 15 samples, a mosaic of normal and leukemic cells was observed in the SP−CD34+CD38− subpopulation whereas in the remaining patient (patient 9), exclusively abnormal cells were detected. In contrast, only normal cells in the SP+CD34+CD38− subpopulation were found in 9 of 15 patients. An additional 3 patients (patients 9, 10, 12) had significantly smaller proportions of leukemic cells in the SP+CD34+CD38− subpopulation than among SP−CD34+CD38− cells; in the remaining 3 patients (patients 2, 5, 8), a similar frequency of abnormal cells was found in both subpopulations. In all patients analyzed, the SP+CD34−CD38−subpopulation contained leukemic blasts. In 9 of 15 patients, a mosaic of normal and leukemic cells was seen, and in 5 patients only leukemic cells were detected by FISH. Interestingly, the latter 5 samples from patients 1, 3, 11, 13, and 16 contained only normal cells in the SP+34+38− subpopulation (Table 3).
Fluorescence in situ hybridization of sorted cells
| Patient . | Cytogenetic marker at diagnosis (% abnormal)3-150 . | SP+CD34+CD38− . | SP−CD34+CD38− . | SP+CD34−CD38− . | |||
|---|---|---|---|---|---|---|---|
| No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | ||
| 1 | inv 16 (72.9) | 0/139 | 0 | 7/29 | 24 | 7/7 | 100 |
| 2 | +8 (100) | 30/30 | 100 | 29/37 | 78.4 | 11/19 | 58 |
| 3 | inv 16 (73) | 0/20 | 0 | 10/16 | 62.5 | 5/5 | 100 |
| 4 | +8 (97.7) | 0/10 | 0 | 7/9 | 78 | 14/117 | 12 |
| 5 | +8 (6) | 5/50 | 10 | 5/47 | 10.6 | 5/21 | 24 |
| 6 | +8 (54.7) | 0/46 | 0 | 4/110 | 3.6 | 2/29 | 7 |
| 7 | +13 (44.5) | 0/15 | 0 | 8/26 | 31 | 15/37 | 41 |
| 8 | +8 (90.4) | 3/9 | 33.3 | 5/26 | 19.2 | 29/39 | 74 |
| 9 | +8 (97.5) | 2/13 | 15.4 | 60/60 | 100 | 4/13 | 31 |
| 10 | inv 16 (93) | 5/57 | 8.8 | 46/57 | 80.7 | NS | NS |
| 11 | inv 16 (84.6) | 0/20 | 0 | 25/28 | 89.3 | 16/16 | 100 |
| 12 | inv 16 (94) | 2/16 | 12.5 | 30/33 | 91 | 9/31 | 29 |
| 13 | del(11)(q14q25)(100) | 0/31 | 0 | 47/74 | 63.5 | 50/50 | 100 |
| 14 | +8 (74) | NS | NS | NS | NS | NS | NS |
| 15 | inv 16 (83) | 0/36 | 0 | 9/36 | 25 | 8/20 | 40 |
| 16 | t(6;11)(q27;q23)(100) | 0/33 | 0 | 29/50 | 58 | 37/37 | 100 |
| Patient . | Cytogenetic marker at diagnosis (% abnormal)3-150 . | SP+CD34+CD38− . | SP−CD34+CD38− . | SP+CD34−CD38− . | |||
|---|---|---|---|---|---|---|---|
| No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | ||
| 1 | inv 16 (72.9) | 0/139 | 0 | 7/29 | 24 | 7/7 | 100 |
| 2 | +8 (100) | 30/30 | 100 | 29/37 | 78.4 | 11/19 | 58 |
| 3 | inv 16 (73) | 0/20 | 0 | 10/16 | 62.5 | 5/5 | 100 |
| 4 | +8 (97.7) | 0/10 | 0 | 7/9 | 78 | 14/117 | 12 |
| 5 | +8 (6) | 5/50 | 10 | 5/47 | 10.6 | 5/21 | 24 |
| 6 | +8 (54.7) | 0/46 | 0 | 4/110 | 3.6 | 2/29 | 7 |
| 7 | +13 (44.5) | 0/15 | 0 | 8/26 | 31 | 15/37 | 41 |
| 8 | +8 (90.4) | 3/9 | 33.3 | 5/26 | 19.2 | 29/39 | 74 |
| 9 | +8 (97.5) | 2/13 | 15.4 | 60/60 | 100 | 4/13 | 31 |
| 10 | inv 16 (93) | 5/57 | 8.8 | 46/57 | 80.7 | NS | NS |
| 11 | inv 16 (84.6) | 0/20 | 0 | 25/28 | 89.3 | 16/16 | 100 |
| 12 | inv 16 (94) | 2/16 | 12.5 | 30/33 | 91 | 9/31 | 29 |
| 13 | del(11)(q14q25)(100) | 0/31 | 0 | 47/74 | 63.5 | 50/50 | 100 |
| 14 | +8 (74) | NS | NS | NS | NS | NS | NS |
| 15 | inv 16 (83) | 0/36 | 0 | 9/36 | 25 | 8/20 | 40 |
| 16 | t(6;11)(q27;q23)(100) | 0/33 | 0 | 29/50 | 58 | 37/37 | 100 |
NS indicates FISH analysis was unsuccessful. See Table 2 for other abbreviations.
As determined on unsorted samples by FISH.
In vitro growth characteristics of SP cells
Cells from the 3 FACS-sorted subpopulations from 10 patient samples were placed in CFC and LTC-IC assays. The number of cells plated varied from 50 to 10 000 cells/mL in the CFC and 85 to 7500 cells/mL in the LTC-IC assay, depending on the subpopulation frequency in the patient sample tested. In the CFC assay, colony growth was observed for 8 of 10 samples from the SP+CD34+CD38− subpopulation and 5 of 9 samples from the SP−CD34+CD38− subpopulation, whereas only 1 of 10 samples (patient 7) had any colony growth from the SP+CD34−CD38− fraction. The relative number of CFC per 1 × 106 unsorted cells ranged from 0 to 1780 for SP+CD34+CD38−cells and from 0 to 15 960 for the SP−CD34+CD38− subpopulation. Although most CFCs were found in the SP−CD34+CD38− fraction from patients 2, 5, 7, and 8, for patients 1, 3, and 13 the CFCs were predominantly SP+CD34+CD38− (Table4).
In vitro growth characteristics of side population cells: colony-forming cell and 5-week long-term culture-initiating cell assays
| Patient . | Total viable cells (%) . | CFC/106 unsorted cells4-150 . | LTC-derived CFC/106unsorted cells4-150 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SP+34+38− . | SP−34+38− . | SP+34−38− . | SP+34+38− . | SP−34+38− . | SP+34−38− . | SP+34+38− . | SP−34+38− . | SP+34−38− . | |
| 1 | 2.2 | 31.6 | 2.6 | 74.8 | 47.4 | 0 | 134 | 0 | 0 |
| 2 | 1.0 | 9.5 | 1.9 | 1780 | 15 960 | 0 | 44 | 57 | 0 |
| 3 | 2.3 | 20.2 | 1.9 | 2.3 | 0 | 0 | 0 | 222 | 0 |
| 4 | 0.08 | 0.1 | 14.3 | 17.6 | ND | 0 | 49.6 | ND | 0 |
| 5 | 0.05 | 0.5 | 7.5 | 1.3 | 15 | 0 | 82.9 | 0 | 0 |
| 6 | 0.09 | 4.7 | 1.6 | 0 | 0 | 0 | 3.8 | 73.8 | 0 |
| 7 | 0.014 | 0.8 | 0.08 | 0.5 | 4 | 2.8 | 0.07 | 0 | 0.04 |
| 8 | 0.048 | 2.9 | 4.3 | 44.8 | 174 | 0 | 0 | 0 | 0 |
| 13 | 0.061 | 26.6 | 0.2 | 3.1 | 0 | 0 | 1.6 | 0 | 0 |
| 14 | 0.018 | 0.3 | 0.6 | 0 | 0 | 0 | 0 | 68.6 | 0 |
| Patient . | Total viable cells (%) . | CFC/106 unsorted cells4-150 . | LTC-derived CFC/106unsorted cells4-150 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SP+34+38− . | SP−34+38− . | SP+34−38− . | SP+34+38− . | SP−34+38− . | SP+34−38− . | SP+34+38− . | SP−34+38− . | SP+34−38− . | |
| 1 | 2.2 | 31.6 | 2.6 | 74.8 | 47.4 | 0 | 134 | 0 | 0 |
| 2 | 1.0 | 9.5 | 1.9 | 1780 | 15 960 | 0 | 44 | 57 | 0 |
| 3 | 2.3 | 20.2 | 1.9 | 2.3 | 0 | 0 | 0 | 222 | 0 |
| 4 | 0.08 | 0.1 | 14.3 | 17.6 | ND | 0 | 49.6 | ND | 0 |
| 5 | 0.05 | 0.5 | 7.5 | 1.3 | 15 | 0 | 82.9 | 0 | 0 |
| 6 | 0.09 | 4.7 | 1.6 | 0 | 0 | 0 | 3.8 | 73.8 | 0 |
| 7 | 0.014 | 0.8 | 0.08 | 0.5 | 4 | 2.8 | 0.07 | 0 | 0.04 |
| 8 | 0.048 | 2.9 | 4.3 | 44.8 | 174 | 0 | 0 | 0 | 0 |
| 13 | 0.061 | 26.6 | 0.2 | 3.1 | 0 | 0 | 1.6 | 0 | 0 |
| 14 | 0.018 | 0.3 | 0.6 | 0 | 0 | 0 | 0 | 68.6 | 0 |
ND indicates not done (no cells were placed in the colony-forming cell [CFC] or long-term culture-initiating cells assays [LTC-IC]). See Table 2 for other abbreviations.
Calculated as described in “Materials and methods.”
LTC-IC activity was detected in 7 of 10 5-week-old cultures LTC-ICs of SP+CD34+CD38− cells and in 4 of 9 LTCs of SP−CD34+CD38−cells. Among SP+CD34−CD38− cells only 1 sample, from patient 7, which also showed growth in the CFC assay, showed LTC-IC activity. The relative number of week 5 LTC-IC–derived CFC per 1 × 106 unsorted cells ranged from 0 to 134 in the SP+CD34+CD38−cells compared to 0 to 223 in the SP−CD34+CD38− subpopulation (Table 4). Although most CD34+CD38− LTC-IC were SP+ in patients 1, 5, and 13, they were primarily SP− in patients 3, 6, and 14. Thus, among the 3 FACS-sorted subpopulations studied, in terms of both CFC and LTC-IC content, the most distinct was the SP+CD34−CD38−fraction from which growth could be obtained for only 1 of 10 samples. The majority of both CFCs and LTC-ICs detected were CD34+CD38−, and the proportion of these progenitors found in the side population varied from sample to sample.
FISH analysis of CFC- and 5-week LTC-IC–derived colonies
FISH analysis was performed on colonies plucked from direct CFC assays and colony assays from 5-week-old LTCs of FACS-sorted AML cells (Table 5). SP+CD34+CD38− CFCs from 6 of 8 samples, in which colony growth was observed and FISH analysis was successful, were entirely normal by FISH. Similarly, LTC-IC assays of SP+CD34+CD38− cells from 6 of 7 analyzable samples yielded colonies that were exclusively normal by FISH. Only SP+CD34+CD38− cells isolated from patient 2 generated CFCs and LTC-ICs that were entirely abnormal by FISH, consistent with the finding that 100% of SP+CD34+CD38− cells from this patient, analyzed directly after FACS sorting, were also cytogenetically abnormal (Table 3).
Fluorescence in situ hybridization of colonies from colony-forming cell and long-term culture-initiating cell assays of sorted cells
| Patient . | CFC . | LTC-IC . | ||||||
|---|---|---|---|---|---|---|---|---|
| SP+CD34+CD38− . | SP−CD34+CD38− . | SP+CD34+CD38− . | SP−CD34+CD38− . | |||||
| No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | |
| 1 | 0/16 | 0 | 3/7 | 42.9 | 0/17 | 0 | NG | — |
| 2 | 20/20 | 100 | 26/26 | 100 | 14/14 | 100 | 10/10 | 100 |
| 3 | 0/1 | 0 | NG | — | NG | — | NS | — |
| 4 | 0/26 | 0 | ND | — | 0/9 | 0 | ND | — |
| 5 | 0/7 | 0 | 0/5 | 0 | 0/18 | 0 | NG | — |
| 6 | NG | — | NG | — | 0/30 | 0 | 0/20 | 0 |
| 7 | 0/4 | 0 | 3/10 | 30 | 0/17 | 0 | NG | — |
| 8 | 0/9 | 0 | 2/9 | 22.2 | NG | — | NG | — |
| 13 | 1/30 | 3 | NG | — | 0/10 | 0 | NG | — |
| 14 | NG | — | NG | — | NG | — | NS | — |
| Patient . | CFC . | LTC-IC . | ||||||
|---|---|---|---|---|---|---|---|---|
| SP+CD34+CD38− . | SP−CD34+CD38− . | SP+CD34+CD38− . | SP−CD34+CD38− . | |||||
| No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | No. abnormal/no. analyzed . | Abnormal (%) . | |
| 1 | 0/16 | 0 | 3/7 | 42.9 | 0/17 | 0 | NG | — |
| 2 | 20/20 | 100 | 26/26 | 100 | 14/14 | 100 | 10/10 | 100 |
| 3 | 0/1 | 0 | NG | — | NG | — | NS | — |
| 4 | 0/26 | 0 | ND | — | 0/9 | 0 | ND | — |
| 5 | 0/7 | 0 | 0/5 | 0 | 0/18 | 0 | NG | — |
| 6 | NG | — | NG | — | 0/30 | 0 | 0/20 | 0 |
| 7 | 0/4 | 0 | 3/10 | 30 | 0/17 | 0 | NG | — |
| 8 | 0/9 | 0 | 2/9 | 22.2 | NG | — | NG | — |
| 13 | 1/30 | 3 | NG | — | 0/10 | 0 | NG | — |
| 14 | NG | — | NG | — | NG | — | NS | — |
In contrast, among the 5 samples in which CFCs were detected among SP−CD34+CD38− cells, 4 contained cytogenetically abnormal colonies. LTC-IC assays of the SP−CD34+CD38− subpopulation yielded colony growth from 4 of 9 patient samples. FISH analysis was successful in 2 of these, with one sample (patient 6) showing exclusively normal colonies whereas colonies from the other sample were entirely abnormal (patient 2) (Table 5).
Engraftment of NOD/SCID mice with sorted subpopulations of cells from patients with AML
After Hoechst 33342 staining and FACS sorting, the SP+CD34+CD38−, SP−CD34+CD38−, and SP+CD34−CD38− subpopulations from 4 patients (patients 1, 4, 7, and 16) were transplanted into cohorts of 1 to 3 NOD/SCID mice. Engraftment was observed in 1 mouse at week 8 and in 2 mice at week 12 after injection of the cells from patient 1 (Table6). The proportion of human cells in mouse bone marrow at week 8 was 0.77% in the mouse injected with 1 × 105SP+CD34+CD38− cells. Among 20 000 cells from this mouse marrow analyzed by FACS, 15 cells co-expressed CD45/CD71 and CD15/CD66b and 20 cells expressed CD19/CD20, consistent with engraftment with both myeloid and B-lymphoid human cells. FISH analysis of 258 sorted CD45+ cells from this animal revealed only cytogenetically normal cells that had lymphoid morphology on Wright-Giemsa staining. The 2 mice that showed engraftment at week 12 were injected with 1 × 105SP+CD34+CD38− and 1 × 106SP−CD34+CD38− cells and showed 0.21% and 0.6% CD45+ cells in mouse marrow, respectively. FISH analysis was normal for all 67 CD45+ cells analyzed from the SP+CD34+CD38− mouse, whereas 26 of 44 (55%) of CD45+ cells from the SP−CD34+CD38− mouse showed the inv(16) rearrangement.
Engraftment of sorted subpopulations in nonobese diabetic-severe combined immunodeficient mice
| Patient . | Subpopulation . | No. cells injected per mouse . | CD45+ wks 8-12 (%) . | FISH . | |
|---|---|---|---|---|---|
| No. cells abnormal/ no. analyzed . | Abnormal (%) . | ||||
| 1 | SP+CD34+CD38− | 2 × 105 | 0 | ||
| 1 × 105 | 0.77, 0.21 | 0/258, 0/67 | 0 | ||
| SP−CD34+CD38− | 1 × 105 | 0 | |||
| 3 × 105 | 0 | ||||
| 1 × 106 | 0.6 | 26/44 | 55 | ||
| SP+CD34−CD38− | 3.7 × 104 | 0 | |||
| 7 | SP+CD34+CD38− | 8.7 × 104 | 0.34 | 0/55 | 0 |
| SP−CD34+CD38− | 8.7 × 104 | 0 | |||
| 1.8 × 105 | 0 | ||||
| SP+CD34−CD38− | 8.7 × 104 | 0 | |||
| Patient . | Subpopulation . | No. cells injected per mouse . | CD45+ wks 8-12 (%) . | FISH . | |
|---|---|---|---|---|---|
| No. cells abnormal/ no. analyzed . | Abnormal (%) . | ||||
| 1 | SP+CD34+CD38− | 2 × 105 | 0 | ||
| 1 × 105 | 0.77, 0.21 | 0/258, 0/67 | 0 | ||
| SP−CD34+CD38− | 1 × 105 | 0 | |||
| 3 × 105 | 0 | ||||
| 1 × 106 | 0.6 | 26/44 | 55 | ||
| SP+CD34−CD38− | 3.7 × 104 | 0 | |||
| 7 | SP+CD34+CD38− | 8.7 × 104 | 0.34 | 0/55 | 0 |
| SP−CD34+CD38− | 8.7 × 104 | 0 | |||
| 1.8 × 105 | 0 | ||||
| SP+CD34−CD38− | 8.7 × 104 | 0 | |||
Among mice injected with cells from patient 7, one animal that received 1 × 105SP+CD34+CD38− cells engrafted at week 12 with 0.34% CD45+ cells. Thirty-one of 20 000 mouse marrow cells analyzed by FACS co-expressed CD45/CD71 and CD15/CD66b, whereas 28 cells expressed CD19/CD20, consistent with lymphomyeloid engraftment. FISH analysis of the sorted CD45+ BM cells of this mouse revealed all 55 cells to be cytogenetically normal (Table 6). The morphology of these cells was lymphoid. No engraftment was achieved with any of the sorted subpopulations from patients 4 and 16.
Discussion
One of the challenges in devising novel therapy for AML is to identify clear differences between normal and leukemic progenitor cells that can be targeted.26,27 It seems logical that the AML progenitors, identified by their ability to maintain long-term malignant hematopoiesis in vitro and in vivo (in immunodeficient mice), would be important targets for antileukemic therapy. Although cell-sorting studies have suggested much similarity between the immunophenotype of the most primitive normal and leukemic progenitors detected in these assays, some differences have been identified. For example, though most SCID leukemia-initiating cells and AML LTC-IC are similar to normal LTC-IC and NOD/SCID CRU in their expression of CD34 and their lack of expression of CD38, they often differ from many of these primitive normal cells in their lack of Thy-1 expression.1,4-6,10,28 However, among AML samples, variability in the success with which these antigens can be used to isolate and purify progenitor cell populations is frequently seen.11,16 In addition, evidence is accumulating to suggest that some markers, such as CD34, may not be as reliable in identifying primitive normal progenitors as was previously thought.19-21,29,30 Thus, it seemed relevant to examine the value of using new characteristics attributed to normal stem cells to study AML progenitors. We chose to analyze the side population of hematopoietic cells, identified by their efflux of Hoechst 33342, because little was known about the functional properties of this population in either normal or leukemic human hematopoiesis.21 Our first observation was that cells with the characteristics of SP cells, as defined by Goodell et al18 in the mouse, were easily identified in the marrow and peripheral blood of patients with newly diagnosed AML. Although the frequency of such cells varied greatly from sample to sample, in general it was much higher (4- to 500-fold) than that seen in normal mouse or human marrow samples.21
One possible explanation for the high frequency of SP+cells we observed might be the overexpression of the multidrug transporter protein, Pgp or MDR-1, in the AML blasts analyzed. High levels of Pgp expression have been well-described in AML, both at the time of diagnosis and at disease recurrence.31-36 Efflux of Hoechst 33342 is blocked by verapamil, which is known to inhibit the Pgp transporter function. Thus, we reasoned that SP cells would probably express high levels of this protein. However, in this study we were unable to demonstrate a correlation between the expression of Pgp in AML cells (as detected by FACS analysis using the UIC2 antibody) and the presence of SP cells in the same population. In fact, for several patient samples, none of the isolated SP cells showed detectable staining with the UIC2 antibody (Table 2). Although it is possible that the flow cytometry technique was insufficiently sensitive to detect low levels of Pgp expression—thus leading to false-negative results—it may be more likely that the mechanism of Hoechst dye efflux is more complex than previously thought, perhaps involving ABC transporter molecules sensitive to verapamil inhibition, such as the multidrug resistance protein or breast cancer-related protein or unknown molecules.37-39 Additional studies designed to elucidate the molecular mechanisms involved in Hoechst dye efflux will be necessary to clarify these issues.
Our second observation was that the side population detected in AML samples is heterogeneous with regard to the expression of other antigens expressed by primitive and mature hematopoietic cells. CD34, CD38, and various markers of lineage determination were expressed at different frequencies among the SP cells. Aberrant expression of antigens associated with hematopoietic differentiation is well recognized in AML and is often used for clinical monitoring of minimal residual disease.40-43 Our finding that aberrant or discordant antigen expression could be detected on the side population from AML samples is consistent with the fact that the SP population contained leukemic cells. However, expression of Thy-1 or c-kit among SP cells was infrequent (0%-0.9%), a finding in keeping with data from SP cells in normal bone marrow and AML progenitors detected in long-term suspension culture and in immunodeficient mice, both of which seem to lack these antigens.10,12 21
When SP+ cells were sorted according to their expression of CD34 and CD38, CD34+CD38−cells from 9 of 15 AML samples were entirely cytogenetically normal, as were most CFC, LTC-IC, and cells engrafting in NOD/SCID mice from this population. In contrast, leukemic cells could be easily detected among SP+CD34−CD38− cells and among CD34+CD38− cells outside the side population. Many CFC, LTC-IC, and, in one case, NOD/SCID-engrafting cells grown from the SP−CD34+CD38− fraction were determined to be abnormal by FISH. Thus, the FISH data obtained directly from sorted AML cells and from cells detected in functional in vitro and in vivo assays are consistent in demonstrating the segregation of normal from leukemic CD34+CD38−cells by Hoechst dye efflux. Interestingly, SP+CD34−CD38− cells from all but one patient contained no detectable CFC or LTC-IC, and in no case did these cells engraft in NOD/SCID mice. The latter data contrast with those from murine systems in which SP cells do not express CD34 but, nevertheless, are enriched for progenitors with multilineage competitive bone marrow repopulating ability.18 Similarly, rhesus monkey CD34−, Lin− SP cells are enriched for LTC-IC and T-cell precursors.21 However, recent data on the phenotypic and functional properties of SP cells isolated from human fetal liver and umbilical cord blood suggest that the expression of CD34 within this population is not uncommon.44,45 Thus, human SP cells isolated from healthy persons and patients with leukemia appear to be more heterogeneous in their expression of CD34 (and other cell surface markers) than SP cells from mice and other species. Nevertheless, among CD34+CD38− human cells, Hoechst dye efflux appears to identify cells enriched for normal LTC-IC and NOD/SCID-repopulating ability (Tables 4-6).45 Thus, this property appears to be a consistent feature of primitive hematopoietic cells isolated from many sources.
Although we could not demonstrate the presence of LTC-IC or NOD/SCID-repopulating cells in the SP+CD34−cell fraction from our AML samples, it remains possible that such cells exist but were not detected in our experiments because of their relatively low frequencies. It is also possible that prestimulation with cytokines would have enhanced the engraftment potential of the CD34− cells from our samples, as has been observed by other investigators.20 Nevertheless, our data clearly indicate that some CD34+ human SP cells have the functional properties expected of primitive hematopoietic progenitors with multilineage potential. Furthermore, the fact that combining Hoechst dye efflux with analysis for CD34 and CD38 expression allowed a sorting strategy to be developed that discriminates between normal and leukemic progenitors in many AML samples is a significant finding that may be exploited to allow further understanding of leukemic stem cell biology. Although the frequency of cytogenetically normal progenitors detected was low in some of these samples, the isolation of these cells as targets for stem cell expansion or gene therapy/gene manipulation may ultimately prove to be clinically relevant.46
We thank Brigitte Gerhard for technical assistance, Gayle Thornbury, Giovanna Cameron, and Rick Zapf for FACS operation, and Christine Kelly for help with manuscript preparation.
Supported by grants from the National Cancer Institute of Canada with funds from the Terry Fox Run (D.E.H.) and by a grant from the Deutsche Krebshilfe, Bonn, Germany (M.F.-B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
D. E. Hogge, Terry Fox Laboratory, British Columbia Cancer Agency, 601 West 10th Ave, Vancouver, British Columbia, Canada V5Z 1L3; e-mail: dhogge@bccancer.bc.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal