Cathelicidins are a family of antimicrobial proteins found in the peroxidase-negative granules of neutrophils. The known biologic functions reside in the C-terminus, which must be cleaved from the holoprotein to become active. Bovine and porcine cathelicidins are cleaved by elastase from the azurophil granules to yield the active antimicrobial peptides. The aim of this study was to identify the physiological setting for cleavage of the only human cathelicidin, hCAP-18, to liberate the antibacterial and cytotoxic peptide LL-37 and to identify the protease responsible for this cleavage. Immunoelectron microscopy demonstrated that both hCAP-18 and azurophil granule proteins were present in the phagolysosome. Immunoblotting revealed no detectable cleavage of hCAP-18 in cells after phagocytosis. In contrast, hCAP-18 was cleaved to generate LL-37 in exocytosed material. Of the 3 known serine proteases from azurophil granules, proteinase 3 was solely responsible for cleavage of hCAP-18 after exocytosis. This is the first detailed study describing the generation of a human antimicrobial peptide from a promicrobicidal protein, and it demonstrates that the generation of active antimicrobial peptides from common proproteins occurs differently in related species.
Introduction
Human polymorphonuclear neutrophilic leukocytes (PMNs) contain a variety of antibiotic proteins.1 These are mainly localized in granules.2 When the granules are mobilized, these proteins are released to the exterior or into the phagolysosome, where the contents of the peroxidase-negative and peroxidase-positive granules of neutrophils meet and cooperate in the killing of microbes.
In human PMNs, most bactericidal proteins are localized in the azurophil granules2—for example, bactericidal/permeability increasing protein (BPI),3 CAP37 (azurocidin),4 and defensins.5 Here they colocalize with the serine proteases, cathepsin G, proteinase 3, and elastase.6 These proteases possess antimicrobial activity independent of their catalytic activity.1
Cathelicidins are a family of antimicrobial and endotoxin-binding proteins found in peroxidase-negative granules of vertebrate neutrophils.7 Members of this protein family share a highly conserved N-terminus of 12 kd, named cathelin after a protein isolated from porcine neutrophils.8
The cathelicidins are synthesized as preproproteins.7After removal of the signal peptide, they are stored in granules as inactive proforms. The active biologic domains of the cathelicidins generally reside in the C-terminus. The C-terminal antibacterial peptides are activated when cleaved from the proforms of the cathelicidins by serine proteases from azurophil granules.9-11 The C-termini of the cathelicidins vary greatly in amino acid sequence and structure, ranging from proline- and arginine-rich sequences to sequences forming amphipathic α-helices.
Porcine and bovine neutrophils contain a variety of cathelicidins, whereas hCAP-18 is the only human cathelicidin.12-16hCAP-18 is a major protein in specific granules of neutrophils,17 but it is also present in subpopulations of lymphocytes and monocytes,18 in squamous epithelia,19 epididymis and seminal plasma,20in the lung,21,22 and in keratinocytes during inflammatory skin diseases.23 Plasma contains a high concentration of hCAP-18 bound to lipoproteins.24 The antibacterial C-terminus of hCAP-18, LL-37, has been isolated from exocytosed material from neutrophils.15 It shows broad antimicrobial activity toward both gram-negative and gram-positive bacteria,25 has synergistic antibacterial effects with the defensins,26 and is a chemotactic agent for neutrophils, monocytes, and T cells using the formyl peptide receptor–like 1 receptor.27 However, LL-37 is also cytotoxic toward mammalian cells.28
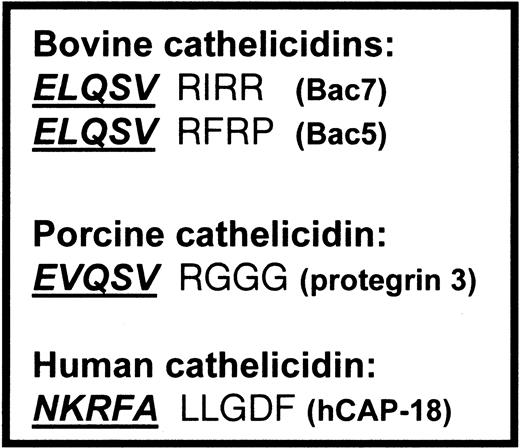
In bovine and porcine neutrophils, the antimicrobial peptides are liberated by elastase-mediated cleavage of cathelicidins.10 11 However, the potential cleavage site of hCAP-18 is different from the cleavage site of bovine and porcine cathelicidins (Figure 1). The aims of this study were to identify the biologic settings in which LL-37 is cleaved from hCAP-18 and to identify the protease responsible for this cleavage.
Cleavage sites of cathelicidins.
Cleavage sites between the cathelin part and the antimicrobial peptide of the bovine and porcine cathelicidins, cleaved by elastase, compared with the cleavage site of hCAP-18. Cathelin parts are shown in boldface italics and are underlined.
Cleavage sites of cathelicidins.
Cleavage sites between the cathelin part and the antimicrobial peptide of the bovine and porcine cathelicidins, cleaved by elastase, compared with the cleavage site of hCAP-18. Cathelin parts are shown in boldface italics and are underlined.
Materials and methods
Materials
Fresh EDTA plasma was collected from healthy donors. Specific polyclonal rabbit anti–hCAP-18 antibodies were generated by immunization of rabbits with recombinant hCAP-18.29Monoclonal antibodies toward LL-37 were generated by immunization of mice with glutaraldehyde–cross-linked synthetic LL-37. Monoclonal antibodies were obtained using conventional hybridoma technology (S.T. et al, manuscript in preparation).
Anti–proteinase 3 antibodies and proteinase 3 were generously provided by Jörgen Wieslander (Wieslab AB, Lund, Sweden). Human leukocyte elastase, cathepsin G, and SLPI were purchased from ICN Biomedicals (Costa Mesa, CA). Antielastase antibodies were from Biodesign International (Kennebunk, ME). All other antibodies were purchased from DAKO A/S (Glostrup, Denmark).
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE)30 and immunoblottting31 were performed with Mini-Protean 3 Cells and Mini Trans-Blot Electrophoretic Transfer Cells according to the instructions given by the manufacturer (Bio-Rad, Hercules, CA). For immunoblotting, polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA) were blocked for 1 hour with 5% skimmed milk in phosphate-buffered saline (PBS) after the transfer of proteins from the 14% polyacrylamide gels. For visualization of hCAP-18, the PVDF membranes were incubated overnight with primary antibodies. The next day, membranes were incubated for 2 hours with horseradish peroxidase–conjugated secondary antibodies (DAKO) and visualized by diaminobenzidine–metal concentrate and stable substrate buffer (Pierce, Rockford, IL).
Isolation of neutrophils from peripheral blood
Human neutrophils were isolated from freshly prepared buffy coats or from healthy donors as described.32 Briefly, after sedimentation with 2% Dextran T-500 (Amersham Pharmacia Biotech, Uppsala, Sweden) in isotonic NaCl, the leukocyte-rich supernatant was pelleted and resuspended in saline for subsequent centrifugation on Lymphoprep (Nycomed Pharma A/S, Oslo, Norway) at 400g for 30 minutes for the removal of lymphocytes and monocytes. Remaining erythrocytes were lysed in ice-cold de-ionized water for 30 seconds. Tonicity was restored by the addition of 1 vol of 1.8% NaCl. Cells were washed once and resuspended in the desired buffer. With the exception of Dextran sedimentation, all steps were carried out at 4°C.
Isolation of exudate neutrophils from skin window chambers
Exudate neutrophils were isolated from skin window chambers placed on the forearms of healthy human donors, as described.33 34 Briefly, chambers with 3 0.6-mL wells covering the lesions were used. They were filled with autologous serum and incubated for 18 hours. Chambers were then emptied, washed, and filled with fresh autologous citrated plasma. Neutrophils were allowed to accumulate in the chambers for 7 hours. Cells were harvested, pelleted by centrifugation, washed once, and resuspended in the desired buffer. More than 95% of the harvested cells were neutrophils.
Purification of hCAP-18 from neutrophils
Neutrophils were disrupted by nitrogen cavitation after the addition of 5 mM di-isopropyl fluorophosphate (Sigma, St Louis, MO). Postnuclear supernatants were loaded on 2-layer gradients (1.05/1.12 g/mL) of Percoll (Amersham Pharmacia Biotech).35 This resulted in 3 visible bands. Starting at the bottom, the bands are designated the α-band, containing azurophil granules; the β-band, containing specific and gelatinase granules; and the γ-band, containing plasma membranes and secretory vesicles.
The β-band containing specific granules was harvested manually, and Percoll was removed by ultracentrifugation. Isolated granules were treated with 5 mM di-isopropyl fluorophosphate. Granules were lysed in PBS containing 1% Triton X-100 (Boehringer Ingelheim, Heidelberg, Germany), 1 mM phenylmethylsulfonyl fluoride (Sigma), 100 kallikrein inhibitory U/mL aprotinin (Bayer, Leverkusen, Germany), 100 μg/mL leupeptin (Sigma), and 1 mM EDTA (Sigma). Membranes were pelleted by centrifugation, and the supernatant containing the specific granule proteins was frozen at −80°C until further use.
Isolated specific granule proteins were subjected to cation exchange chromatography on a MonoS column using ÄKTA-FPLC (Amersham Pharmacia Biotech AB). Most of the bound material was eluted with 1 M NaCl, 9.5 mM phosphate, pH 7.4. hCAP-18 was subsequently eluted with 10 mM NaOH, 140 mM NaCl. Immunoblotting with anti–hCAP-18 antibodies of the eluted hCAP-18 showed one band of the appropriate molecular mass.
For cleavage experiments with purified proteases and amino acid sequence analysis, hCAP-18 was purified from specific granules on an anti–hCAP-18 antibody column as previously described.24
Isolation of azurophil granule proteins from neutrophils
Neutrophils were subjected to nitrogen cavitation and subcellular fractionation as described above but without protease inhibitors. After the removal of Percoll from the α-band containing the azurophil granules, the granules were freeze-thawed 5 times in 1 M NaCl. Membranes were pelleted by ultracentrifugation, and the supernatant containing the matrix proteins of azurophil granules was harvested and stored at −80°C until further use.
Exocytosis and phagocytosis experiments
Isolated neutrophils, freshly prepared from peripheral blood or skin windows of healthy donors, were resuspended in Krebs Ringer phosphate (10 mM NaH2PO4/Na2HPO4, 130 mM NaCl, 5 mM KCl, 0.95 mM CaCl2, 5 mM glucose) at a concentration of 107 cell/mL. Cells were preincubated at 37°C for 5 minutes and then stimulated with 1 μM ionomycin (Calbiochem, La Jolla, CA), 10−8 M formyl methionyleucylphenylalanine (fMLP; Sigma), or IgG-coated latex beads for 20 minutes at 37°C. Stimulation was stopped by the addition of 2 vol ice-cold buffer and subsequent pelleting by centrifugation. The supernatant containing the exocytosed material was analyzed by enzyme-linked immunosorbent assay (ELISA) or immunoblotting.
After stimulation, aliquots of the cells were either used for quantification of granule proteins by ELISA or resuspended to a concentration of 1 × 106 cells/mL and precipitated with 5% trichloroacetic acid (final concentration). The pellet was washed 5 times with acetone and resuspended in Laemmli sample buffer for analysis by SDS-PAGE and immunoblotting. Remaining cells were fixed for electron microscopy.
Preparation of exocytosed material for cleavage experiments
Neutrophils (3 × 107 cells/mL) were stimulated to exocytosis by 1 μM ionomycin as described above. After stimulation, the cells were placed on ice for 10 minutes and subsequently pelleted by centrifugation. The supernatant was frozen at −20°C until further experiments. Endogenous hCAP-18 was subsequently removed from the exocytosed material by affinity chromatography on an anti–hCAP-18 antibody column. After affinity chromatography, the exocytosed material was immediately used as a source of proteases for cleavage of hCAP-18.
Cleavage experiments
Intact hCAP-18 isolated from specific granules byÄKTA-FPLC was incubated with exocytosed material from neutrophils, azurophil granule proteins, or purified proteases at 37°C for 30 minutes. The sample was subsequently boiled in Laemmli sample buffer and run on a SDS-PAGE followed by immunoblotting.
Amino acid sequence analysis
Amino acid sequence was analyzed on the PVDF-blotted protein in a 494 A Procise Protein Sequencer (PerkinElmer, Palo Alto, CA) using the blot cartridge and PVDF cycles. All reagents and solvents were supplied by PerkinElmer.
Immunoprecipitation
Antibodies against elastase, cathepsin G, proteinase 3, α1-antitrypsin, and normal rabbit immunoglobulins were incubated with Protein A Sepharose (Pharmacia) for 30 minutes at room temperature in PBS (pH 7) with 0.5 M NaCl. Sepharose particles were subsequently washed 7 times in PBS with 0.5 M NaCl to remove unbound antibodies; this was followed by incubation with exocytosed material at 4°C for 2 hours. Sepharose particles were pelleted by centrifugation. Supernatants were aspirated and immediately used for cleavage experiments.
Immunoelectron microscopy
Cells were fixed in a mixture of 0.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.2) for 2 hours at room temperature. They were then stored in 4% paraformaldehyde at 4°C until they were processed for ultrathin cryosectioning. For single labeling, cryosections were incubated with rabbit anti–hCAP-18; this was followed by 10-nm protein A–conjugated colloidal gold. For double labeling, the sections were first incubated with mouse monoclonal anti–human myeloperoxidase (CLB; Amsterdam, The Netherlands) followed by rabbit anti–mouse IgG and 5-nm protein A gold, and then they were treated with 1% glutaraldehyde for 10 minutes to prevent interference between the different antibody gold complexes in the sections.36 They were further incubated with rabbit anti–hCAP-18 followed by 10-nm protein A–conjugated colloidal gold (5-nm and 10-nm protein A–conjugated gold; EM Laboratory, Utrecht University, The Netherlands). After immunolabeling, the cryosections were embedded in a mixture of methylcellulose and uranyl acetate and examined with a Philips CM 10 electron microscope (Eindhoven, The Netherlands). For controls, the primary antibody was replaced by a nonrelevant murine or rabbit antiserum, respectively.
Quantitation of proteins
Myeloperoxidase, hCAP-18, and gelatinase were measured by ELISA as previously described.29,37,38α1-Antitrypsin, elastase, cathepsin G, and proteinase 3 were quantitated by semiquantitative ELISA. Anti–proteinase 3 and anti–cathepsin G antibodies were isolated from antiserum usingÄKTA-FPLC. All the antibodies were biotinylated as described.39
Samples were diluted in 50 mM Na2CO3/NaHCO3 buffer, pH 9.6, and incubated in 96-well flat-bottom immunoplates (Nunc, Roskilde, Denmark) overnight at room temperature. Unspecific binding was blocked by incubation with 200 μL/well dilution buffer (0.5 M NaCl, 3 mM KCl, 8 mM Na2HPO4/KH2PO4, 1% BSA (Sigma), 1% Triton X-100, pH 7.2) for 1 hour. Biotinylated antibodies against the above-mentioned antigens were diluted in dilution buffer and incubated for 1 hour. Horseradish peroxidase–labeled avidin (DAKO) was diluted 1500-fold in dilution buffer and incubated for 1 hour. Plates were washed 3 times in washing buffer (0.5 M NaCl, 3 mM KCl, 8 mM Na2H4/KH2PO4, 1% Triton X-100, pH 7.2) after each incubation using a SkanWasher 410 (Skatron, Roskilde, Denmark). Plates were washed once in substrate buffer (0.1 M sodium phosphate, 0.1 M citric acid, pH 5.0) before color development and then incubated with substrate buffer containing 0.04% o-phenyl-diamine (Kem-En-Tec, Copenhagen, Denmark) and 0.03% H2O2. Unless otherwise stated, 100 μL was added to each well at each incubation step. Color development was stopped by the addition of 100 μL 1 M H2SO4, absorbance measured at 492 nm in a Multiscan Plus ELISA Reader (Labsystems, Helsinki, Finland). A standard curve of serial dilutions of exocytosed material from neutrophils was used.
Activity of exocytosed elastase and cathepsin G
Freshly prepared exocytosed material from ionomycin-stimulated neutrophils (5 × 107 cells/mL) was incubated with specific nitroanilide substrates for elastase (N-methoxysuccinyl-ala-ala-pro-val p-nitroanilide; Sigma) or cathepsin G (N-methoxysuccinyl-ala-ala-pro-met p-nitroanilid; Sigma). The amount of free nitroanilide was quantitated by measurement of the absorbance at 410 nm.
Preparation of lipoprotein-bound hCAP-18
Purified hCAP-18 was incubated with plasma for 2 hours at 37°C. Plasma was then subjected to molecular-sieve chromatography on a Superose 12 column using ÄKTA-FPLC. The high molecular peak fraction of hCAP-18 containing lipoprotein-bound hCAP-18, as previously described,24 was used for further cleavage experiments.
Results
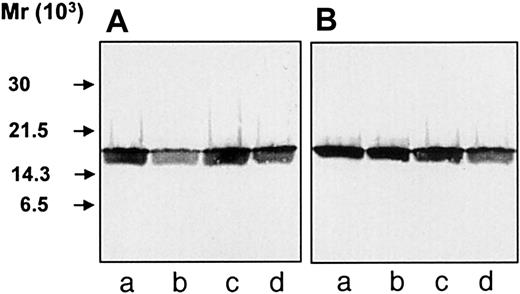
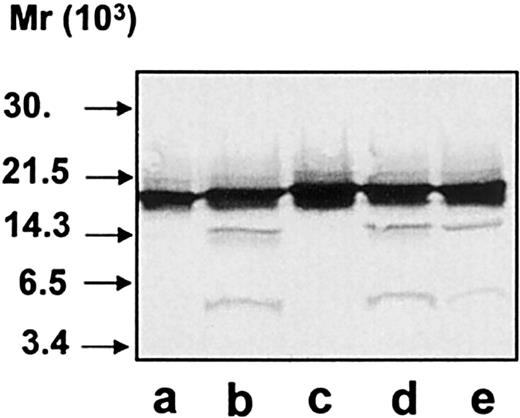
To investigate whether hCAP-18 is cleaved in the phagocytic vacuole, neutrophils from peripheral blood and from skin windows were isolated and stimulated to phagocytosis by immunoglobulin-coated latex beads. After phagocytosis, the cells were fixed for electron microscopy or pelleted and resuspended in 0.9% NaCl followed by TCA-precipitation. Immunoelectron microscopy demonstrated that the PMNs had phagocytosed the latex beads and that hCAP-18 was found both in the specific granules and in the phagolysosomes (Figure2A). In single sections from 106 exudate neutrophils from skin windows, phagolysosomes were found in 103 cells. In blood neutrophils, phagolysosomes were found in only 1 of 103 cells. Double-immunogold labeling of hCAP-18 and myeloperoxidase was performed to demonstrate that both azurophil and specific granules had fused with the phagolysosome (Figure 2B). Thirty-three phagolysosomes were examined for the presence of myeloperoxidase and hCAP-18 in 7 sections from different exudate neutrophils from skin windows. Twenty-seven phagolysosomes were labeled with both myeloperoxidase and hCAP-18. Three were labeled only for myeloperoxidase and 3 only for hCAP-18. Because only one section was examined for each phagolysosome, it cannot be ruled out that those positive only for one marker would have been positive for both markers in another section of the same phagolysosome. Thus, most phagolysosomes contained both hCAP-18 and azurophil granule proteins. TCA precipitates from neutrophils after phagocytosis were analyzed by SDS-PAGE, and then they were immunoblotted with anti–hCAP-18 antibodies. Despite the “priming” of phagocytosis in cells from skin windows, no intracellular cleavage of hCAP-18 was found after phagocytosis of latex beads (Figure 3A, lane c). The same result was found in blood neutrophils (Figure 3B, lane c). As expected, unperturbed cells and cells stimulated to exocytosis by fMLP and ionomycin showed no cleavage of hCAP-18 (see Figure 3A-B, lanes a, b, and d). Control experiments ascertained that TCA-precipitation did not influence the detection of the low-molecular-weight fragments by immunoblotting and that the cleavage of hCAP-18 by serine proteases from azurophil granules was not inhibited by immunoglobulin-coated latex beads (data not shown). To validate that the lack of detectable cleavage of hCAP-18 in cell lysates after phagocytosis did not result from insufficient degranulation of hCAP-18 into the phagocytic vacuole, the immunogold-labeled hCAP-18 was counted in granules and phagolysosomes. More than 50% of the labeled hCAP-18 was present in the phagolysosome in the neutrophils harvested from skin windows and stimulated to phagocytosis by latex beads (Table 1). Although this was a semiquantitative measure of degranulation into the phagolysosome, it demonstrated that a substantial part of the hCAP-18 in these cells was localized to the phagolysosome. Phagocytosis experiments with serum-treated zymosan particles performed with neutrophils from skin windows and peripheral blood gave similar results (data not shown). Thus, cleavage of hCAP-18 was not detectable in the phagocytic vacuole.
Electron microscopy of neutrophils from skin windows after phagocytosis of latex beads.
(A) Cryosection incubated with anti–hCAP-18 and 10-nm protein A-gold. Neutrophil with phagolysosomes (p) containing latex beads. (Inset) Higher magnification of the marked area showing a granule containing hCAP-18 (large arrow) and a phagolysosome also labeled for hCAP-18 (small arrows). Bars, 400 nm; inset, 100 nm. (B) Double-immunogold labeling of hCAP-18 (as a marker of specific granules) with 10-nm gold particles and myeloperoxidase (MPO, as a marker of azurophil granules) with 5-nm gold particles demonstrated that both azurophil and specific granules fused with the phagolysosome. Bar, 100 nm.
Electron microscopy of neutrophils from skin windows after phagocytosis of latex beads.
(A) Cryosection incubated with anti–hCAP-18 and 10-nm protein A-gold. Neutrophil with phagolysosomes (p) containing latex beads. (Inset) Higher magnification of the marked area showing a granule containing hCAP-18 (large arrow) and a phagolysosome also labeled for hCAP-18 (small arrows). Bars, 400 nm; inset, 100 nm. (B) Double-immunogold labeling of hCAP-18 (as a marker of specific granules) with 10-nm gold particles and myeloperoxidase (MPO, as a marker of azurophil granules) with 5-nm gold particles demonstrated that both azurophil and specific granules fused with the phagolysosome. Bar, 100 nm.
Immunoblotting of cell lysates.
Neutrophils were TCA-precipitated, and the precipitates were run on SDS-PAGE and analyzed by immunoblotting with anti–hCAP-18 antibodies. (A) Precipitates of neutrophils harvested from skin windows (107 cells/mL), unstimulated cells (lane a), or cells stimulated with fMLP (lane b), IgG-coated latex beads (lane c), or ionomycin (lane d). (B) Precipitates of neutrophils isolated from peripheral blood (107 cells/mL), unstimulated cells (lane a) or cells stimulated with fMLP (lane b), IgG-coated latex beads (lane c), or ionomycin (lane d).
Immunoblotting of cell lysates.
Neutrophils were TCA-precipitated, and the precipitates were run on SDS-PAGE and analyzed by immunoblotting with anti–hCAP-18 antibodies. (A) Precipitates of neutrophils harvested from skin windows (107 cells/mL), unstimulated cells (lane a), or cells stimulated with fMLP (lane b), IgG-coated latex beads (lane c), or ionomycin (lane d). (B) Precipitates of neutrophils isolated from peripheral blood (107 cells/mL), unstimulated cells (lane a) or cells stimulated with fMLP (lane b), IgG-coated latex beads (lane c), or ionomycin (lane d).
Degranulation of hCAP-18 into the phagolysosome
| . | Total labeled hCAP-18 . | hCAP-18 labeled in granules . | hCAP-18 labeled phagolysosomes . | Labeled hCAP-18 in phagolysosomes (%) . |
|---|---|---|---|---|
| Donor 1 (micrograph) | ||||
| 1 | 202 | 149 | 53 | 26 |
| 2 | 218 | 35 | 183 | 84 |
| 3 | 140 | 83 | 57 | 41 |
| 4 | 259 | 97 | 162 | 63 |
| Donor 2 (micrograph) | ||||
| 1 | 188 | 32 | 156 | 83 |
| 2 | 90 | 22 | 68 | 76 |
| 3 | 87 | 51 | 36 | 41 |
| 4 | 196 | 72 | 124 | 63 |
| 5 | 94 | 31 | 63 | 67 |
| . | Total labeled hCAP-18 . | hCAP-18 labeled in granules . | hCAP-18 labeled phagolysosomes . | Labeled hCAP-18 in phagolysosomes (%) . |
|---|---|---|---|---|
| Donor 1 (micrograph) | ||||
| 1 | 202 | 149 | 53 | 26 |
| 2 | 218 | 35 | 183 | 84 |
| 3 | 140 | 83 | 57 | 41 |
| 4 | 259 | 97 | 162 | 63 |
| Donor 2 (micrograph) | ||||
| 1 | 188 | 32 | 156 | 83 |
| 2 | 90 | 22 | 68 | 76 |
| 3 | 87 | 51 | 36 | 41 |
| 4 | 196 | 72 | 124 | 63 |
| 5 | 94 | 31 | 63 | 67 |
Degranulation of hCAP-18 into the phagolysosome in exudate neutrophils from skin chamber stimulated to phagocytosis by latex beads. Immunogold-labeled hCAP-18 present in granules and phagolysosomes were counted to determine the degree of degranulation of hCAP-18 into the phagolysosome. Labeled hCAP-18 was counted from 2 donors. Each micrograph represents a different cell.
Exocytosis experiments
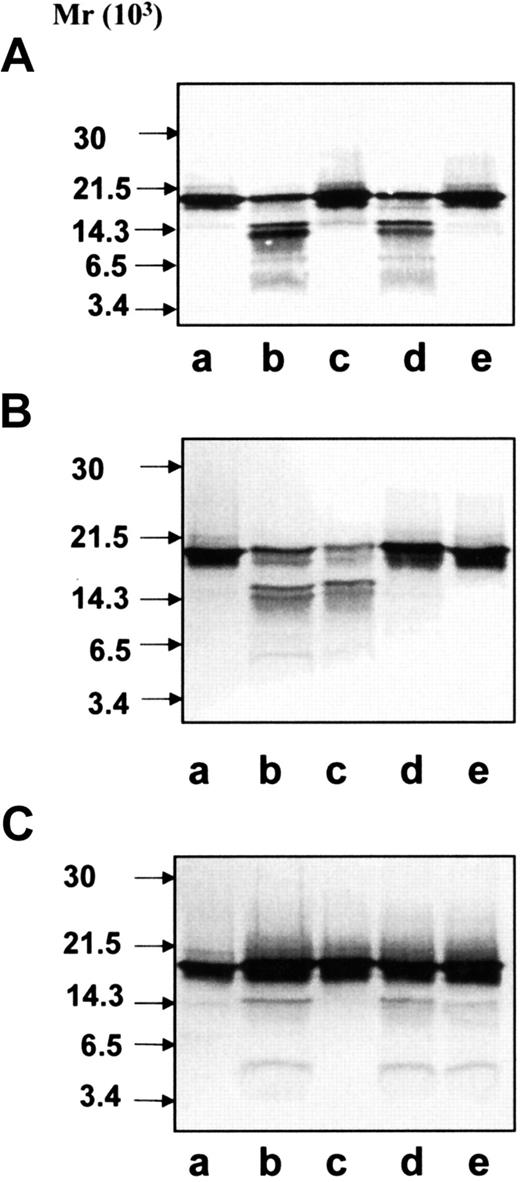
Neutrophils from peripheral blood or from skin chamber windows were stimulated to exocytosis by different secretagogues (Table2). The exocytosed material was analyzed by immunoblotting with anti–hCAP-18 antibodies. Significant cleavage of hCAP-18 was only detected in the exocytosed material from ionomycin-stimulated neutrophils (Figure 4A-B, lane d). We have previously demonstrated that the 14-kd fragment of hCAP-18 in the exocytosed material is cathelin and that the 4-kd fragment represents the noncathelin C-terminus of hCAP-18.24 The absolute concentrations of azurophil granule proteins were highest in the exocytosed material from ionomycin-stimulated cells (in particular from blood neutrophils) (Table 2). The absolute concentration of azurophil markers correlated with the degree to which hCAP-18 was cleaved. This indicates that the concentration of protease in the medium determines whether hCAP-18 is cleaved. Prolonged incubation (1 hour) of neutrophils did not give rise to further cleavage of hCAP-18 in the exocytosed material (data not shown). When neutrophils were stimulated by fMLP at a cell concentration of 3 × 108 cells/mL, the hCAP-18 in the exocytosed material was cleaved (Figure 4C). Thus, cleavage occurs even after stimulation with weak secretagogues if the cell concentration is high enough, indicating that hCAP-18 cleavage may take place during the accumulation of neutrophils in acute inflammation.
Exocytosis of granule constituents in response to stimulation
| . | Control neutrophils . | Exudate neutrophils . | ||||
|---|---|---|---|---|---|---|
| MPO . | hCAP-18 . | Gelatinase . | MPO . | hCAP-18 . | Gelatinase . | |
| No addition | 0.8 (34) | 0.9 (29) | 1.6 | 4.2 (187) | 13.4 (316) | 24.1 |
| fMLP (10−8M) | 2.3 (66) | 5.2 (153) | 31.0 | 9.6 (274) | 15.5 (387) | 49.9 |
| Immunoglobulin-coated latex beads | 14.3 (354) | 10.4 (272) | 45.3 | 17.1 (268) | 27.8 (309) | 63.7 |
| Ionomycin (1 μM) | 34.9 (924) | 64.3 (2415) | 88.8 | 11.9 (560) | 65.5 (1658) | 92.9 |
| . | Control neutrophils . | Exudate neutrophils . | ||||
|---|---|---|---|---|---|---|
| MPO . | hCAP-18 . | Gelatinase . | MPO . | hCAP-18 . | Gelatinase . | |
| No addition | 0.8 (34) | 0.9 (29) | 1.6 | 4.2 (187) | 13.4 (316) | 24.1 |
| fMLP (10−8M) | 2.3 (66) | 5.2 (153) | 31.0 | 9.6 (274) | 15.5 (387) | 49.9 |
| Immunoglobulin-coated latex beads | 14.3 (354) | 10.4 (272) | 45.3 | 17.1 (268) | 27.8 (309) | 63.7 |
| Ionomycin (1 μM) | 34.9 (924) | 64.3 (2415) | 88.8 | 11.9 (560) | 65.5 (1658) | 92.9 |
Isolated neutrophils were incubated with or without stimulus. Exocytosis of granule proteins was determined by ELISA measurements. MPO was chosen as a marker for azurophil granules, hCAP-18 for specific granules, and gelatinase for gelatinase granules. Exocytosis is expressed as percentage of total amounts in the cells and medium. Absolute concentrations (ng/mL) of MPO and hCAP-18 are given in parentheses. It should be noted that although the release of MPO from ionomycin-stimulated exudate neutrophils was lower in terms of percentage of total amount than that from exudate cells stimulated with latex beads, the release of MPO was highest in the ionomycin-stimulated cells in terms of absolute concentration of released MPO.
MPO indicates myeloperoxidase; fMLP, formyl methionyleucylphenylalanine; ELISA, enzyme-linked immunosorbent assay.
Immunoblotting of exocytosed material.
After stimulation, the neutrophils were pelleted and the supernatant containing the exocytosed material was analyzed by SDS-PAGE and immunoblotting with anti–hCAP-18 antibodies. (A) Exocytosed material from neutrophils harvested from skin windows (107cells/mL), unstimulated cells (lane a) or cells stimulated with fMLP (lane b), IgG-coated latex beads (lane c), or ionomycin (lane d). (B) Exocytosed material from neutrophils isolated from peripheral blood (107 cells/mL), unstimulated cells (lane a) or cells stimulated with fMLP (lane b), IgG-coated latex beads (lane c), or ionomycin (lane d). (C) Exocytosed material from fMLP-stimulated neutrophils (3 × 108 cells/mL) from peripheral blood.
Immunoblotting of exocytosed material.
After stimulation, the neutrophils were pelleted and the supernatant containing the exocytosed material was analyzed by SDS-PAGE and immunoblotting with anti–hCAP-18 antibodies. (A) Exocytosed material from neutrophils harvested from skin windows (107cells/mL), unstimulated cells (lane a) or cells stimulated with fMLP (lane b), IgG-coated latex beads (lane c), or ionomycin (lane d). (B) Exocytosed material from neutrophils isolated from peripheral blood (107 cells/mL), unstimulated cells (lane a) or cells stimulated with fMLP (lane b), IgG-coated latex beads (lane c), or ionomycin (lane d). (C) Exocytosed material from fMLP-stimulated neutrophils (3 × 108 cells/mL) from peripheral blood.
Cleavage experiments with serine proteases
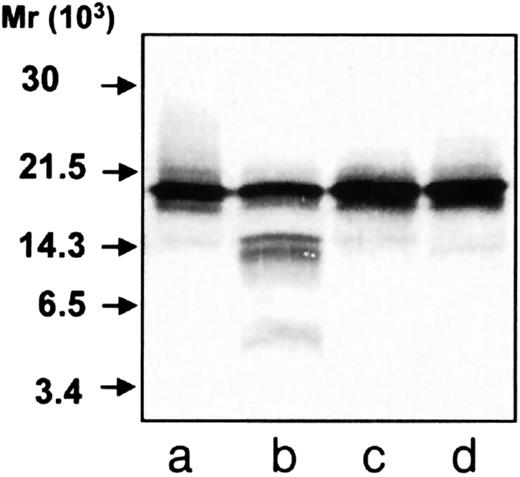
Immunoblotting of TCA-precipitated cells showed that hCAP-18 exists intracellularly as a holoprotein, as previously described,17 indicating that cleavage of hCAP-18 is performed by a protease not present in the same subcellular compartment as hCAP-18. Thus, it seemed likely that hCAP-18 was cleaved by a serine protease from azurophil granules, as described for bovine and porcine cathelidicins.10 11 Incubation with azurophil granule proteins resulted in the cleavage of hCAP-18 (Figure5, lane b), which could be inhibited both by phenylmethylsulfonyl fluoride and by aprotinin (Figure 5, lanes c, d), showing that serine proteases were responsible for the cleavage of hCAP-18 by the azurophil granule proteins. However, the cleavage of hCAP-18 by azurophil granule proteins did not resemble the cleavage observed after exocytosis. There were clearly 2 bands of approximately 14 kd rather than only one band in the exocytosed material.
Immunoblotting of hCAP-18 after incubation with azurophil granule proteins.
Purified hCAP-18 was incubated with azurophil granule proteins. Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies. Purified hCAP-18 (lane a) incubated with azurophil granule proteins (lanes b-d) and phenylmethylsulfonyl fluoride (lane c) or aprotinin (lane d).
Immunoblotting of hCAP-18 after incubation with azurophil granule proteins.
Purified hCAP-18 was incubated with azurophil granule proteins. Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies. Purified hCAP-18 (lane a) incubated with azurophil granule proteins (lanes b-d) and phenylmethylsulfonyl fluoride (lane c) or aprotinin (lane d).
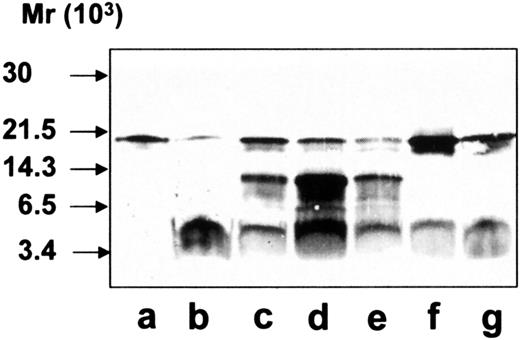
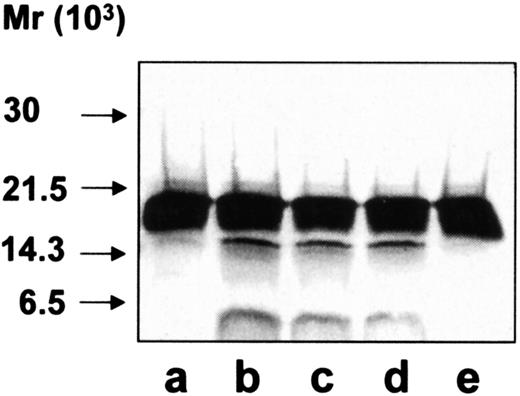
To further characterize the cleavage pattern of hCAP-18 by azurophil granule proteases, immunoblotting was performed with a monoclonal antibody toward the antimicrobial domain of hCAP-18, LL-37 (Figure6). Immunoblotting of exocytosed material from neutrophils showed one band of 18 kd (the holoprotein) and one band of 4 kd (LL-37) (Figure 6, lane b). As anticipated, the 14-kd band of cathelin was not recognized by the monoclonal antibody. In contrast, a band of 14 kd was detected by the monoclonal antibody against LL-37 when hCAP-18 was cleaved by extracts of azurophil granules (Figure 6, lane c). Thus, at least some of the cleavage of hCAP-18 by serine proteases from azurophil granules occurs at a location different from that between the cathelin part and LL-37, resulting in a 14-kd fragment that contains parts of the cathelin and of the LL-37 moiety.
Immunoblotting with monoclonal anti–LL-37 antibody.
All samples were run on SDS-PAGE followed by immunoblotting with monoclonal anti–LL-37 antibody. Lane a, purified hCAP-18; lane b, exocytosed material from ionomycin-stimulated neutrophils; lane c, purified hCAP-18 incubated with azurophil granule proteins; lane d, elastase; lane e, cathepsin G; lane f, proteinase 3; lane g, with exocytosed material from ionomycin-stimulated neutrophils after depletion of the endogenous hCAP-18.
Immunoblotting with monoclonal anti–LL-37 antibody.
All samples were run on SDS-PAGE followed by immunoblotting with monoclonal anti–LL-37 antibody. Lane a, purified hCAP-18; lane b, exocytosed material from ionomycin-stimulated neutrophils; lane c, purified hCAP-18 incubated with azurophil granule proteins; lane d, elastase; lane e, cathepsin G; lane f, proteinase 3; lane g, with exocytosed material from ionomycin-stimulated neutrophils after depletion of the endogenous hCAP-18.
hCAP-18 was then incubated with each of the 3 known serine proteases in azurophil granules, and immunoblotting was performed with the monoclonal anti–LL-37 antibody. All 3 proteases were capable of cleaving hCAP-18 (Figure 6, lanes d-f). Cleavage of hCAP-18 by elastase and cathepsin G resulted in clearly visible bands at 14 kd not seen in the exocytosed material. Incubation of hCAP-18 with different concentrations of elastase or cathepsin G did not give rise to the cleavage pattern observed in the exocytosed material (data not shown). Cleavage by proteinase 3 gave rise only to LL-37 (Figure 6, lane f), similar to what was observed in the exocytosed material.
Inhibition and immunoprecipitation experiments with exocytosed material from neutrophils
Because the pattern of hCAP-18 cleavage by azurophil granule extracts was different from the cleavage pattern of endogenous hCAP-18 in the exocytosed material, experiments were performed with the exocytosed material from ionomycin-stimulated neutrophils to identify the protease responsible for cleaving hCAP-18. Endogenous hCAP-18 in the exocytosed material was removed by affinity chromatography on an anti–hCAP-18 antibody column (in the presence of 0.5 M NaCl to prevent unspecific absorption to the column), and the exocytosed material was then incubated with purified hCAP-18. This resulted in cleavage of hCAP-18 that was similar to that of endogenous hCAP-18, originally observed in the exocytosed material, when immunoblotting was performed with monoclonal antibody (Figure 6, lane g) and with polyclonal antibodies (Figure 7, lane b). The cleavage of hCAP-18 by proteins in the exocytosed material was totally inhibited by the elastase inhibitor (N-methoxy-succinyl-ala-ala-pro-val chloromethyl ketone [CMK]), but not by chymostatin (an inhibitor of chymotrypsin-like proteases such as cathepsin G) or secretory leukocyte protease inhibitor (SLPI) (a known inhibitor of elastase and cathepsin G) (Figure 7A, lanes c-e).
Cleavage experiment with exocytosed material.
Exocytosed material from ionomycin-stimulated neutrophils was depleted of endogenous hCAP-18 and subsequently incubated with purified hCAP-18 with or without protease inhibitors. Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies. Purified hCAP-18 (lane a) incubated with exocytosed material (lanes b-e). This resulted in cleavage of hCAP-18 (lane b). This cleavage was inhibited by the addition of CMK (lane c) but not by chymostatin (lane d) or SLPI (lane e).
Cleavage experiment with exocytosed material.
Exocytosed material from ionomycin-stimulated neutrophils was depleted of endogenous hCAP-18 and subsequently incubated with purified hCAP-18 with or without protease inhibitors. Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies. Purified hCAP-18 (lane a) incubated with exocytosed material (lanes b-e). This resulted in cleavage of hCAP-18 (lane b). This cleavage was inhibited by the addition of CMK (lane c) but not by chymostatin (lane d) or SLPI (lane e).
We then examined the susceptibility of purified serine proteases to these inhibitors. The cleavage of hCAP-18 by elastase (Figure8A, lane b) was totally inhibited by CMK and SLPI but not by chymostatin (Figure 8A, lanes c-e). Cleavage by cathepsin G (Figure 8B, lane b) was totally inhibited by chymostatin and SLPI but not by CMK (Figure 8B, lanes c-e). Cleavage by proteinase 3 (Figure 8C, lane b) was totally inhibited by CMK but not by chymostatin or SLPI (Figure 8C, lanes c-e).
Differential inhibition of hCAP-18 cleavage by serine proteases from azurophil granules.
Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies. (A) hCAP-18 (lane a) was incubated with elastase alone (lane b) or together with the inhibitors CMK (lane c), chymostatin (lane d), or SLPI (lane e). (B) hCAP-18 (lane a) was incubated with cathepsin G alone (lane b) or together with the inhibitors CMK (lane c), chymostatin (lane d,) or SLPI (lane e). (C) hCAP-18 (lane a) was incubated with proteinase 3 alone (lane b) or together with the inhibitors CMK (lane c), chymostatin (lane d), or SLPI (lane e).
Differential inhibition of hCAP-18 cleavage by serine proteases from azurophil granules.
Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies. (A) hCAP-18 (lane a) was incubated with elastase alone (lane b) or together with the inhibitors CMK (lane c), chymostatin (lane d), or SLPI (lane e). (B) hCAP-18 (lane a) was incubated with cathepsin G alone (lane b) or together with the inhibitors CMK (lane c), chymostatin (lane d,) or SLPI (lane e). (C) hCAP-18 (lane a) was incubated with proteinase 3 alone (lane b) or together with the inhibitors CMK (lane c), chymostatin (lane d), or SLPI (lane e).
Thus, both the cleavage pattern of hCAP-18 and the results of the inhibition experiments in the exocytosed material are similar to those obtained with purified proteinase 3. To validate the experiments with exocytosed material, the serine proteases were quantitated by ELISA. The removal of hCAP-18 by affinity chromatography did not increase the concentration of proteinase 3 relative to the concentrations of elastase and cathepsin G (data not shown).
Proteinase 3 was then immunoprecipitated from the exocytosed material before incubation with hCAP-18. Immunoprecipitation with preimmune rabbit antibodies, antielastase antibodies, or anti–cathepsin G antibodies did not inhibit the cleavage of hCAP-18 (Figure9, lanes b-d) in the exocytosed material, whereas there was no cleavage of hCAP-18 after immunoprecipitation of proteinase 3 (Figure 9, lane e). Measurements in the supernatants after immunoprecipitation showed specific immunoprecipitation of proteinase 3 but no precipitation of elastase or cathepsin G after immunoprecipitation of proteinase 3. Proteinase 3 was not precipitated by antielastase or anti–cathepsin G antibodies (data not shown). The specificity of the anti–proteinase 3 antibodies was validated by immunoblotting. Before immunoblotting with anti–proteinase 3 antibodies, 1.25 μg purified elastase, cathepsin G, and proteinase 3 were run in separate lanes on SDS-PAGE. Reactivity was found only in the lane with proteinase 3 (data not shown). Thus, proteinase 3 was solely responsible for the cleavage of hCAP-18 in the exocytosed material.
Cleavage of hCAP-18 by exocytosed material after the immunoprecipitation of individual serine proteases.
Endogenous hCAP-18 fragments were deleted from the exocytosed material. Individual serine proteases were removed from the exocytosed material by immunoprecipitation before incubation with purified hCAP-18. Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies. Purified hCAP-18 (lane a) was incubated with the exocytosed material after immunoprecipitation with preimmune rabbit antibodies (lane b), antielastase antibodies (lane c), anti–cathepsin G antibodies (lane d), and anti–proteinase 3 antibodies (lane e).
Cleavage of hCAP-18 by exocytosed material after the immunoprecipitation of individual serine proteases.
Endogenous hCAP-18 fragments were deleted from the exocytosed material. Individual serine proteases were removed from the exocytosed material by immunoprecipitation before incubation with purified hCAP-18. Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies. Purified hCAP-18 (lane a) was incubated with the exocytosed material after immunoprecipitation with preimmune rabbit antibodies (lane b), antielastase antibodies (lane c), anti–cathepsin G antibodies (lane d), and anti–proteinase 3 antibodies (lane e).
Because of the in vitro activity of elastase and cathepsin G toward hCAP-18, we examined whether these proteases were inhibited in vivo after exocytosis by 2 inhibitors reported to be exocytosed from human neutrophils.
SLPI is reported to be a major protein in the neutrophil cytosol and to be exocytosed from human neutrophils.40 It inhibits elastase and cathepsin G but not proteinase 3. Thus, exocytosed SLPI could prevent the cleavage of hCAP-18 by elastase or cathepsin G in the exocytosed material from neutrophils. However, we were not able to detect any significant amounts of SLPI in unperturbed neutrophils or in the exocytosed material (O.E.S., N.B., P.S.H., unpublished observation, July 1999).
α1-Antitrypsin is expressed in neutrophils,41 and the association constant for α1-antitrypsin and proteinase 3 is one order of magnitude less than that between elastase and α1-antitrypsin.42 By immunoblotting and ELISA, we found that α1-antitrypsin was present in the exocytosed material from neutrophils and that α1-antitrypsin could inhibit the cleavage of hCAP-18 by all 3 serine proteases (data not shown). In immunoprecipitation experiments with exocytosed material from neutrophils, none of the 3 serine proteases—elastase, cathepsin G, or proteinase 3—were co-precipitated when α1-antitrypsin was immunoprecipitated. Furthermore, in gel filtration experiments with excytosed material, α1-antitrypsin eluted as free monomeric protein and did not co-localize with any of the 3 serine proteases (data not shown). Thus, the lack of in vivo activity of elastase and cathepsin G toward hCAP-18 was not due to the inhibition by SLPI or α1-antitrypsin.
The activities of elastase and cathepsin G were then measured in the exocytosed material from neutrophils (5 × 108 cells/mL) using specific nitroanilide substrates. These experiments were performed in the presence and absence of SLPI to validate that the measured activity was not caused by proteinase 3. Absorbance measured in the presence of the elastase substrate was 3.23 compared to 0.43 when SLPI was added before incubation with the substrate; the corresponding values in the experiment with cathepsin C substrate were 1.42 and 0.26. Measured activities in these experiments were greater than those necessary in the in vitro experiments for the cleavage of hCAP-18 by isolated elastase or cathepsin G. Thus, both elastase and cathepsin G are present as active enzymes in the exocytosed material from neutrophils.
Identification of the C-terminal fragments after cleavage of hCAP-18
To further validate that LL-37 was liberated by proteinase 3– mediated cleavage of hCAP-18, purified hCAP-18 was cleaved by incubation with proteinase 3. The sample was run on SDS-PAGE and blotted to a PVDF membrane, and the low-molecular-mass fragment was analyzed by N-terminal amino acid sequencing of the first 10 residues. These were identified as (L)LGDFFRKSK, consistent with LL-37. Because of contamination, the identity of the first residue could not be unequivocally determined.
Influence of binding to lipoproteins
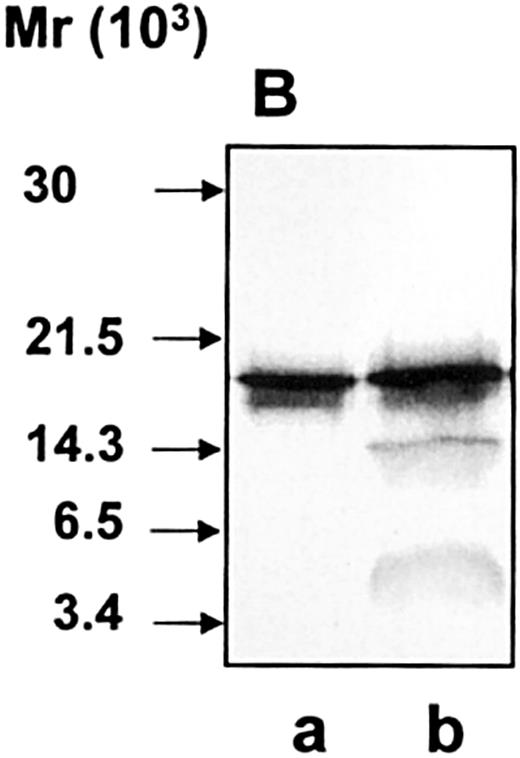
We have previously shown that hCAP-18 circulates in plasma in a high concentration bound to lipoproteins.24 We therefore investigated whether lipoprotein-bound hCAP-18 was susceptible to cleavage by proteinase 3. Plasma was incubated with purified hCAP-18 and was subjected to gel filtration. After gel filtration, the peak fraction of hCAP-18 bound to lipoproteins of very low density and low density was incubated with proteinase 3. The lipoprotein-bound hCAP-18 was still susceptible to cleavage by proteinase 3 (Figure10). Thus, the association to lipoproteins does not prevent the cleavage of hCAP-18.
Susceptibility of lipoprotein-bound hCAP-18 to cleavage by proteinase 3.
Lipoprotein-bound hCAP-18 (lane a) was incubated with proteinase 3 (lane b). Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies.
Susceptibility of lipoprotein-bound hCAP-18 to cleavage by proteinase 3.
Lipoprotein-bound hCAP-18 (lane a) was incubated with proteinase 3 (lane b). Samples were run on SDS-PAGE followed by immunoblotting with anti–hCAP-18 antibodies.
Discussion
The antibacterial peptide LL-37 is cleaved from the human cathelicidin hCAP-18 between an alanyl and a leucyl residue. This site differs from the cleavage sites in the bovine and porcine cathelicidins, which are cleaved by elastase at elastase-cleavage sites (Figure 1). Most notably, the basic arginyl residue after the cleavage site is substituted with the small aliphatic leucyl residue, and the traditional valyl residue just before the cleavage site is substituted with an alanyl residue. Leukocyte elastase prefers to cleave at a valyl rather than at an alanyl residue.43 Proteinase 3, on the other hand, prefers to cleave between 2 small aliphatic amino acids such as Ala-Leu,42 as found in the cleavage site of hCAP-18. The cleavage of hCAP-18 by proteinase 3 is a specific cleavage between the antimicrobial peptide and the cathelin part with no further degradation of the cathelin part. Similar specific cleavage of the porcine cathelicidin protegrin 3 is mediated by elastase.11
In contrast to the porcine cathelicidins, not all the bovine cathelicidins contain a valyl residue at the putative cleavage site.7 Thus, it remains to be seen whether some of the bovine cathelicidins are cleaved by proteases other than elastase.
In mice44,45 and rabbits,46 the putative cleavage sites of the cathelicidins do not resemble those in human, porcine, or bovine cathelicidins. The specific proteases responsible for cleavage of these cathelicidins remain to be characterized.
Cathelicidin genes are composed of 4 exons and 3 introns. There is great similarity between the first 3 exons encoding the conserved cathelin part between different cathelicidins but no homology in the fourth exon encoding the active antimicrobial domain and the putative cleavage site.15,16 47-50 Cleavage of hCAP-18 by proteinase 3 demonstrates that the cleavage site is a functional variable part of the cathelicidins, together with the antimicrobial domain, and that the members of the cathelicidin family are activated by different proteases in related species. Thus, during evolution the variable biologic functions of the cathelicidins have been changed solely by alterations in the fourth exon.
The 3 known serine proteases in azurophil granules—elastase, cathepsin G, and proteinase 3—cleave many of the same substrates, and hCAP-18 was susceptible to cleavage by all 3 serine proteases in vitro. However, proteinase 3 was found to be solely responsible for the cleavage of hCAP-18 after exocytosis, even though all 3 serine proteases were found in the exocytosed material. This was clearly demonstrated by the fact that the cleavage of hCAP-18 was totally abolished after specific immunoprecipitation of proteinase 3 from the exocytosed material and that immunoblotting with the monoclonal anti–LL-37 antibody showed a cleavage pattern of hCAP-18 after exocytosis consistent only with cleavage by proteinase 3. Both elastase and cathepsin G were found in the exocytosed material, and both enzymes were found to be active. Thus, in a biologic setting, hCAP-18 is a specific substrate for proteinase 3. Interestingly, elastase, cathepsin G, and proteinase 3 also have well-documented in vitro affinity for α1-antitrypsin, but this was not reflected by complex formation in the exocytosed material.
In our in vitro experiments, proteinase 3 did not seem to be as active toward hCAP-18 as elastase is toward bovine10 and porcine cathelicidins.11 The interesting question is whether hCAP-18 is processed extracellularly in vivo to a lesser extent by proteinase 3 than the bovine and porcine cathelicidins processed by elastase. Comparison is difficult because of different experimental conditions used (including different types of antibodies). Our polyclonal anti–hCAP-18 antibodies seemed to overestimate the amount of holoprotein, and the monoclonal anti–LL37 seemed to overestimate the amount of LL-37. However, we have previously blocked the binding of the polyclonal antibodies to the cathelin part of hCAP-18 by adding recombinant cathelin to the primary antibodies24; this is probably the best way to estimate the amount of LL-37 compared to holoprotein. In an experiment in which 1.4 × 107neutrophils/mL was stimulated with ionomycin, we estimated that approximately 95% of the holoprotein was processed to LL-37.24 Even though hCAP-18 seemed to be activated extracellularly to a lesser extent than the bovine and porcine cathelicidins in vivo,9,11 51 most of the secreted hCAP-18 was processed to LL-37 if a sufficient amount of proteinase 3 was present extracellularly.
Cleavage by proteinase 3 may be functionally significant for the hCAP-18 expressed in nonhematopoietic tissues, such as lung,21,22 skin,23 and epididymis.20 Pulmonary monocytes from patients with cystic fibrosis express proteinase 3,52 and the levels of proteinase 3 activity are greater than those of elastase in the sputum from patients with cystic fibrosis who have chronic lung infections.53 SLPI is assumed to play an important role in the protection against the leukocyte proteases, elastase, and cathepsin G and in the mucosa at various sites—eg, in airway fluid,54 semen,55 and keratinocytes.56 The presence of SLPI in these tissues will not interfere with proteinase 3–mediated cleavage of hCAP-18, as would have been the case if elastase or cathepsin G cleaved hCAP-18.
Conditions in the phagocytosis experiments were made optimal to positively demonstrate a cleavage of hCAP-18 in the phagolysosome. After phagocytosis, the cells were TCA-precipitated to avoid cleavage of hCAP-18 during further processing of the cells for immunoblotting. Control experiments with exocytosed material from neutrophils demonstrated that TCA precipitation did not influence the detection of low-molecular-weight fragments after the cleavage of hCAP-18. Furthermore, we used exudate neutrophils from skin windows. These “primed” cells are the closest experimental correlate to the neutrophils active in the tissues.33,34 Indeed, exudate neutrophils from skin windows were significantly more active in phagocytosing the latex beads than neutrophils from peripheral blood. Immunoglobulins were used as opsonizing ligands because they optimize the incorporation of specific granules (and, thus, hCAP-18) into the phagolysosome.57 Immunoglobulin-coated latex particles were found not to inhibit the cleavage of hCAP-18 by the serine proteases from azurophil granules. Furthermore, a substantial amount of the hCAP-18 in the cells was present in the phagolysosome, and azurophil granule constituents were found together with hCAP-18 in most examined phagolysosomes. Even very limited degranulation of azurophil granules into the phagolysosome would generate much higher concentrations of azurophil granule proteases in the phagolysosome than found extracellularly because of the much smaller volume of the phagolysosome. Yet, no cleavage of hCAP-18 was observed. Additional experiments were performed with serum-treated zymosan particles with similar results. We cannot completely rule out that small undetectable amounts of LL-37 are generated in the phagolysosome, but our data do show that phagocytosis, during which specific and azurophil granules fuse with the phagolysosome, is insufficient for the generation of significant amounts of LL-37.
The main function of hCAP-18, therefore, seems to be extracellular, where LL-37 also acts as a chemotactic agent for neutrophils, monocytes, and T cells.27 In contrast to the bovine cathelicidins it is unknown whether the porcine cathelicidins are processed in the phagolysosome. Extracellular inhibition of elastase in wound fluids from pigs, which prevents activation of the porcine cathelicidins, impairs the clearance of bacteria from the wounds in vivo.58 Thus, cathelicidins seem to be important mediators of the extracellular antibacterial activity generated by neutrophils.
In summary, we found that the human cathelicidin hCAP-18 is processed extracellularly to the antimicrobial peptide LL-37 by proteinase 3. This is the first detailed description of the generation of a human antimicrobial peptide from a promicrobicidal protein, and it demonstrates that the generation of active antimicrobial peptides from common proproteins occurs differently in related species.
We thank Hanne Kidmose, Allan Kastrup, Hans Janssen, and Nico Ong for their expert technical assistance. We thank Karsten Lollike, Jack B. Cowland, Kim Theilgaard-Mönch, Malene Bjerregaard, Daniel Carter, and Lene Udby for critical review of the manuscript, and we thank Veronique Witko-Sarsat for useful discussions.
Supported by grants from the Danish Medical Research Council and The Amalie Jørgensen Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ole E. Sørensen, Dept of Hematology, Granulocyte Research Laboratory, L-9322, Rigshospitalet, 9 Blegdamsvej, DK-2100 Copenhagen, Denmark; e-mail: olesoeren@rh.dk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal