Abstract
Using flow cytometric and RNase protection assays, this study examined the expression of chemokine receptors in nonactivated natural killer (NK) cells and compared this expression with NK cells activated with interleukin (IL)-2, which either adhered to plastic flasks (AD) or did not adhere (NA). None of the NK cell subsets expressed CXCR2, CXCR5, or CCR5. The major differences between these cells include increased expression of CXCR1, CCR1, CCR2, CCR4, CCR8, and CX3CR1 in AD when compared to NA or nonactivated NK cells. The chemotactic response to the CXC and CC chemokines correlated with the receptor expression except that all 3 populations responded to GRO-α, despite their lack of CXCR2 expression. Pretreatment of these cells with anti-CXCR2 did not inhibit the chemotactic response to GRO-α. In addition, nonactivated and NA cells responded to fractalkine, although they lack the expression of CX3CR1. This activity was not inhibited by anti-CX3CR1. Viral macrophage inflammatory protein (vMIP)-I, I-309, and TARC competed with the binding of 125I-309 to AD cells with varying affinities. Transforming growth factor (TGF)-β1 but not any other cytokine or chemokine examined including interferon (IFN)-γ, MIP-3β, macrophage-derived chemokine (MDC), thymus and activation-regulated chemokine (TARC) or I-309, up-regulated the expression of CXCR3 and CXCR4 on NK cell surface. This is correlated with increased chemotaxis of NK cells treated with TGF-β1 toward stromal cell–derived factor (SDF)-1α and interferon-inducible protein-10 (IP-10). Messenger RNA for lymphotactin, RANTES, MIP-1α, and MIP-1β, but not IP-10, monocyte chemotactic protein (MCP)-1, IL-8, or I-309 was expressed in all 3 NK cell subsets. Our results may have implications for the dissemination of NK cells at the sites of tumor growth or viral replication.
Introduction
Natural killer (NK) cells were first discovered because of the ability of a small population of blood lymphocytes (also known as large granular lymphocytes, or LGL) to kill tumor cells (reviewed in reference 1). Although it was difficult to isolate highly purified NK cells based on the technologies available, Vujanovic et al successfully purified LGL from rat spleens.2 These cells are generated by adherence to plastic flasks after activation with interleukin (IL)-2 and are designated as adherent lymphokine-activated killer (AD-LAK) cells. Because of their potential application in the treatment of cancer patients, we examined their in vivo tissue distribution and reported that these cells have restricted tissue localization.3 Also, another population of killer cells was recovered from cells cultured with IL-2. These do not adhere to plastic flasks and are designated as nonadherent lymphokine-activated killer (NA-LAK) cells. On examining their cytolytic behavior, the NA cells show lower NK cell activity than the AD cells.4,5Later work showed that AD cells accumulate at the sites of pulmonary and hepatic B16 melanoma in mice.6 In a more related work, Vujanovic et al7 showed that AD cells migrate significantly better than NA cells into the spheroids of human squamous cell carcinoma of the head and neck or breast carcinoma, emphasizing the antitumor therapeutic potential of adherent IL-2–activated NK cells. Subsequent studies showed that AD cells in conjunction with IL-2 eliminate liver metastases in a xenogeneic tumor model, where nude mice bear human gastric carcinoma.8,9 Also, it was observed that some of these cells establish a cell-to-cell contact with metastatic cells.6-9 However, the mechanisms contributing to the elimination of tumors, and to the close contact between adherent cells and tumor cells, are to a large extent not known.
Most of the above studies were conducted before the surge of chemokines. These low-molecular-weight molecules are divided into 4 subfamilies: CXC or α, CC or β, C or γ, and CX3C or δ (reviewed in reference 10). Also several chemokine receptors including CXCR1 to CXCR5, CCR1 to CCR10, XCR1, and CX3CR1 have been cloned. Another classification based on function and stage of maturation or activation was developed by Sallusto et al.11 In this, chemokines and their receptors have been divided into 2 categories—constitutive and inflammatory. Recent results showed that NK or IL-2–activated NK cells can be attracted in vitro toward the concentration gradients of various chemokines (reviewed in reference 12). Also, we reported that the CXC chemokine IL-8 influences the chemokinesis of these cells.13 These results suggest that freshly isolated NK cells or IL-2–activated NK cells express receptors for chemokines. Morohashi et al,14using polyclonal antibodies, reported that CXCR1 and CXCR2 are expressed on nonactivated NK cells. Freshly isolated NK cells are also reported to express CX3CR1.15 Expression of CXCR3 on the surface of a small proportion of NK cells has been shown by 2 groups.16,17 Also, messenger RNA (mRNA) for CXCR3 is present in cloned NK cells.18 The expression of CCR8 was reported on the NK 3.3 cell line but not on other NK cell lines.19 Using flow cytometry and RNase protection assay (RPA) we recently reported that AD NK cells express CCR4 and CCR8.20 On the other hand, CCR6 was not expressed on NK cell clones.21
Although these results are informative, they do not provide a uniform source of identifying chemokine receptors on NK cells. In addition, the effect of activating NK cells with IL-2 on the expression of chemokine receptors has not been addressed in these studies. Here, we systematically investigated the expression of chemokine receptors in nonactivated NK cells and compared this expression with the expression of these receptors in IL-2–activated (adherent “AD” versus nonadherent “NA”) cells. Nonactivated T cells are included because the expression of chemokine receptors in these cells is well documented.
Materials and methods
Culture medium and reagents
Culture medium (CM) consisted of RPMI 1640 supplemented with 10% human AB serum (Ullevål Hospital, Oslo, Norway), 10 U/mL penicillin, 100 μg/mL streptomycin, 1 mM l-glutamine, 1% nonessential amino acids (all from Life Technologies, Paisley, UK), and 5 × 10−5 M 2-ME (Sigma Chemical, St Louis, MO). Monoclonal antibodies to CCR1, CCR2, CCR3, CCR5, CCR6, CXCR1, CXCR2, CXCR3, CXCR4, and CXCR5 were obtained from Celltek Biotechnologies (Cap Rouge, Quebec, Canada). Polyclonal antibodies to the carboxy terminal of CCR4 and CCR7 were purchased from Research Diagnostics (Flanders, NJ). Rabbit antibody to CX3CR1 was purchased from Terrey Pines Biolabs (San Diego, CA). Affinity-purified goat antihuman CCR8 was purchased from Alexis Biochemicals (Laufelfingen, Switzerland). Interferon (IFN)-γ, transforming growth factor (TGF)-β1, monocyte chemotactic protein (MCP)-1, Eotaxin, macrophage inflammatory protein (MIP)-3α, macrophage-derived chemokine (MDC), MIP-3β, TARC, stromal cell–derived factor (SDF)-1α, IP-10, GRO-α, UMIP-I and fractalkine (FK), as well as monoclonal antibodies to CXCR1, CXCR2, CCR1, and CCR5 were purchased from R&D Systems Europe (Abingdon, Oxon, UK). I-309 was purchased from PeproTech (Rocky Hill, NJ).
Preparation of NK and IL-2–activated NK cells
The IL-2–activated NK cells were prepared by adherence to plastic flasks of nylon wool column-nonadherent cells (2 × 106/mL), generated from buffy coats of human volunteers (Ullevål Hospital), in the presence of 1000 IU/mL IL-2 as previously described.22 Twenty-four hours later, 2 populations were separated, those that are plastic adherent (AD cells) and those that are nonadherent (NA cells). These were grown separately for an additional 8 to 10 days. Both populations were depleted of T cells by binding 2 times to M-450 CD3 Dynabeads (Dynal, Oslo, Norway). Nonactivated NK cells were collected after nylon wool column separation and were treated with CD3 Dynabeads as described above. This procedure resulted in more than 97% of the different NK cell subsets expressing CD56 and CD16 cell surface markers. T cells were collected from nylon wool column-nonadherent cells. They were more than 95% CD3+.
Chemotaxis assay
Blind well chemotaxis chambers with a lower well volume of 200 μL were used. A maximum volume of 200 μL RPMI medium containing 1% bovine serum albumin was placed in the lower wells in the presence or absence of chemokines. Cells (4 × 105) were placed in the upper compartments of Boyden chambers above the filters. The chambers were incubated for 2 hours at 37°C in a 5% CO2incubator. The filters were then removed, dehydrated, and stained with 15% Giemsa stain for 7 minutes and then mounted on glass slides. Cells in 10 high-power fields from 2 filters were counted and averaged for each sample. Migration index was calculated as the number of cells migrating toward the concentration gradient of chemokines divided by the number of cells migrating toward medium only.
Flow cytometric analysis
For the detection of CCR4 or CCR7, the cells were permeabilized before introduction of antibodies, which detect the carboxy terminal of these receptors, as recently described.20 For the detection of the other chemokine receptors, cells (1 × 106/mL) were incubated with 1 μg/mL monoclonal or polyclonal antichemokine receptors for 1 hour at 4°C, washed, and then incubated with 1:100 dilution of F(ab′)2 fluorescein isothiocyanate (FITC)-conjugated rabbit antimouse for 45 minutes at 4°C, washed 3 times, and then examined. For CX3CR1, the secondary antibody was FITC-conjugated F(ab′)2 goat antirabbit (Sigma). For CCR8, the secondary antibody was FITC-conjugated F(ab′)2 rabbit antigoat. Regulation of chemokine receptor expression by cytokines or chemokines was determined by incubating nonactivated NK cells (1 × 106/mL) with culture medium, 15 μg/mL IFN-γ, or 20 ng/mL of TGF-β1, MIP-3β, SDF-1α, MDC, TARC, or I-309 for 5 days at 37°C. These concentrations have been found to be optimal for activating NK cells.20 22-24 The cells were washed and examined for the presence of surface chemokine receptors, as described above.
Analysis of mRNA expression by multiprobe RPA
Total RNA was prepared by guanidium thiocyanate/cesium chloride gradients following standard protocols. The specific mRNA for chemokine receptors was detected by the hCR5 and hCR6 multiprobe template sets (RiboQuant, Pharmingen, San Diego, CA), which contain templates for various chemokine receptors. Detection of mRNA for chemokines was done by using the hCK5 multiprobe template set (Pharmingen), which detects mRNA for various chemokines. In brief, antisense RNA probes were generated from DNA templates using T7 DNA-dependent RNA polymerase, in the presence of α-32P]UTP (Amersham Pharmacia Biotech, specific activity 10 μCi/μL). Labeled probes were hybridized with total RNA (10 μg) overnight at 56°C. Unhybridized RNA was digested with RNase according to Pharmingen's supplied procedures. RNase-protected probes were resolved on denaturing 5% polyacrylamide gel. The gels were dried and exposed to film (BIOMAX MS, Eastman Kodak, Rochester, NY) at −70°C for 3 hours.
Radioligand binding assay
The AD cells (3 × 105) were resuspended in a binding buffer containing 50 mM Hepes, 1 mM CaCl2, 5 mM MgCl2, 0.5% BSA, pH 7.0, and increasing concentrations of125I-309 (specific activity 2000 Ci/mmol/L; Amersham Pharmacia Biotech, Buckinghamshire, UK) and cold I-309, in a final volume of 0.1 mL for 4 hours at room temperature with continuous shaking. In the competition for binding experiments, 100 pmol/L of125I-309 and increasing concentrations of cold I-309, vMIP-I , or TARC were added to the cells in 0.1 mL. The cells were harvested into glass fiber filters using Skatron combi cell harvester (Skatron, Suffolk, UK), and washed with washing buffer containing 500 mM NaCl. After drying, scintillation fluids were added to the filters and counted in a Packard liquid scintillation analyzer. All assays were done in duplicate. Data were analyzed by nonlinear regression using Prism GraphPad (GraphPad Software, San Diego, CA).
Statistical analysis
Significant values were determined using the 2-tailed Student t test.
Results
Analysis of CXC chemokine receptor expression in NK and IL-2–activated NK cells
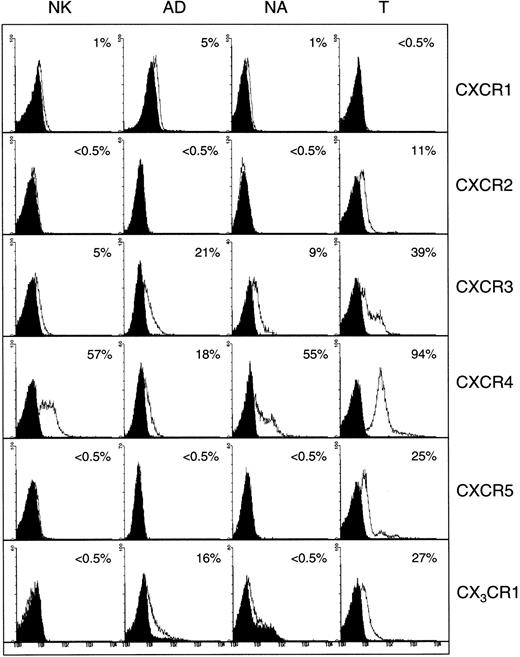
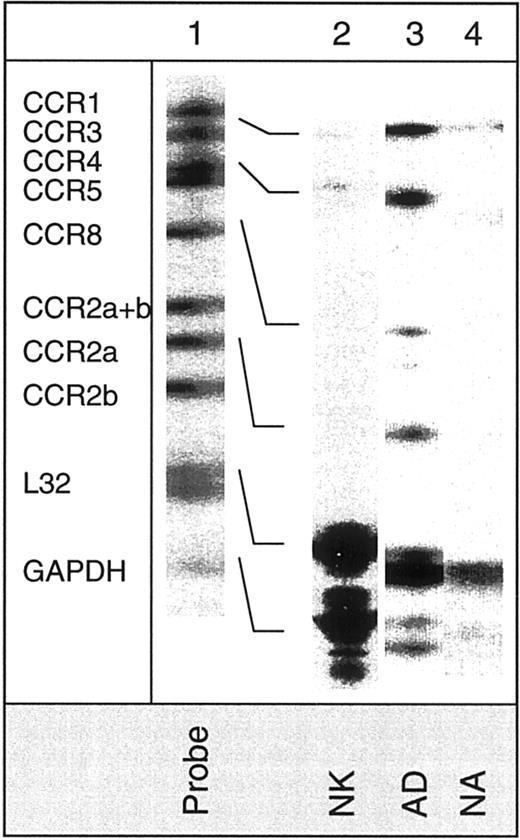
Figure 1 shows the expression of CXC chemokine receptors on nonactivated NK, AD, NA, and T cells. About 5% of AD and only 1% of nonactivated or NA cells expressed CXCR1, as determined by antibody to CXCR1. Using RPA, CXCR1 was shown in AD cells but not in other cell types (Figure 2). Using 2 different monoclonal antibodies, one shown here and the other from R&D Systems, none of the NK cell subsets expressed surface CXCR2. Only T cells (about 11%) expressed this receptor. The lack of expression of CXCR2 in nonactivated NK or IL-2–activated NK (AD and NA) cells is also shown in the RPA assay (Figure 2). CXCR3 was expressed on the surface of 21% AD, 9% NA, 5% to 12% nonactivated NK, and 39% T cells (Figure 1). The proportion of nonactivated T and NK cells expressing this receptor resembles those observed by Qin et al.16 The mRNA for CXCR3 was observed in these cells only after longer exposure of the gels (longer than 3 days; not shown).
Detection of surface CXC and CX3C chemokine receptors in NK cells.
Expression of CXC and CX3C chemokine receptors on the surface of nonactivated, adherent, and nonadherent cells (NK, AD, and NA, respectively). Expression of these receptors on nonactivated T cells is also shown. Numbers indicate the percentage of positive cells. Background controls in the presence of control antibody (mouse IgG) and FITC-conjugated secondary antibodies are shown in black.
Detection of surface CXC and CX3C chemokine receptors in NK cells.
Expression of CXC and CX3C chemokine receptors on the surface of nonactivated, adherent, and nonadherent cells (NK, AD, and NA, respectively). Expression of these receptors on nonactivated T cells is also shown. Numbers indicate the percentage of positive cells. Background controls in the presence of control antibody (mouse IgG) and FITC-conjugated secondary antibodies are shown in black.
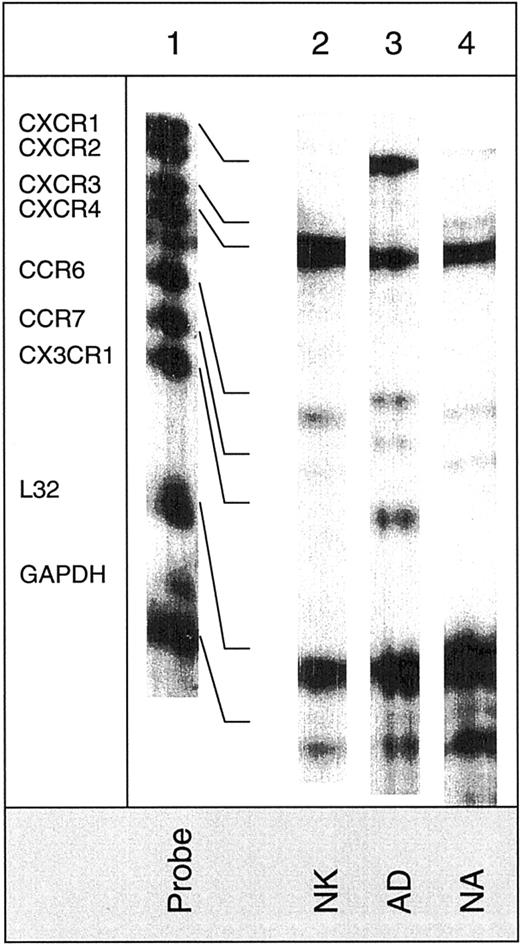
Detection of mRNA for chemokine receptors in various NK cell subsets.
The hCR6 multitemplate kit, which detects mRNA for CXCR1, CXCR2, CXCR3, CXCR4, CCR6, CCR7, and CX3CR1, was used. Lane 1 shows mRNA of RNase-unprotected probes. Lanes 2, 3, and 4 show mRNA of protected probes in nonactivated (NK), adherent (AD), and nonadherent (NA) cells. Arrows indicate the presence of mRNA for the particular chemokine receptor.
Detection of mRNA for chemokine receptors in various NK cell subsets.
The hCR6 multitemplate kit, which detects mRNA for CXCR1, CXCR2, CXCR3, CXCR4, CCR6, CCR7, and CX3CR1, was used. Lane 1 shows mRNA of RNase-unprotected probes. Lanes 2, 3, and 4 show mRNA of protected probes in nonactivated (NK), adherent (AD), and nonadherent (NA) cells. Arrows indicate the presence of mRNA for the particular chemokine receptor.
Figure 1 also shows that CXCR4 was expressed on all cell types including nonactivated NK, NA, and AD NK cells (57%, 55%, and 18%, respectively). T cells highly expressed this receptor (94%). Messenger RNA for this receptor was also present in the 3 NK cell preparations (Figure 2). CXCR5 was reported to be expressed on B cells,25 and it binds the chemokine B-cell–attracting chemokine 1 (BCA-1), which is responsible for the migration of B cells into the B-cell areas in the spleen. Here, we observed that none of the NK cell preparations expressed this receptor (Figure 1). In contrast, about 25% of T cells expressed CXCR5.
CX3CR1, the receptor for CX3C chemokine,15,26 is expressed on AD (16%) and T cells (27%), but not on nonactivated or NA NK cells (Figure 1). Similarly, mRNA for CX3CR1 was detected only in AD but not in nonactivated or NA NK cells (Figure 2). In contrast to our results, Imai et al15 showed mRNA, as well as receptor expression for CX3CR1, in nonactivated NK cells. The reason for these differences is not known, but it could be due to the process of preparing NK cells. In our hands, nonactivated NK cells were highly purified and contained more than 97% CD16+ cells. In addition, our method of negatively selecting NK cells ensured that they were not nonspecifically activated during the purification procedures.
Effect of CXC chemokines on the chemotaxis of NK cells
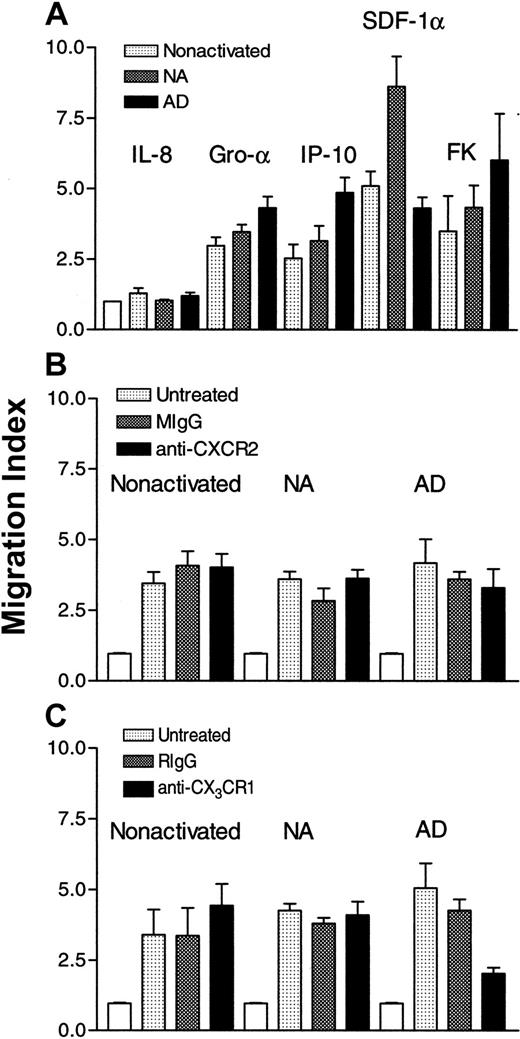
To correlate the expression of CXC and CX3C chemokine receptors with functional properties, we examined the effect of CXC and CX3C chemokines on the in vitro chemotaxis of the 3 different subsets of NK cells. Higher concentrations of IP-10 and fractalkine were used on nonactivated and NA cells than AD cells. Results in Figure 3A show that IL-8 was without effect. This is not surprising because IL-8 was previously found to be chemokinetic but not chemotactic for AD NK cells.13 IP-10 was chemoattractant for the 3 subsets (P < .003, < .005 and <. 0001, for nonactivated, NA, and AD, respectively, as compared to cells migrating in the absence of chemokines). Also SDF-1α was chemotactic for these cells (P < .001, < .0001, and < .001). Although both nonactivated and NA NK cells highly expressed CXCR4 (> 50% of the cells were positive), the effect of this chemokine was more robust for NA cells, suggesting that SDF-1α may have higher affinity for NA cells than the other NK cell populations. Surprisingly, GRO-α was chemotactic for the 3 NK cell subsets (P < .0001), although the receptor for this chemokine, CXCR2, was absent from these cells. In addition, FK was chemotactic for nonactivated, NA, and AD cells (P < .006, < .001, and < .0009, respectively), although nonactivated and NA NK cells lacked the expression of CX3CR1. To address the issue of the effect of GRO-α and fractalkine, we treated the 3 subsets with antibodies to CXCR2 or CX3CR1. The cells (5 × 106/mL) were pretreated with 0.1, 1, or 10 μg/mL of the antibodies for 1 hour at 4°C. Only results obtained with 10 μg/mL are shown. Figure 3B shows that pretreatment of nonactivated, NA, or AD NK cells with mouse IgG or monoclonal anti-CXCR2 did not affect their chemotaxis toward GRO-α. However, the same antibody inhibited more than 80% of GRO-α–induced T-cell chemotaxis (data not shown). These results suggest that this chemokine must use receptors other than CXCR2 to induce the chemotaxis of NK cells. Similarly, pretreatment of nonactivated or NA NK cells with rabbit IgG or rabbit anti-CX3CR1 did not affect their chemotaxis toward fractalkine (Figure 3C). In contrast, pretreatment of AD cells with rabbit anti-CX3CR1 inhibited their migration toward fractalkine (P < .03 when compared to untreated cells that migrate toward this chemokine). However, anti-CX3CR1 pretreated cells migrated significantly higher than control cells (cells migrating in the absence of chemokine;P < .005), indicating that anti-CX3CR1 does not completely inhibit AD NK cell chemotaxis toward fractalkine.
Effect of CXC chemokines on the chemotaxis of NK cells.
In panel A, IL-8 (100 ng/mL), GRO-α (50 ng/mL), IP-10 (25 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), SDF-1α (1 ng/mL), or fractalkine (1 ng/mL for nonactivated and NA and 100 pg/mL for AD cells), was placed in the lower chambers, whereas cells (4 × 105) were placed in the upper wells of Boyden chambers. In panel B, cells (5 × 106/mL) were either untreated or pretreated with 10 μg/mL of either mouse IgG (MIgG) or monoclonal anti-CXCR2 for 1 hour at 4°C. The cells were extensively washed and then placed (4 × 105) in the upper wells, whereas 50 ng/mL of GRO-α was placed in the lower wells of the chambers. In panel C, cells (5 × 106/mL) were either left untreated or were pretreated with 10 μg/mL of rabbit IgG (RIgG) or rabbit anti-CX3CR1 for 1 hour at 4°C, washed extensively, and then placed (4 × 105) in the upper wells. In the lower wells, fractalkine (1 ng/mL for nonactivated and NA, and 0.1 ng/mL for AD cells) was added. Migration index was calculated by dividing the number of cells migrating in the presence of chemokines by those migrating toward medium only (control: white columns).
Effect of CXC chemokines on the chemotaxis of NK cells.
In panel A, IL-8 (100 ng/mL), GRO-α (50 ng/mL), IP-10 (25 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), SDF-1α (1 ng/mL), or fractalkine (1 ng/mL for nonactivated and NA and 100 pg/mL for AD cells), was placed in the lower chambers, whereas cells (4 × 105) were placed in the upper wells of Boyden chambers. In panel B, cells (5 × 106/mL) were either untreated or pretreated with 10 μg/mL of either mouse IgG (MIgG) or monoclonal anti-CXCR2 for 1 hour at 4°C. The cells were extensively washed and then placed (4 × 105) in the upper wells, whereas 50 ng/mL of GRO-α was placed in the lower wells of the chambers. In panel C, cells (5 × 106/mL) were either left untreated or were pretreated with 10 μg/mL of rabbit IgG (RIgG) or rabbit anti-CX3CR1 for 1 hour at 4°C, washed extensively, and then placed (4 × 105) in the upper wells. In the lower wells, fractalkine (1 ng/mL for nonactivated and NA, and 0.1 ng/mL for AD cells) was added. Migration index was calculated by dividing the number of cells migrating in the presence of chemokines by those migrating toward medium only (control: white columns).
Analysis of CC chemokine receptor expression in NK and IL-2–activated NK cells
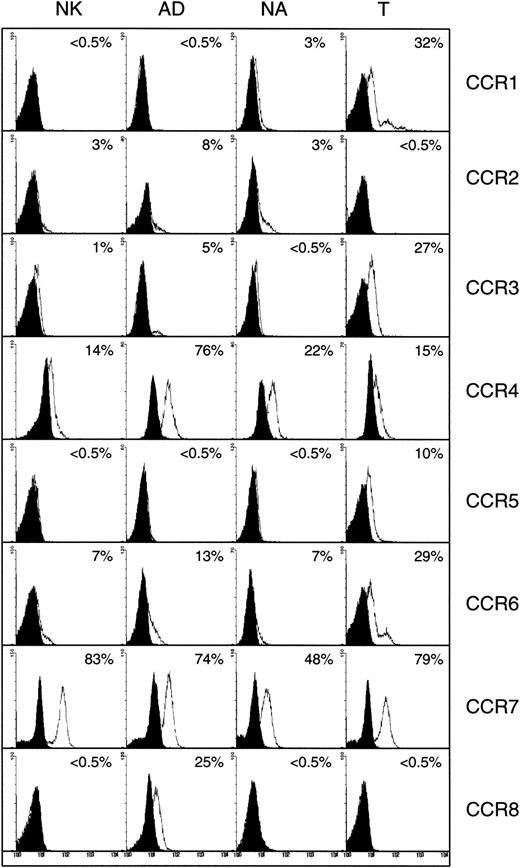
The expression of CC chemokine receptors examined in the flow cytometer is shown in Figure 4. Two different antibodies, one shown here and the other supplied by R&D Systems, did not detect surface expression of CCR1 (3% on NA, and < 0.5% on AD or nonactivated cells). In contrast, mRNA for this receptor was present in AD, and to a lower extent in NA or nonactivated cells (Figure 4). It is not clear why these cells did not show higher expression of surface CCR1. It is either that the protein is truncated or the antibody does not bind the epitopes expressed on the surface of these cells, although the same antibody stained CCR1 on the surface of T cells (Figure 4). Alternatively, because these cells secrete MIP-1α and RANTES, the ligands for this receptor (see below), these chemokines may induce the internalization of CCR1. Hence, we were unable to detect this receptor by flow cytometric analysis. It is worth noting that AD cells respond to MIP-1α and RANTES,27 implicating that IL-2–activated NK cells must express receptors for these chemokines, which could be CCR1, because AD cells lack the expression of CCR5, the other receptor for these chemokines.
Detection of surface CC chemokine receptors.
Expression of CC chemokine receptors on the surface of nonactivated (NK), adherent (AD), nonadherent (NA), and T cells. Numbers indicate the percentage of positive cells. Background controls in the presence of control antibody (mouse or goat IgG) and FITC-conjugated secondary antibody are shown in black.
Detection of surface CC chemokine receptors.
Expression of CC chemokine receptors on the surface of nonactivated (NK), adherent (AD), nonadherent (NA), and T cells. Numbers indicate the percentage of positive cells. Background controls in the presence of control antibody (mouse or goat IgG) and FITC-conjugated secondary antibody are shown in black.
The expression of CCR2 was higher on AD cells (8%), than on nonactivated or NA NK cells (3% each). Also, mRNA for this receptor was present in AD cells (Figure 5). Naive T cells did not express surface CCR2 (Figure 4). About 5% of AD NK cells express CCR3, as compared to less than 1% on NA or nonactivated NK cells (Figure 4). The mRNA for CCR3 was difficult to reveal in the RPA assay (Figure 5). Expression of CCR4 was higher in AD cells (76%) when compared to NA (22%) or nonactivated NK cells (14%). All 3 NK cell subsets expressed mRNA for CCR4. However, this expression was more pronounced in AD cells than the other 2 cell populations (Figure 5).
Demonstration of mRNA expression for various CC chemokine receptors.
Expression of mRNA in nonactivated (NK), adherent (AD), and nonadherent (NA) cells using the hCR5 multitemplate kit, which detects mRNA for CCR1, CCR2, CCR3, CCR4, CCR5, and CCR8.
Demonstration of mRNA expression for various CC chemokine receptors.
Expression of mRNA in nonactivated (NK), adherent (AD), and nonadherent (NA) cells using the hCR5 multitemplate kit, which detects mRNA for CCR1, CCR2, CCR3, CCR4, CCR5, and CCR8.
Using 2 different anti-CCR5, one shown in Figure 4, and the other obtained from R&D Systems (data not shown), we failed to detect any surface CCR5 expression on the 3 subsets of NK cells. Using hCR5, mRNA for this receptor was also absent in nonactivated, AD, and NA NK cells (Figure 5). Bleul et al28 also noted that human CD56+ NK cells do not express surface CCR5. The lack of CCR5 expression supports our earlier findings showing that AD NK cells do not respond to the concentration gradients of MIP-1β, the ligand for this receptor.27
Surface CCR6 was expressed on AD (13%), as well as on nonactivated or NA NK cells (7% each), whereas about 29% of naive T cells expressed this receptor (Figure 4). The mRNA for CCR6 was present in the 3 NK cell subsets (Figure 2). CCR7 was expressed both on the surface (Figure4) and at the mRNA level (Figure 2) in all 3 NK cell subsets examined. Interestingly, about 25% to 30% of AD cells expressed CCR8, which was lacking on either NA or nonactivated NK cells (< 0.5%). Heterogeneous naive T cells also lacked the expression of this receptor (Figure 4). In addition, only AD NK cells showed mRNA for this receptor (Figure 5).
Effect of CC chemokines on the chemotaxis of NK cells
To compare the expression of CC chemokine receptors with the ability of CC chemokines to induce the chemotaxis of these cells, we examined the chemotactic activity of the ligands for these receptors. Because CCR1 and CCR5 are not expressed on the surface of these cells, we did not examine the ligands for these receptors in the chemotaxis assay. Except for MIP-3β, higher concentrations of chemokines were used for nonactivated and NA cells than for AD cells. Results in Figure6A show that MCP-1 induced the chemotaxis of nonactivated and NA cells (P < .01) and was a robust chemoattractant for AD cells (P < .0001). Allavena et al29 also observed that nonactivated NK cells respond to MCP-1. Eotaxin, a CCR3 ligand, did not induce the chemotaxis of nonactivated or NA cells, but was a chemoattractant for AD cells, supporting the flow cytometric analysis data (Figure 4). MDC was chemotactic for the 3 NK cell subsets (P < .002). This correlates with the expression of CCR4 on the surface of these subsets (Figure 4). MIP-3α and MIP-3β were also chemotactic for nonactivated, NA, and AD NK cells (P < .002, < .001, and < .0001 for MIP-3α, and P < .0001 for MIP-3β, respectively). Surprisingly, MIP-3β is not more potent than MIP-3α, although CCR7 is highly expressed on the 3 NK cell subsets, when compared to the expression of CCR6 (Figure 4). The concentration of MIP-3α (25 ng/mL) used to induce the chemotaxis of nonactivated and NA cells was higher than that used for MIP-3β (1 ng/mL). Therefore, the difference in the response may be related to the affinity of these chemokines for these cells. Others also observed that NK cells respond to the ligands for CCR7.30
Effect of CC chemokines on the in vitro motility of various preparations of NK cells.
In panel A, MCP-1 (25 ng/mL for nonactivated [▩] and NA [▧] and 100 pg/mL for AD [▪] cells), Eotaxin (10 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), MDC (10 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), MIP-3α (25 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), MIP-3β (1 ng/mL for nonactivated, NA, and AD cells), or I-309 (10 ng/mL for nonactivated and NA and 1 ng/mL for AD cells) was placed in the lower wells of Boyden chambers, whereas 4 × 105 (nonactivated, AD, or NA) NK cells were added to the upper wells. In panel B, various concentrations (1 pg/mL, ░; 10 pg/mL, ▩; 100 pg/mL, ▪; 1000 pg/mL, ▤; 10 000 pg/mL, ▥) of vMIP-I were examined. Migration index was calculated as the number of cells migrating in the presence of chemokines divided by the number of cells migrating in the presence of medium only (controls, shown in white columns). Mean ± SD of 6 to 9 experiments. Panel C shows competition binding analysis of 100 pmol/L 125I-309 to AD cells, by cold I-309, TARC, or vMIP-I. Log concentrations of these chemokines are shown in the X-axis, whereas the total count of binding is shown in the Y-axis. IC50 for cold ligands is also shown.
Effect of CC chemokines on the in vitro motility of various preparations of NK cells.
In panel A, MCP-1 (25 ng/mL for nonactivated [▩] and NA [▧] and 100 pg/mL for AD [▪] cells), Eotaxin (10 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), MDC (10 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), MIP-3α (25 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), MIP-3β (1 ng/mL for nonactivated, NA, and AD cells), or I-309 (10 ng/mL for nonactivated and NA and 1 ng/mL for AD cells) was placed in the lower wells of Boyden chambers, whereas 4 × 105 (nonactivated, AD, or NA) NK cells were added to the upper wells. In panel B, various concentrations (1 pg/mL, ░; 10 pg/mL, ▩; 100 pg/mL, ▪; 1000 pg/mL, ▤; 10 000 pg/mL, ▥) of vMIP-I were examined. Migration index was calculated as the number of cells migrating in the presence of chemokines divided by the number of cells migrating in the presence of medium only (controls, shown in white columns). Mean ± SD of 6 to 9 experiments. Panel C shows competition binding analysis of 100 pmol/L 125I-309 to AD cells, by cold I-309, TARC, or vMIP-I. Log concentrations of these chemokines are shown in the X-axis, whereas the total count of binding is shown in the Y-axis. IC50 for cold ligands is also shown.
In contrast, only AD cells (P < .001, as compared to cells migrating in the absence of chemokine), but not nonactivated or NA NK cells responded to I-309 (Figure 6). These results confirm the RPA and the flow cytometric analysis results regarding the expression of CCR8 in AD cells only. To clearly establish the ability of AD cells to respond to CCR8 ligands, we examined the effect of vMIP-I, another ligand for CCR8.31 32 Our results show that as low as 10 pg/mL of vMIP-I induced the chemotaxis of AD cells (P < .002, as compared to cells migrating in the absence of chemokine). Also, 100, 1000 and 10 000 pg/mL concentrations of this chemokine were chemoattractants for AD cells (P < .001, < .002 and < .03, respectively, Figure 6B).
Binding of radioligand I-309 to AD NK cells
To further examine the presence of CCR8 on AD cells, we performed the radioligand-binding assay using 125I-309. Three different saturation binding experiments showed that I-309 binds to AD cells with a dissociation constant (Kd) of 213 ± 21 pmol/L (mean ± SE). This is higher than the affinity of this chemokine for Th2 cells,33 but is comparable to its affinity for IL-2–activated T cells.31 Next we investigated whether the binding of radiolabeled I-309 can be inhibited by chemokines reported to bind CCR8 transfectant cells.31,32 34 Results in Figure 6C show that cold I-309 and vMIP-I competed with the binding of 125I-309 to AD NK cells. The inhibitory concentration of 50% (IC50) for I-309 is 558 pmol/L, which is comparable to its Kd. Surprisingly, we observed that vMIP-I competed with higher affinity for CCR8 (IC50 is 123 pmol/L) than I-309. In addition TARC competed with I-309 for binding to AD NK cells with IC50 of 1640 pmol/L.
Regulation of chemokine receptor expression on NK cells by various cytokines and chemokines
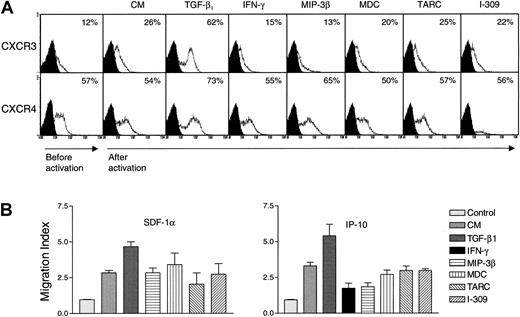
The effect of various cytokines on the expression of chemokine receptors in NK cells has not been previously examined. It was reported that IL-2 up-regulates the expression of CCR2 in NK cells.35 However, NK cells are under the control of various cytokines and chemokines.1,24,36 Therefore, it was important to study whether proinflammatory and other regulatory cytokines such as TGF-β1 and IFN-γ may influence the expression of chemokine receptors in highly purified nonactivated NK cells. The effect of constitutive chemokines (eg, MIP-3β and MDC) and inflammatory chemokines (eg, TARC and I-309) on the expression of various chemokine receptors in these cells was also examined. There was no effect by these cytokines on the expression of chemokine receptors 24 hours after stimulation (data not shown). Except for a low induction of CCR1, none of the chemokines or cytokines affected the expression of CCR2, CCR3, CCR5, CCR6, and CCR8 (because of the negative results, these data are not shown). Results of the expression of CXCR3 and CXCR4 are shown in Figure 7A, because these receptors are the only ones that proved to be regulated by cytokines in NK cells. We observed that NK cells acquired the expression of CXCR3 after in vitro incubation. Before culture, between 5% and 12% of NK cells expressed CXCR3, and this was increased to about 26% after culture (Figure 7A). The reason for this increase is unknown, but it could be due to the effect of stress on the cells, removed from their in vivo natural environment and placed in the in vitro cultures. This increase was inhibited by IFN-γ and MIP-3β, suggesting that under these conditions, these factors control the expression of CXCR3. More important, TGF-β1 increased the expression of CXCR3. Similarly, TGF-β1 increased the level of CXCR4, when compared to the proportion of cells incubated with medium only (Figure 7A). No effect with MDC, TARC, or I-309 was observed on the expression of either CXCR3 or CXCR4. Because TGF-β1 does not induce the proliferation of NK cells,24 the increase in the percentage of positive cells induced by TGF-β1 must be due to an increase in the receptor level, rather than an increase in the number of cells expressing these receptors.
Regulation of chemokine receptor expression by various cytokines and chemokines.
In panel A, highly purified (> 97% CD16+) NK cells (1 × 106/mL) were incubated with either culture medium (CM), 15 μg/mL IFN-γ, or 20 ng/mL TGF-β, MIP-3β, SDF-1α, MDC, TARC, or I-309 for 5 days at 37°C. The cells were extensively washed and then examined for the presence of the specific chemokine receptor in the flow cytometer. Numbers indicate the percentage of positive cells expressing either CXCR3 or CXCR4. The expression of these receptors prior to activation, and 5 days after activation with the various ligands, is shown. In panel B, either 1 ng/mL of SDF-1α (left panel) or 25 ng/mL of IP-10 (right panel) was placed in the lower wells of Boyden chambers. NK cells (4 × 105) incubated for 5 days with CM, 15 μg/mL IFN-γ, or 20 ng/mL TGF-β1, MIP-3β, MDC, TARC, or I-309, were placed in the upper wells. Migration index was determined by dividing the cells migrating in the presence of chemokines or cytokines by those migrating in the absence of ligands (control).
Regulation of chemokine receptor expression by various cytokines and chemokines.
In panel A, highly purified (> 97% CD16+) NK cells (1 × 106/mL) were incubated with either culture medium (CM), 15 μg/mL IFN-γ, or 20 ng/mL TGF-β, MIP-3β, SDF-1α, MDC, TARC, or I-309 for 5 days at 37°C. The cells were extensively washed and then examined for the presence of the specific chemokine receptor in the flow cytometer. Numbers indicate the percentage of positive cells expressing either CXCR3 or CXCR4. The expression of these receptors prior to activation, and 5 days after activation with the various ligands, is shown. In panel B, either 1 ng/mL of SDF-1α (left panel) or 25 ng/mL of IP-10 (right panel) was placed in the lower wells of Boyden chambers. NK cells (4 × 105) incubated for 5 days with CM, 15 μg/mL IFN-γ, or 20 ng/mL TGF-β1, MIP-3β, MDC, TARC, or I-309, were placed in the upper wells. Migration index was determined by dividing the cells migrating in the presence of chemokines or cytokines by those migrating in the absence of ligands (control).
To correlate the increase in the expression of CXCR3 and CXCR4 with the response of these receptors to their ligands, we examined the chemotactic response of NK cells incubated for 5 days with medium (CM) or with various chemokines and cytokines, toward IP-10 (the ligand for CXCR3) or SDF-1α (the ligand for CXCR4). Results in Figure 7B show that NK cells responded to IP-10 or SDF-1α. Incubating NK cells with TGF-β1 for 5 days resulted in enhanced chemotactic response to SDF-1α or IP-10 (P < .001 or P < .05, respectively, compared to the chemotaxis of cells incubated with CM only). However, incubation with IFN-γ or MIP-3β for 5 days inhibited the response to IP-10 (P < .03 orP < .02, respectively, as compared to the chemotactic response of cells incubated for 5 days with CM only). In contrast, no effect on chemotaxis was observed by incubating the cells with MDC, TARC, or I-309 (P not significant). These results correlate the increase in receptor expression by TGF-β1 with increased motility toward the ligands for these chemokine receptors. In contrast, reduced expression of CXCR3 by IFN-γ or MIP-3β results in reduced chemotaxis. The effect of IFN-γ–treated NK cells on their chemotaxis toward SDF-1α was variable between donors and is not shown here.
Analysis of mRNA for chemokines in various subsets of NK cells
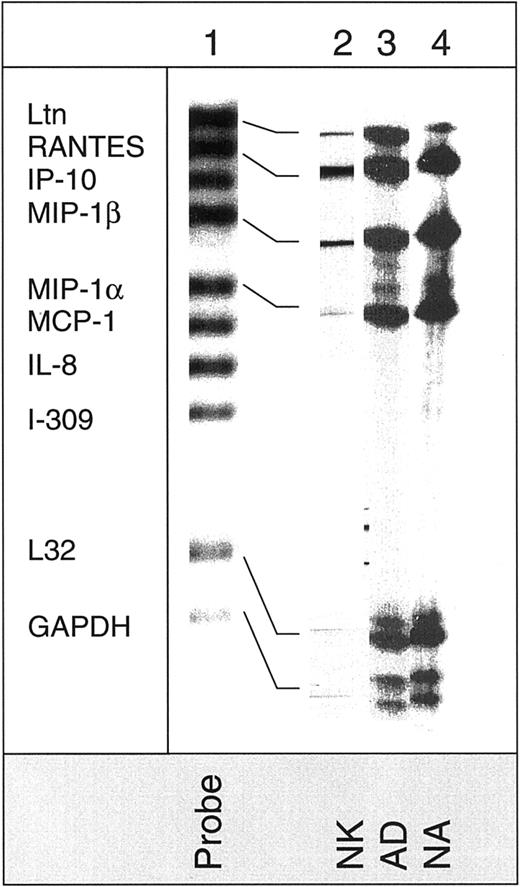
Finally, we examined chemokine production by the various NK cell subsets. Figure 8 shows RPA analysis of the mRNA expression for various chemokines. All 3 NK cell subsets expressed mRNA for lymphotactin (Ltn), RANTES, MIP-1α, and MIP-1β but not IP-10, MCP-1, IL-8, or I-309. This expression was similar in IL-2–activated (AD and NA) and nonactivated NK cells. Although the intensities of the chemokine bands in IL-2–activated cells were higher than nonactivated cells, mRNA for the housekeeping L32 and GAPDH was also low in the nonactivated NK cells when compared to IL-2–activated cells.
Detection of chemokine mRNA expression in NK cells.
Expression of mRNA in nonactivated (NK), adherent (AD), and nonadherent (NA) cells was determined for various chemokines using the hCK5 multitemplate kit. The mRNA for these chemokines is examined: lymphotactin (Ltn), RANTES, IP-10, MIP-1α, MIP-1β, IL-8, MCP-1, and I-309.
Detection of chemokine mRNA expression in NK cells.
Expression of mRNA in nonactivated (NK), adherent (AD), and nonadherent (NA) cells was determined for various chemokines using the hCK5 multitemplate kit. The mRNA for these chemokines is examined: lymphotactin (Ltn), RANTES, IP-10, MIP-1α, MIP-1β, IL-8, MCP-1, and I-309.
Discussion
Natural killer cells and activated NK cells respond to various chemokines (reviewed in Maghazachi and Al-Aoukaty12) suggesting that they express receptors for these chemokines. Here, we examined the expression of chemokine receptors in nonactivated NK cells, as well as in IL-2 activated NK cells that either adhere to plastic flasks (adherent, AD) or those that do not adhere (nonadherent, NA). It was important to examine these cell types because of their differential tendency to migrate toward the sites of tumor growth.
Except for the constitutive chemokine receptors, CXCR4 and CCR7, which are expressed more in nonactivated or NA than in AD NK cells, all other chemokine receptors that are detected are expressed higher in the AD than in nonactivated or NA NK cells. These results indicate that NK cells are not uniform in terms of their expression of chemokine receptors and that this expression depends on their activation pattern. The most important differences are: (1) increased surface expression and mRNA for CXCR1, CCR4, CCR8 and CX3CR1, and (2) increased mRNA for CCR1 after activation with IL-2, specifically on NK cells that adhered to plastic flasks. Other receptors show this pattern: CXCR3 (AD > NA > nonactivated), CXCR4 (nonactivated = NA > AD), CCR2 (AD > NA = nonactivated), CCR3 (AD > NA = nonactivated), CCR6 (AD > NA = nonactivated), and CCR7 (nonactivated > AD > NA). All 3 subsets of NK cells lack the expression of CXCR2, CXCR5, and CCR5. Despite the low expression of certain chemokine receptors on nonactivated or NA NK cells, these cells migrate toward the respective chemokines, when these are used at high concentrations. For example, AD cells migrate toward 100 pg/mL of the inflammatory chemokine MCP-1, whereas 25 ng/mL induced the migration of nonactivated and NA cells, although only 3% of nonactivated and NA NK cells express CCR2. It is plausible that an additional receptor may be expressed on the surface of NK cells that facilitates the chemoattraction toward MCP-1. Similarly, higher concentrations of IP-10, MDC, and MIP-3α induced the chemotaxis of nonactivated and NA NK cells, when compared to the lower concentrations of these chemokines used for the chemotaxis of AD NK cells.
Surprisingly, nonactivated and NA cells respond to fractalkine despite their lack of CX3CR1 expression both at the protein and at the transcriptional levels. This effect of fractalkine was not inhibited by anti-CX3CR1, suggesting that fractalkine may use another receptor in these cells to induce their chemotaxis. However, the same antibody inhibited the chemotactic response of AD NK cells more than 50%, indicating that in these cells fractalkine uses 2 different receptors; one of them is CX3CR1. In addition, all 3 NK cell subsets respond to the CXC chemokine, GRO-α, although they lack the expression of CXCR2, the receptor for this chemokine. Two different anti-CXCR2 antibodies failed to inhibit GRO-α–induced nonactivated, NA, or AD cell chemotaxis. These results suggest that another receptor must exist on these cells that facilitates their chemotaxis toward this chemokine.
Because CXCR3 is implicated in the recruitment of cytotoxic T lymphocytes toward the sites of murine renal adenocarcinoma RENCA,37 we assumed that this might also be true for NK cells. We have previously reported that the ligand for CXCR3, IP-10, induces the in vitro migration of AD cells.38 Also, Mahalingam et al39 reported that the murine homologues of IP-10, MuMig and Crg-2, recruit NK cells toward the sites of vaccinia virus infection resulting in decreased infection with this virus. TGF-β1 highly increased the expression of CXCR3 on NK cells suggesting that during inflammation, this regulatory cytokine may influence the migration of NK cells toward IP-10. In fact, treatment of NK cells for 5 days with TGF-β1 increased their chemotaxis toward IP-10, which is a known antiangiogenic chemokine.40Therefore, increased expression of CXCR3 on NK cells by TGF-β1 may have a role in inhibition of angiogenesis by this cytokine. In addition, recent work showed that NK cells and the ligands for CXCR3 play a major role in controlling viral infection.39,41Hence, the finding that TGF-β1 increased the expression of CXCR3 may have implications for controlling viral infection. The expression of CXCR4 on NK cells suggests that NK cells may be targets for infection with the syncytium forming XR4 virus, which uses CXCR4 as a coreceptor, and in some cases infects cells via CXCR4 in the absence of CD4 molecules.42 The expression of CXCR4 on NK cells may provide an explanation of how these cells are infected with human immunodeficiency virus (HIV)-1.43 The ability of TGF-β1 to increase the expression of CXCR4 on NK cells may influence the susceptibility of these cells to HIV-1 infection. In conclusion, TGF-β1 may have both beneficial and detrimental effects for NK cells.
The expression of CCR8 on AD cells supports our recent findings showing that these cells express this receptor.20 The importance of the current study is that this receptor is found to be exclusively expressed in AD but not in NA or nonactivated NK cells. Hence, a combination of IL-2 activation and adherence to plastic flasks results in the expression of this receptor, which is detected on about 25% to 30% of the AD cells 10 days after stimulation with IL-2. AD cells but not nonactivated or NA NK cells respond chemotactically to I-309, supporting the flow cytometric and RPA results. The etiologic agent of Kaposi sarcoma (KS), known as human herpes virus 8 (HH8), has been shown to encode at least 3 chemokines known as vMIP-I, vMIP-II, and vMIP-III.44 Using either CCR8 transfectants or IL-2–activated T cells, 2 groups observed that CCR8 is the receptor for vMIP-I.31,32 Our results show that vMIP-I is a potent chemoattractant for AD cells. In fact, it induces the chemotaxis of these cells at a much lower concentration than I-309. Competition for the binding of 125I-309 showed that vMIP-I has a higher affinity for AD cells than I-309. In addition, we observed that TARC competes efficiently with radiolabeled I-309 for binding AD cells. These results support those of others showing that TARC binds CCR8 in Jurkat cell line,34 and ours showing that TARC desensitizes the calcium flux response induced by I-309 in AD NK cells,20 but are in contrast to those reported in Th2 cells.33
The pattern of chemokine production by the various NK cell subsets is the same. AD, NA, or nonactivated NK cells produce lymphotactin, RANTES, MIP-1β, and MIP-1α, but not IP-10, MCP-1, IL-8, or I-309. These results suggest that the differential expression of chemokine receptors in nonactivated versus AD or NA NK cells is not due to the presence of the ligands, which may differentially regulate the expression of these receptors in the different NK cell subsets. A support for this hypothesis comes from the failure of inflammatory and constitutive chemokines to up-regulate the expression of chemokine receptors on the surface of NK cells. However, it is noteworthy that the receptors for the secreted chemokines MIP-1α, MIP-1β, and RANTES, that is, CCR1 and CCR5 (XCR1, the receptor for lymphotactin was not examined), are not present in any of the NK cell subsets examined. In contrast, the receptors for the chemokines that are not secreted (eg, IL-8, IP-10, MCP-1, and I-309) are present, at least on the surface of AD cells. These include CXCR1, CXCR3, CCR2, and CCR8. Collectively, these observations suggest that chemokines do not up-regulate the expression of chemokine receptors, but they may induce the internalization of their corresponding inflammatory chemokine receptors. In summary, we have identified chemokine receptors in different NK cell subsets and compared this expression with the functional activities of these receptors. Table1 summarizes these results.
Expression and function of chemokine receptors in different subsets of NK cells
| Receptor . | Expression . | Function . | ||||
|---|---|---|---|---|---|---|
| NK . | NA . | AD . | NK . | NA . | AD . | |
| CXCR1 | no | no | low* | no | no | no |
| CXCR2 | no | no | no | no | no | no |
| CXCR3‡ | mod | mod* | high* | active | active | active† |
| CXCR4 | high | high | high | active | active | active |
| CXCR5 | no | no | no | NT | NT | NT |
| CX3CR1 | no | no | high | no | no | active |
| CCR11-153 | no | no | no | NT | NT | NT |
| CCR2 | low | low | mod | active | active | active |
| CCR3 | no | no | low | no | no | active |
| CCR4 | high | high | high | active | active | active |
| CCR5 | no | no | no | NT | NT | NT |
| CCR6 | mod | mod | mod | active | active | active |
| CCR7 | high | high | high | active | active | active |
| CCR8 | no | no | high | no | no | active |
| Receptor . | Expression . | Function . | ||||
|---|---|---|---|---|---|---|
| NK . | NA . | AD . | NK . | NA . | AD . | |
| CXCR1 | no | no | low* | no | no | no |
| CXCR2 | no | no | no | no | no | no |
| CXCR3‡ | mod | mod* | high* | active | active | active† |
| CXCR4 | high | high | high | active | active | active |
| CXCR5 | no | no | no | NT | NT | NT |
| CX3CR1 | no | no | high | no | no | active |
| CCR11-153 | no | no | no | NT | NT | NT |
| CCR2 | low | low | mod | active | active | active |
| CCR3 | no | no | low | no | no | active |
| CCR4 | high | high | high | active | active | active |
| CCR5 | no | no | no | NT | NT | NT |
| CCR6 | mod | mod | mod | active | active | active |
| CCR7 | high | high | high | active | active | active |
| CCR8 | no | no | high | no | no | active |
This table summarizes the results obtained regarding the surface expression of chemokine receptors as determined by flow cytometric assay, and functional analysis as determined by in vitro chemotaxis assay. NK cell populations used were nonactivated (designated as NK), those that are activated with IL-2 but did not adhere to plastic flasks (designated as NA), and those that are activated with IL-2 and adhered to plastic flasks (designated as AD).
The expression of chemokine receptors was classified as low (when the proportion of the positive cells is 5% or less); moderate (mod; when the proportion of positive cells is 5-10%); and high (when the proportion of positive cells is higher than 10%).
Active designation means that the receptor supports the chemotaxis response.
The expression of CXCR3 on NK cells is considered moderate because between 5% and 12% of the cells were positive.
Although flow cytometric analysis shows no expression of CCR1, mRNA for this receptor is expressed in the three NK cell subsets. The lack of surface expression may be due to CCR1 internalization.
No indicates no expression and/or no function; NT, not tested.
Supported by grants from the Norwegian Research Council, the Norwegian Cancer Society, and Anders Jahres Foundation. A.A.M. is a Senior Scientist of the Norwegian Cancer Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Azzam A. Maghazachi, Department of Anatomy, University of Oslo, POB 1105 Blindern, N-0317 Oslo, Norway; e-mail: azzam.maghazachi@basalmed.uio.no.

![Fig. 6. Effect of CC chemokines on the in vitro motility of various preparations of NK cells. / In panel A, MCP-1 (25 ng/mL for nonactivated [▩] and NA [▧] and 100 pg/mL for AD [▪] cells), Eotaxin (10 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), MDC (10 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), MIP-3α (25 ng/mL for nonactivated and NA and 1 ng/mL for AD cells), MIP-3β (1 ng/mL for nonactivated, NA, and AD cells), or I-309 (10 ng/mL for nonactivated and NA and 1 ng/mL for AD cells) was placed in the lower wells of Boyden chambers, whereas 4 × 105 (nonactivated, AD, or NA) NK cells were added to the upper wells. In panel B, various concentrations (1 pg/mL, ░; 10 pg/mL, ▩; 100 pg/mL, ▪; 1000 pg/mL, ▤; 10 000 pg/mL, ▥) of vMIP-I were examined. Migration index was calculated as the number of cells migrating in the presence of chemokines divided by the number of cells migrating in the presence of medium only (controls, shown in white columns). Mean ± SD of 6 to 9 experiments. Panel C shows competition binding analysis of 100 pmol/L 125I-309 to AD cells, by cold I-309, TARC, or vMIP-I. Log concentrations of these chemokines are shown in the X-axis, whereas the total count of binding is shown in the Y-axis. IC50 for cold ligands is also shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/2/10.1182_blood.v97.2.367/5/m_h80210588006.jpeg?Expires=1769095905&Signature=Eel4OPCHpsq-CCjkLjquSVSFttT9YE~0VSVrW-npzCgfBAq1lxFEphQ-cd1P8o9C8Omx1PuBv2-foXWGdQzYzHzk4kN7JwGQjJPVzFRO9QB1bzAPoonJA9TTZkT0fLN-F26ycuUEJqm2qDjAf7cBhMHlcPNVILiUk-7faebbS5IkZcCejqNVSzzZ9NlyFDrj~u~g8C4sulD~hR3yyEHaaR~klZ2QYkmbWuhOn7r43PvFZhT1qEyITReWvoaMP1RrPzxJKr3md0VkCsB7TlbmiiTmhqp6WwbADDi4NgY~sV2J5lccACX5Sx3lHvdu0lWvYMWybNc09gNrJqG4O0OwrA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal