Abstract
Regulation of allelic and isotypic exclusion of human immunoglobulin (Ig) light-chain genes was studied in 113 chronic B-cell leukemias as a “single-cell” model that allowed complete analysis of each light chain allele. Our data show that monospecific Ig light chain expression is in about 90% of cases determined by ordered recombination: Igκ gene (IGK) rearrangements, followed byIGK deletions and Igλ gene (IGL) rearrangements, resulting in the presence of only one functional Ig light chain rearrangement. In about 10% (10 cases), 2 functional Ig light chain rearrangements (IGK/IGL or IGL/IGL, but not IGK/IGK) were identified. This might be explained by the fact that regulation of the ordered recombination process is not fully strict, particularly when the IGL locus is involved. Unfavorable somatic mutations followed by receptor editing might have contributed to this finding. Eight of these 10 cases indeed contained somatic mutations. In cases with 2 functional Ig light chain rearrangements, both alleles were transcribed, but monospecific Ig expression was still maintained. This suggests that in these cases allelelic exclusion is not regulated at the messenger RNA level but either at the level of translation or protein stability or via preferential pairing of Ig light and Ig heavy chains. Nevertheless, ordered rearrangement processes are the main determinant for monospecific Ig light chain expression.
Introduction
In normal and malignant human B cells, functional expression of immunoglobulin κ (Igκ) genes (IGK) occurs more frequently than functional expression of Igλ genes (IGL), resulting in an Igκ/λ distribution of approximately 1.4.1 The observed ratio between Igκ- and Igλ-expressing B cells in mice is significantly higher (by about 10).2 The difference in κ/λ ratio between mice and man might be related to differences in the organization of theIGK and IGL loci. Although the human IGKlocus is organized like the murine IGK locus, the organization of the IGL loci differs.3,4 Mice only have 3 Vλ and 4 Jλ segments, each followed by a Cλ segment, which are arranged in 2 clusters.5,6 In human beings, about 30 functional Vλ gene segments are located upstream of 4 functional J-Cλ clusters.7 8 Thus, the theoretical combinatorial repertoire of Vλ-Jλrearrangements in humans is 10-fold higher than in mice, which might explain the difference in κ/λ ratio.
Two models have been proposed for explaining the relative “overrepresentation” of IGK genes in both species: the ordered model and the stochastic model.9-12 The ordered model proposes that IGK genes rearrange prior to IGL;the stochastic model postulates that, in principle, the 2 types of Ig light chain genes rearrange totally independently but that other factors render IGL gene rearrangements more difficult.13 14 The latter would imply that IGLgene rearrangements can occur in the absence of IGK gene rearrangements and vice versa.
Alt and Baltimore first postulated the ordered model of Ig gene rearrangements in mice.15,16 They suggested that the Ig gene rearrangement process starts within the Ig heavy chain gene (IGH) locus and that the rearrangement process is terminated as soon as a productive (in-frame) IgH chain is expressed on a pre-B cell, resulting in allelic exclusion of the IGHlocus.16 They also postulated the ordered model for Ig light chain gene rearrangements, starting with IGK gene recombination, only followed by rearrangement in the IGLlocus if no functional combination is formed.15 The ordered model assumes a feedback mechanism, which implies that the V(D)J recombinase system is down-regulated upon surface Ig expression. This feedback mechanism explains the establishment of allelic exclusion (expression of one heavy chain and one Ig light chain), including isotypic exclusion (Igκ or Igλ expression).
In mice, monoallelic IGK demethylation ensures the ordered process of IGK rearrangements, thereby establishing allelic exclusion of the IGK alleles.17 By analogy, Engel et al described in their murine hit-and-run model thatIGK and IGL are activated for recombination at consecutive developmental stages.3 Also, in humans it is believed that IGK genes rearrange prior toIGL,9,18 which is supported by recent studies in a human immature B-cell line showing that the IGKenhancer, but not the IGL enhancer, is accessible for DNaseI.19 However, it is known that an IGL gene rearrangement can occasionally be present while theIGK genes are in germline configuration.20
Until recently, allelic exclusion was generally regarded to be a safe mechanism that guarantees the expression of a single type of antigen receptor on each lymphocyte. However, during the last few years, several reports have indicated that dual receptor expression might occur in B lymphocytes as well as in T lymphocytes because of ongoing rearrangements after a functional receptor gene has been formed and expressed.21-26 It was found that a single T lymphocyte might express 2 different T-cell receptor (TCR)β chains and 2 different TCRα chains, indicating that both TCRB alleles as well as both TCRA alleles are functionally rearranged and expressed.24,25 By analogy, Giachino et al demonstrated that in 0.2% to 0.5% of human B lymphocytes dual expression of Igκ and Igλ occurs.21,22 27 This would imply that dual Ig light chain expression might be even higher owing to dual Igκ/Igκ and dual Igλ/Igλ expression, which would theoretically occur in 0.3% to 0.6% and about 0.1%, respectively. Although it will be difficult to prove the presence of dual Igκ/Igκ and dual Igλ/Igλ expression by immunophenotyping due to lack of Vκ- and Vλ-specific antibodies, the estimated dual Ig light chain expression might thus even be as high as 1%.
Here we have studied human chronic B-cell leukemias (CBLs) for the presence of functional IGK and IGL gene rearrangements. Clonal leukemic proliferations allow a complete and extensive analysis of both IGK and both IGLalleles so that CBL can be regarded as the ideal “single-cell model” of human B cells, in which regulation of allelic and isotypic exclusion of light chain genes can be studied in detail.
Materials and methods
Cell samples, immunophenotyping, and DNA and RNA isolation
Mononuclear cells (MNCs) were obtained from peripheral blood or bone marrow samples by Ficoll-Paque (density 1.077 g/L; Pharmacia, Uppsala, Sweden) centrifugation from a series of 113 patients with CBL, including 107 B-cell chronic leukemia (B-CLL), 5 B-cell prolymphocytic leukemia, and a single hairy cell leukemia. MNCs were used for detailed immunophenotyping.28 29 In a few cases, lymph node suspensions were used. In all samples the tumor load was at least 75%.
DNA and RNA were isolated from MNCs or lymph node cells, as described.30 Complementary DNA was prepared from RNA using either AMV reverse transcriptase (Promega, Madison, WI) or Superscript RT enzyme (Life Technologies, Paisley, UK) according to the manufacturer's instructions.
Southern blot analysis
Fifteen micrograms of DNA was digested with the appropriate restriction enzymes (Life Technologies, Rockville, MD), separated in 0.7% agarose gels, and transferred by vacuum blotting to Nytran-13N nylon membranes (Schleicher and Schuell, Dassel, Germany).30 The filters were hybridized with32P-labeled probes. Probes specific for either Jκ (IGKJ5), Cκ (IGKC), and kappa-deleting element (Kde) (IGKDE) (Dako, Carpinteria, CA) were used in combination with BglII and BamHI/HindIII restriction enzyme digests to determine the IGK gene configuration.31 For the IGL alleles, isotype-specific probes (IGLC1D, IGLJ2, IGLC2D, IGLC4D, IGLJ5, IGLJ6, IGLJ7) were used in combination with HindIII,BglII, BamHI/HindIII, andEcoRI/HindIII digests.32
Reverse transciptase–polymerase chain reactionheteroduplex analysis
The reverse transciptase–polymerase chain reaction (RT-PCR) mixture of 100 μL contained 0.2 mM deoxyribonucleoside triphosphate (Pharmacia), 13 pmol of each primer, 1 unit AmpliTaq Gold polymerase in Buffer II (Applied Biosystems, Foster City, CA), 1.5 mM MgCl2, and 100 ng genomic DNA or 5 μL complementary DNA (derived from 0.25 μg total RNA). The family-specific Vκ and Vλ primers, the Jκ and Jλ primers, and the Cκ and Cλexon primers are listed in Table 1. PCR conditions were 10 minutes at 94°C followed by 40 cycles of 1 minute at 94°C, 1 minute at 72°C, and a final extension of 7 minutes at 72°C. PCR products were further analyzed by heteroduplex analysis to determine whether the PCR products were derived from clonal or polyclonal rearrangements.33
Primers for polymerase chain reaction analysis ofIGK and IGL gene rearrangements
| Primers . | Sequence (5′-3′) . |
|---|---|
| IGKlocus | |
| LeaderVκI+II | GGTCCCCGCTCAGCTCCT |
| LeaderVκIV | TCTCTGTTGCTCTGGATCTCT |
| VκI | GTAGGAGACAGAGTCACCATCACT |
| VκII | TGGAGAGCCGGCCTCCATCTC |
| VκIII | GGGAAAGAGCCACCCTCTCCTG |
| VκIV | GGCGAGAGGGCCACCATCAAC |
| VκV | CCAGGAGACAAAGTCAACATCTCC |
| VκVI | CTGTGACTCCAAAGGAGAAAGTC |
| VκVII | AGGACAGAGGGCCACCATCACC |
| Jκ1,2,4 | CCCTGGTTCCACCTCTAGTTTGCA |
| Jκ3 | GGGACCAAAGTGGATATCAAACGT |
| Jκ5 | GGGACACGACTGGAGATTAAACGT |
| Cκ | ACTTTGGCCTCTCTGGGATA |
| IGLlocus | |
| LeaderVλI | GCCCAGTCTGTGCTGAC |
| LeaderVλII | CTGGGCTCTGCTCCTCCT |
| LeaderVλIII | GTGACCTCCTATGTGCTGACT |
| VλI | GGCAGAGGGTCACCATCTC |
| VλII | ATCTCCTGCACTGGAACCA |
| VλI+II | ATTCTCTGGCTCCAAGTCTGGCA |
| VλIII | ATTCTCTGGCTCCAACTCTGGGAA |
| Jλ1 | AGGCTGGGAAAGGTTGAG |
| Jλ2,3 | AGAGGGGAGAAGAGACTCAC |
| Cλ | TTGACGGGGCTGCTATCT |
| Primers . | Sequence (5′-3′) . |
|---|---|
| IGKlocus | |
| LeaderVκI+II | GGTCCCCGCTCAGCTCCT |
| LeaderVκIV | TCTCTGTTGCTCTGGATCTCT |
| VκI | GTAGGAGACAGAGTCACCATCACT |
| VκII | TGGAGAGCCGGCCTCCATCTC |
| VκIII | GGGAAAGAGCCACCCTCTCCTG |
| VκIV | GGCGAGAGGGCCACCATCAAC |
| VκV | CCAGGAGACAAAGTCAACATCTCC |
| VκVI | CTGTGACTCCAAAGGAGAAAGTC |
| VκVII | AGGACAGAGGGCCACCATCACC |
| Jκ1,2,4 | CCCTGGTTCCACCTCTAGTTTGCA |
| Jκ3 | GGGACCAAAGTGGATATCAAACGT |
| Jκ5 | GGGACACGACTGGAGATTAAACGT |
| Cκ | ACTTTGGCCTCTCTGGGATA |
| IGLlocus | |
| LeaderVλI | GCCCAGTCTGTGCTGAC |
| LeaderVλII | CTGGGCTCTGCTCCTCCT |
| LeaderVλIII | GTGACCTCCTATGTGCTGACT |
| VλI | GGCAGAGGGTCACCATCTC |
| VλII | ATCTCCTGCACTGGAACCA |
| VλI+II | ATTCTCTGGCTCCAAGTCTGGCA |
| VλIII | ATTCTCTGGCTCCAACTCTGGGAA |
| Jλ1 | AGGCTGGGAAAGGTTGAG |
| Jλ2,3 | AGAGGGGAGAAGAGACTCAC |
| Cλ | TTGACGGGGCTGCTATCT |
Sequencing analysis
Clonal RT-PCR products were directly sequenced on an ABI 377 fluorescent cycle sequencer (Applied Biosystems) with Dye Terminator mix or Big Dyes (Applied Biosystems) according to the manufacturer's instructions. Vκ, Jκ, Vλ, and Jλ segments were identified using DNAplot software (W. Müller, H-H. Althaus, University of Cologne, Germany) via VBASE and IMGT databases (http://imgt.cnusc.fr:8104).34Subsequently, the frame of the rearrangement and the mutation status were determined. A case was classified as somatically mutated if the involved V gene segment had less than 98% homology with the most related V gene segment.
Results
Southern blot analysis of the CBL samples
To study the complete and exact configuration of bothIGK and both IGL alleles in a cohort of 113 CBLs, detailed Southern blot analysis was performed. In previous studies, the configuration of the IGK alleles (ie, germline, Vκ-Jκ rearrangement, or deletion of Jκ and/or Cκ) and the IGL alleles of most CBLs were determined.31 32 The data of the Igκ+ and Igλ+ CBLs are summarized in Table2. In our series of CBL, half of the Igκ+ CBLs (25 of 53) had one rearranged IGKallele, and the others had biallelic IGK gene rearrangements (11 of 53) or one rearranged and one deleted IGKallele (17 or 53). A total of 94% (50 of 53) of them showed bothIGL genes in germline configuration (Table 2). On the other hand, all 60 Igλ+ CBLs had at least one deletedIGK allele and 88.3% (53 of 60) of them even had either both IGK alleles deleted or one deletedIGK allele with the other in germline configuration. This supports the general idea that IGL gene rearrangements are preceded by IGK gene deletions. Table 2 demonstrates the hierarchic order in Ig light chain gene rearrangements: The light chain gene rearrangement process starts at the IGK locus, followed by IGK deletion and subsequent IGL gene rearrangement.
IGK and IGL gene configurations in 113 chronic B-cell leukemias (53 immunoglobulin κ+ and 60 immunoglobulin λ+) as determined by Southern blot analysis
| IGK gene configuration . | Igκ+CBL . | Igλ+ CBL . | ||||
|---|---|---|---|---|---|---|
| IGL gene configuration . | ||||||
| G/G . | G/R . | R/R . | G/G . | G/R . | R/R . | |
| G/G | — | — | — | — | — | — |
| R/G | 47% | — | — | — | — | — |
| (25/53) | ||||||
| R/R | 19% | 2% | — | — | — | — |
| (10/53) | (1/53) | |||||
| D/R | 28% | 2% | 2% | — | 10% | 1.5% |
| (15/53) | (1/53) | (1/53) | (6/60) | (1/60) | ||
| D/D | — | — | — | — | 57% | 21.5% |
| (34/60) | (13/60) | |||||
| G/D | — | — | — | — | 7% | 3% |
| (4/60) | (2/60) | |||||
| IGK gene configuration . | Igκ+CBL . | Igλ+ CBL . | ||||
|---|---|---|---|---|---|---|
| IGL gene configuration . | ||||||
| G/G . | G/R . | R/R . | G/G . | G/R . | R/R . | |
| G/G | — | — | — | — | — | — |
| R/G | 47% | — | — | — | — | — |
| (25/53) | ||||||
| R/R | 19% | 2% | — | — | — | — |
| (10/53) | (1/53) | |||||
| D/R | 28% | 2% | 2% | — | 10% | 1.5% |
| (15/53) | (1/53) | (1/53) | (6/60) | (1/60) | ||
| D/D | — | — | — | — | 57% | 21.5% |
| (34/60) | (13/60) | |||||
| G/D | — | — | — | — | 7% | 3% |
| (4/60) | (2/60) | |||||
Ig indicates immunoglobulin; G, germline configuration; R, rearranged allele; D, deletion of Jκ and/or Cκ gene segments.
Because the presence of a rearranged band in Southern blot analysis does not necessarily imply the presence of a V-J joining, no distinction was possible between physiologic V-J rearrangements and other events like translocations. Moreover, Southern blot analysis cannot discriminate between functional and nonfunctional V-J rearrangements. Therefore, CBLs with 2 or more rearranged Ig light chain alleles were studied in more detail by PCR heteroduplex analysis and sequencing.
Igκ+ CBL with IGL gene rearrangements and Igλ+CBL with IGK gene rearrangements
Six percent (3 of 53) of Igκ+ CBLs containedIGL gene rearrangements, and 11.7% (7 of 60) of the Igλ+ CBLs had IGK gene rearrangements based on Southern blot analysis. The configurations of the IGK andIGL genes of these 10 cases are summarized in Table3. The IGK and IGLalleles of the 3 Igκ+ B-CLL and 7 Igλ+B-CLL were further analyzed by PCR heteroduplex analysis and sequencing to determine the frame of the rearrangements, the involved gene segments, and the mutation status of the V genes.
Combined Southern blot and sequence data of the immunoglobulin κ+ chronic B-cell leukemia withIGL gene rearrangements and immunoglobulin λ+chronic B-cell leukemia with IGK gene rearrangements
| Igκ+ CBL . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | IGK gene configuration3-150 . | IGL gene configuration . | IGLalleles . | Somatic mutations3-155 . | |||||||
| Vλ . | Junctional region3-151 . | Jλ . | Frame3-152 . | RT-PCR3-153 . | IGK . | IGL . | |||||
| 1 | R/VJ-Kde | R/G | Vλ3-21 | 0 | GGGG | −3 | Jλ1 | + | ND | + | − |
| 2 | R/VJ-Kde | R/R | 1st Vλ2-33 | −3 | AAA | −2 | Jλ2 | −3-154 | ND | + | − |
| 2nd not found | |||||||||||
| 3 | R/R# | R/G | Vλ1-40 | 0 | − | 0 | Jλ3 | + | + | NI | + |
| Igκ+ CBL . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | IGK gene configuration3-150 . | IGL gene configuration . | IGLalleles . | Somatic mutations3-155 . | |||||||
| Vλ . | Junctional region3-151 . | Jλ . | Frame3-152 . | RT-PCR3-153 . | IGK . | IGL . | |||||
| 1 | R/VJ-Kde | R/G | Vλ3-21 | 0 | GGGG | −3 | Jλ1 | + | ND | + | − |
| 2 | R/VJ-Kde | R/R | 1st Vλ2-33 | −3 | AAA | −2 | Jλ2 | −3-154 | ND | + | − |
| 2nd not found | |||||||||||
| 3 | R/R# | R/G | Vλ1-40 | 0 | − | 0 | Jλ3 | + | + | NI | + |
| Igλ+ CBL . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | IGK gene configuration . | IGL gene configuration . | IGKalleles . | Somatic mutations . | |||||||
| Vκ . | Junctional region . | Jκ . | Frame . | RT-PCR . | IGK . | IGL . | |||||
| 4 | R/VJ-Kde | R/R3-160 | 1st not found | NI | + | ||||||
| 2nd Vκ1-39 | −6 | GA | −9 | Jκ4 | − | − | |||||
| 5 | R/V-Kde | R/G | Vκ2-30 | −10 | TGACC | −4 | Jκ2 | − | ND | − | NI |
| 6 | R/V-Kde | R/G | Vκ2-28 | −4 | TCACT | −6 | Jκ4 | − | ND | − | − |
| 7 | R/V-Kde | R/G | Vκ1-33 | −1 | ACC | −6 | Jκ4 | − | ND | − | − |
| 8 | R/V-Kde | R/G | Vκ4-1 | −3 | CCTG | −2 | Jκ2 | + | + | − | − |
| 9 | R/VJ-Kde | R/G | 1st Vκ1-33 | −3 | ATC | −2 | Jκ3 | + | + | − | − |
| 2nd Vκ4-1 | −3 | CT | 0 | Jκ4 | − | − | |||||
| 10 | R/VJ-Kde | R/G | 1st Vκ1-12 | −3 | − | 0 | Jκ3 | + | + | + | + |
| 2nd Vκ1-33 | −3 | AT | 0 | Jκ3 | − | − | |||||
| Igλ+ CBL . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | IGK gene configuration . | IGL gene configuration . | IGKalleles . | Somatic mutations . | |||||||
| Vκ . | Junctional region . | Jκ . | Frame . | RT-PCR . | IGK . | IGL . | |||||
| 4 | R/VJ-Kde | R/R3-160 | 1st not found | NI | + | ||||||
| 2nd Vκ1-39 | −6 | GA | −9 | Jκ4 | − | − | |||||
| 5 | R/V-Kde | R/G | Vκ2-30 | −10 | TGACC | −4 | Jκ2 | − | ND | − | NI |
| 6 | R/V-Kde | R/G | Vκ2-28 | −4 | TCACT | −6 | Jκ4 | − | ND | − | − |
| 7 | R/V-Kde | R/G | Vκ1-33 | −1 | ACC | −6 | Jκ4 | − | ND | − | − |
| 8 | R/V-Kde | R/G | Vκ4-1 | −3 | CCTG | −2 | Jκ2 | + | + | − | − |
| 9 | R/VJ-Kde | R/G | 1st Vκ1-33 | −3 | ATC | −2 | Jκ3 | + | + | − | − |
| 2nd Vκ4-1 | −3 | CT | 0 | Jκ4 | − | − | |||||
| 10 | R/VJ-Kde | R/G | 1st Vκ1-12 | −3 | − | 0 | Jκ3 | + | + | + | + |
| 2nd Vκ1-33 | −3 | AT | 0 | Jκ3 | − | − | |||||
Ig indicates immunoglobulin; RT-PCR, reverse transcriptase–polymerase chain reaction.
R indicates IGK gene rearrangement; V-Kde, Vκ segment rearranged to kappa-deleting element; VJ-Kde, Vκ-Jκ and intron RSS-Kde rearrangement on the same allele.
The junctional region is described as the number of bases lost at the 3′ end of the V segment followed by randomly inserted nucleotides and the number of bases lost from the 5′ end of the J segment.
Frame of the rearrangement: +, in-frame, without stop codons in the junctional region; −, out-of-frame.
ND indicates not determined; +, present; −, absent.
+ indicates V segments with more than 2% somatic mutations; −, less than 2% somatic mutations; NI, not identified, because the rearrangement could not be amplified.
A deletion of 36 nucleotides was found, starting from the last 2 codons of the Jλ2 gene segment up to 30 nucleotides in the J-Cλ intron, thereby deleting the Jλ splice site.
#For biallelic IGK gene rearrangements, see Table 4.
For biallelic IGL gene rearrangements, see Table 5.
In 2 of the 3 Igκ+ CBLs (patients 1 and 3), an in-frameIGL rearrangement was detected at the DNA level. The junctional regions of all in-frame rearrangements were analyzed for the presence of stop codons. In patient 3, a functional transcript of theIGL rearrangement was detected by RT-PCR. Lack of material precluded further analysis in patient 1. In patient 2, only one out-of-frame IGL rearrangement was found. The second rearrangement could not be identified, which might be caused by the inability of the used primers to recognize the involved gene segments or by a chromosomal aberration involving chromosome 22.
The 7 Igλ+ B-CLL (patients 4-10) had one rearranged and one deleted IGK allele (Table 3). In patients 4, 9, and 10, the Kde was rearranged to the intron RSS (recombination signal sequence), thereby deleting the Cκ exon but retaining the Vκ-Jκ rearrangement on the same allele (VJ-Kde).31 This rearrangement does not give rise to a functional transcript, because of deletion of the Cκexon. Therefore, in these 4 patients, 2 Vκ-Jκ rearrangements can be detected at the DNA level, but only the IGK rearrangement on the allele without the Cκ deletion can be detected by RT-PCR. In patients 5 to 8, a Vκ-Kde deletion was detected, which resulted from a rearrangement of a Vκ segment to the Kde in which the Jκ and Cκ segments are deleted together with the preexisting Vκ-Jκrearrangement.
In patient 4, only one Vκ-Jκ rearrangement was found. To determine whether this rearrangement was located at the nondeleted or deleted allele, a primer was designed overlapping the junctional region. Long-range PCR analysis using the junction-specific primer and the Kde primer (and the Cκ primer as control) showed that the identified Vκ-Jκrearrangement was located on the allele with the Cκdeletion. This rearrangement was indeed not found by RT-PCR. The identified Vκ-Jκ rearrangements of patients 5 to 7 were out-of-frame. Patients 8 to 10 had a functional (in-frame)IGK rearrangement on the rearranged allele, as confirmed by detection of Vκ-Cκ transcripts and sequencing.
Somatic mutations in the V genes were determined. Patients 1, 2, 3, 4, and 10 were somatically mutated. Three of the 5 patients with 2 in-frame light chain rearrangements (patients 1, 3, and 10) carried somatic mutations either on one or both alleles against 2 of the 5 patients with a single in-frame light chain rearrangement (patients 2 and 4). In patient 3, the functional rearrangement was not found, but the IGL rearrangement was mutated although not expressed on the membrane.
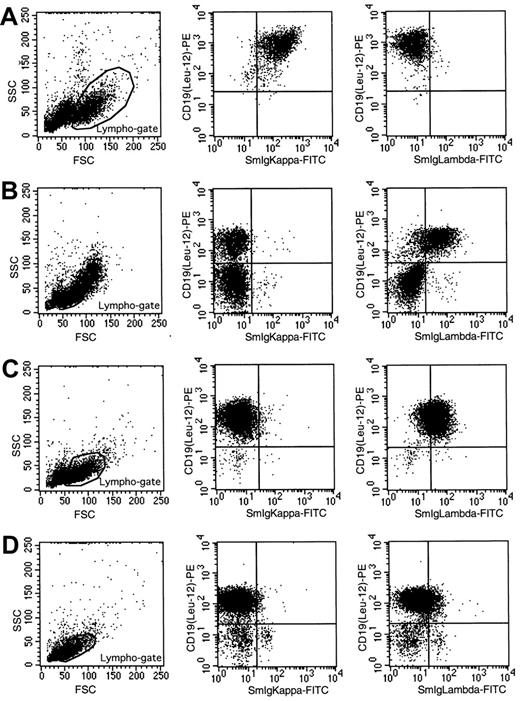
In patients 3, 8, 9, and 10 presenting with double in-frame IGKand IGL gene rearrangements at both the DNA and RNA level, flow cytometric analysis of surface membrane Ig light chains was repeated. No dual Ig light chain expression was observed in the 4 patients (Figure 1). The Igλ expression of patient 10 was lower than the Ig light chain expression in the other 3 patients (Figure 1D), but no Igκ expression was found and, therefore, this CBL was concluded to be monospecific. No remaining cells were available for reanalysis of Ig light chain expression in patient 1, but the original data did not show dual Ig light chain expression in the Igκ+ CBLs.
Flow cytometric immunophenotyping of lymphocytes of 4 B-CLL patients with an in-frame
IGK and IGL gene rearrangement. (A) CD19+ B lymphocytes of patient 3 express Igκ but not Igλ. Patient 8 (B) and patient 9 (C) express Igλ on CD19+ B lymphocytes, but no Igκ expression is detectable. (D) Patient 10 has low Igλ expression, but Igκ expression is absent.
Flow cytometric immunophenotyping of lymphocytes of 4 B-CLL patients with an in-frame
IGK and IGL gene rearrangement. (A) CD19+ B lymphocytes of patient 3 express Igκ but not Igλ. Patient 8 (B) and patient 9 (C) express Igλ on CD19+ B lymphocytes, but no Igκ expression is detectable. (D) Patient 10 has low Igλ expression, but Igκ expression is absent.
To further study whether isotypic exclusion is regulated at the translational level or by posttranslational modification, the cytoplasmic Ig expression of the 4 CBLs was analyzed on cytocentrifuge preparations.29 However, the amount of cytoplasmic Ig in CBL was too low to be detectable; even the isotype expressed on the cell surface could not be detected in the cytoplasm by fluorescence microscopy (data not shown).
Analysis of CBL with biallelic IGK or biallelicIGL rearrangements
In this series of CBL, 19% (10 of 53) of Igκ+ CBLs had biallelic IGK gene rearrangements with IGLgenes in germline configuration (Table 2), and 25% (15 of 60) of the Igλ+ CBLs had biallelic IGL rearrangements with IGK genes deleted or in germline configuration (Table2). In addition, patient 3 (Igκ+ CBL) and patient 4 (Igλ+ CBL) had biallelic IGK andIGL gene rearrangements, respectively, in combination with a rearrangement of the other isotype (Table 3). Sufficient cell material for RT-PCR heteroduplex analysis and sequencing was available for 10 of 11 Igκ+ CBLs with 2 IGK rearrangements and 14 of 16 Igλ+ CBLs with 2 IGL rearrangements. The results are shown in Tables 4 and5, respectively.
Chronic B-cell leukemia with potentially functional biallelic Vκ-Jκ rearrangements as determined by Southern blotting
| Patient . | Vκ . | Junctional region4-150 . | Jκ . | Frame4-151 . | RT-PCR‡ . | Somatic mutations4-153 . | ||
|---|---|---|---|---|---|---|---|---|
| 3 | Vκ1-37 | −3 | CT | −6 | Jκ4 | − | + | − |
| NI | NI | |||||||
| 11 | Vκ1-5 | − | − | − | Jκ4 | + | + | − |
| NA4-155 | NA | |||||||
| 12 | Vκ2-30 | − | − | −3 | Jκ1 | + | + | − |
| Vκ1-39 | −3 | GG | −3 | Jκ2 | − | − | − | |
| 13 | Vκ1-5 | −3 | − | −3 | Jκ1 | + | + | − |
| NI | NI | |||||||
| 14 | Vκ1-5 | −3 | CGC | −1 | Jκ2 | + | + | − |
| Vκ2-28 | − | GA | −4 | Jκ1 | − | + | − | |
| 15 | Vκ3-20 | −1 | G | −1 | Jκ2 | + | + | + |
| Vκ3-11 | − | T | − | Jκ4 | − | − | − | |
| 16 | Vκ1-39 | 0 | A | −1 | Jκ1 | + | ND | + |
| Vκ1-5 | −13 | − | − | Jκ4 | − | ND | − | |
| 17 | Vκ3-20 | − | − | − | Jκ5 | + | + | − |
| Vκ1-37 | − | TT | − | Jκ3 | − | − | − | |
| 18 | Vκ3-20 | −1 | GA | −4 | Jκ1 | + | + | − |
| Vκ4-1 | − | C | −5 | Jκ4 | − | + | − | |
| 19 | Vκ1-27 | −3 | − | − | Jκ1 | + | + | − |
| NI | NI | |||||||
| Patient . | Vκ . | Junctional region4-150 . | Jκ . | Frame4-151 . | RT-PCR‡ . | Somatic mutations4-153 . | ||
|---|---|---|---|---|---|---|---|---|
| 3 | Vκ1-37 | −3 | CT | −6 | Jκ4 | − | + | − |
| NI | NI | |||||||
| 11 | Vκ1-5 | − | − | − | Jκ4 | + | + | − |
| NA4-155 | NA | |||||||
| 12 | Vκ2-30 | − | − | −3 | Jκ1 | + | + | − |
| Vκ1-39 | −3 | GG | −3 | Jκ2 | − | − | − | |
| 13 | Vκ1-5 | −3 | − | −3 | Jκ1 | + | + | − |
| NI | NI | |||||||
| 14 | Vκ1-5 | −3 | CGC | −1 | Jκ2 | + | + | − |
| Vκ2-28 | − | GA | −4 | Jκ1 | − | + | − | |
| 15 | Vκ3-20 | −1 | G | −1 | Jκ2 | + | + | + |
| Vκ3-11 | − | T | − | Jκ4 | − | − | − | |
| 16 | Vκ1-39 | 0 | A | −1 | Jκ1 | + | ND | + |
| Vκ1-5 | −13 | − | − | Jκ4 | − | ND | − | |
| 17 | Vκ3-20 | − | − | − | Jκ5 | + | + | − |
| Vκ1-37 | − | TT | − | Jκ3 | − | − | − | |
| 18 | Vκ3-20 | −1 | GA | −4 | Jκ1 | + | + | − |
| Vκ4-1 | − | C | −5 | Jκ4 | − | + | − | |
| 19 | Vκ1-27 | −3 | − | − | Jκ1 | + | + | − |
| NI | NI | |||||||
RT-PCR indicates reverse transcriptase–polymerase chain reaction.
The junctional region is described as the number of bases lost at the 3′ end of the segment followed by randomly inserted nucleotides and the number of bases lost from the 5′ end of the J segment.
Frame of the junctional region: +, in-frame, without stop codons in the junctional region; −, out-of-frame.
ND indicates not determined; +, present; −, absent.
+ indicates V segments with more than 2% somatic mutations; −, less than 2% somatic mutations; NI, not identified, because the rearrangement could not be amplified.
NA indicates not applicable, because only one intact chromosome 2 was present, although 2 rearrangements seemed to be present in Southern blot analysis (see text).
Chronic B-cell leukemia with potentially functional biallelic Vλ-Jλ rearrangements as determined by Southern blotting
| Patient . | Vλ . | Junctional region5-150 . | Jλ . | Frame5-151 . | RT-PCR5-152 . | Somatic mutations5-153 . | ||
|---|---|---|---|---|---|---|---|---|
| 4 | Vλ1-44 | 0 | G | −1 | Jλ3 | + | + | + |
| Vλ2-55-155 | −4 | TGG | −1 | Jλ3 | − | + | + | |
| 20 | Vλ1-40 | −4 | TCTA | 0 | Jλ3 | + | + | + |
| Vλ3-12 | 0 | − | −3 | Jλ3 | + | + | + | |
| 21 | Vλ3-21 | 0 | − | 0 | Jλ3 | + | ND | − |
| Vλ1-47 | 0 | A | 0 | Jλ3 | − | ND | − | |
| 22 | Vλ2-11 | −15 | 26 nt of Vλ2-14 | −7 | Jλ3 | + | + | + |
| Vλ1-47 | −4 | C | 0 | Jλ3 | + | + | + | |
| 23 | Vλ1-40 | −2 | CGAG | −2 | Jλ3 | + | + | − |
| Vλ3-21 | −2 | − | −2 | Jλ3 | − | + | − | |
| 24 | Vλ3-21 | 0 | CCCT | −1 | Jλ3 | + | + | − |
| Vλ2-55-155 | −4 | − | −1 | Jλ2 | (+)5-155 | + | − | |
| 25 | Vλ3-21 | 0 | − | 0 | Jλ3 | + | ND | − |
| Vλ1-51 | −11 | CCC | 0 | Jλ3 | − | ND | − | |
| 26 | Vλ1-40 | −5 | AG | 0 | Jλ3 | + | ND | + |
| Vλ3-10 | −6 | GGGA | −4 | Jλ1 | − | ND | + | |
| 27 | Vλ3-21 | 0 | − | Jλ3 | + | + | − | |
| NI | Jλ35-154 | NI | NI | |||||
| 28 | Vλ2-18 | −3 | GGT | 0 | Jλ1 | + | + | + |
| Vλ4-60 | −2 | − | −1 | Jλ3 | + | + | + | |
| 29 | Vλ2-11 | −3 | CA | −3 | Jλ3 | + | + | + |
| Vλ1-44 | −2 | − | 0 | Jλ3 | + | + | − | |
| 30 | Vλ2-14 | −3 | GG | −3 | Jλ1 | + | + | + |
| NI | Jλ35-154 | NI | NI | |||||
| 31 | Vλ1-47 | −6 | − | −3 | Jλ3 | + | + | − |
| NI | Jλ25-154 | NI | NI | |||||
| 32 | Vλ3-21 | 0 | − | 0 | Jλ3 | + | + | + |
| Vλ1-47 | −2 | − | −1 | Jλ3 | + | + | + | |
| Patient . | Vλ . | Junctional region5-150 . | Jλ . | Frame5-151 . | RT-PCR5-152 . | Somatic mutations5-153 . | ||
|---|---|---|---|---|---|---|---|---|
| 4 | Vλ1-44 | 0 | G | −1 | Jλ3 | + | + | + |
| Vλ2-55-155 | −4 | TGG | −1 | Jλ3 | − | + | + | |
| 20 | Vλ1-40 | −4 | TCTA | 0 | Jλ3 | + | + | + |
| Vλ3-12 | 0 | − | −3 | Jλ3 | + | + | + | |
| 21 | Vλ3-21 | 0 | − | 0 | Jλ3 | + | ND | − |
| Vλ1-47 | 0 | A | 0 | Jλ3 | − | ND | − | |
| 22 | Vλ2-11 | −15 | 26 nt of Vλ2-14 | −7 | Jλ3 | + | + | + |
| Vλ1-47 | −4 | C | 0 | Jλ3 | + | + | + | |
| 23 | Vλ1-40 | −2 | CGAG | −2 | Jλ3 | + | + | − |
| Vλ3-21 | −2 | − | −2 | Jλ3 | − | + | − | |
| 24 | Vλ3-21 | 0 | CCCT | −1 | Jλ3 | + | + | − |
| Vλ2-55-155 | −4 | − | −1 | Jλ2 | (+)5-155 | + | − | |
| 25 | Vλ3-21 | 0 | − | 0 | Jλ3 | + | ND | − |
| Vλ1-51 | −11 | CCC | 0 | Jλ3 | − | ND | − | |
| 26 | Vλ1-40 | −5 | AG | 0 | Jλ3 | + | ND | + |
| Vλ3-10 | −6 | GGGA | −4 | Jλ1 | − | ND | + | |
| 27 | Vλ3-21 | 0 | − | Jλ3 | + | + | − | |
| NI | Jλ35-154 | NI | NI | |||||
| 28 | Vλ2-18 | −3 | GGT | 0 | Jλ1 | + | + | + |
| Vλ4-60 | −2 | − | −1 | Jλ3 | + | + | + | |
| 29 | Vλ2-11 | −3 | CA | −3 | Jλ3 | + | + | + |
| Vλ1-44 | −2 | − | 0 | Jλ3 | + | + | − | |
| 30 | Vλ2-14 | −3 | GG | −3 | Jλ1 | + | + | + |
| NI | Jλ35-154 | NI | NI | |||||
| 31 | Vλ1-47 | −6 | − | −3 | Jλ3 | + | + | − |
| NI | Jλ25-154 | NI | NI | |||||
| 32 | Vλ3-21 | 0 | − | 0 | Jλ3 | + | + | + |
| Vλ1-47 | −2 | − | −1 | Jλ3 | + | + | + | |
RT-PCR indicates reverse transcriptase–polymerase chain reaction.
The junctional region is described as the number of bases lost at the 3′ end of the V segment followed by randomly inserted nucleotides and the number of bases lost from the 5′ end of the J segment.
Frame of the junctional region: +, in-frame, without stop codons in the junctional region; −, out-of-frame.
ND, not determined; NI, not identified; +, present; −, absent.
+ indicates V segments with more than 2% somatic mutations; −, less than 2% somatic mutations; NI, not identified, because the rearrangement could not be amplified.
Vλ2-5 is a pseudogene and, therefore, not functional.
Based on Southern blot data.
In none of the 10 Igκ+ CBLs were 2 in-frame IGKrearrangements detected. In patient 3, no in-frame IGKrearrangement could be identified, although the CBL clearly showed Igκ positivity. In patient 11, only 1 rearrangement could be detected by PCR, although 2 rearrangements seemed to be present in Southern blot analysis. By karyotyping of the clonal cells of patient 11, only 1 intact chromosome 2 was detected, which might explain why only one Vκ-Jκ rearrangement could be amplified. Apparently, part of the other chromosome 2 with the IGKlocus was still present in the genome. Also, in patients 13 and 19, only one rearrangement could be identified, possibly due to primer mismatching or a chromosome aberration.
The RT-PCR results showed that out-of-frame IGKrearrangements can be transcribed (3 of 6 cases tested). In 2 of the 6 completely analyzed cases (patients 15 and 16), the V gene segment of the in-frame rearrangement was mutated.
Five of the Igλ+ CBLs with biallelic IGL gene rearrangements (patients 4, 21, 23, 25, and 26) had 1 in-frame and 1 out-of-frame IGL junctional region (Table 5). In 6 other Igλ+ CBLs (patients 20, 22, 24, 28, 29, and 32), 2 in-frame IGL rearrangements were found. The presence of both transcripts was demonstrated by RT-PCR in all 6 patients. All used Vλ segments were functional except for the second allele of patient 24. This Vλ2-5 segment is a pseudogene,35 implying that this patient has only one functional IGL gene rearrangement. In patients 27, 30, and 31, only one in-frame rearrangement could be identified by Vλ-Jλ PCR analysis. Somatic mutations were found in 7 of the 11 completely analyzed cases. Five of them (patients 20, 22, 28, 29, and 32) had 2 in-frame IGL gene rearrangements. Patients 20, 22, and 32 carried mutations on both alleles. The V regions of both IGL rearrangements of patient 26 were mutated, and the out-of-frame rearrangement contained a stop codon in the junctional region, which might be the result of a somatic mutation, although this cannot be proven.
Discussion
Ordered Ig light chain gene rearrangements
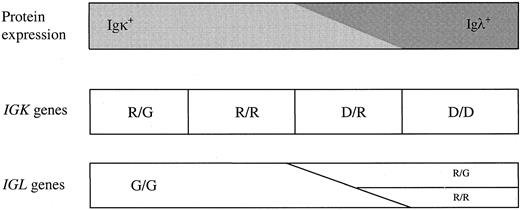
The configuration of the IGK and IGL genes was investigated in a series of 113 CBLs (53 Igκ+ and 60 Igλ+) to study the mechanism of allelic and isotypic exclusion of human Ig light chain genes. CBLs were chosen as a “single-cell” model because large clonal cell populations allow complete analysis of both alleles of the IGK andIGL genes. Our data confirm the hypothesis of the hierarchic order in human Ig light chain gene rearrangements: The gene rearrangement process starts at the IGK locus, followed byIGK deletion and subsequent IGL gene rearrangement (Table 2).12 Because not a single CBL had 1 or 2 rearranged IGL alleles with both IGK genes in germline configuration, the data completely fit with the ordered model. However, the ordered model was not stringent in all cases, because in some cases IGL gene rearrangements apparently had started before both IGK alleles were deleted. Curiously, 3 Igκ+ CBL cases had IGL rearrangements: 1 case in the group with biallelic IGK rearrangements and 2 cases in the group with 1 rearranged and 1 deleted IGK allele. One of the latter 2 cases even had biallelic IGL rearrangements. To include these cases in the ordered model, it must be slightly adapted, although the principle of the ordered Ig light chain rearrangement processes is retained: rearrangement of 1 IGKallele (R/G) → further IGK gene rearrangements (R/R) → 1 IGK allele deleted (D/R) and occasionally 1IGL allele rearranged (R/G) → both IGK alleles deleted (D/D) and 1 or 2 IGL rearrangements (R/G or R/R) (Figure 2). In other words, the original ordered model applies to IGK gene rearrangements, but IGL gene recombination processes seem to be less strictly controlled.
Schematic diagram of Ig light chain protein expression and Ig light chain gene rearrangements according to the ordered model.
Ig light chain gene rearrangements start with rearrangement of 1IGK allele (R/G) → further IGK gene rearrangements (R/R) → 1 IGK allele deleted (D/R) and occasionally 1 IGL allele rearranged (R/G) → bothIGK alleles deleted (D/D) and 1 or 2 IGLrearrangements (R/G or R/R). Because the ordered rearrangement process is not fully strict, IGK andIGL rearrangements might coexist (in 6% of Igκ+ cases and in 12% of Igλ+ cases of the presented CBL series). G indicates germline; R, rearrangement; D, deletion.
Schematic diagram of Ig light chain protein expression and Ig light chain gene rearrangements according to the ordered model.
Ig light chain gene rearrangements start with rearrangement of 1IGK allele (R/G) → further IGK gene rearrangements (R/R) → 1 IGK allele deleted (D/R) and occasionally 1 IGL allele rearranged (R/G) → bothIGK alleles deleted (D/D) and 1 or 2 IGLrearrangements (R/G or R/R). Because the ordered rearrangement process is not fully strict, IGK andIGL rearrangements might coexist (in 6% of Igκ+ cases and in 12% of Igλ+ cases of the presented CBL series). G indicates germline; R, rearrangement; D, deletion.
Regulation of isotypic exclusion in cases with 2 in-frame Ig light chain rearrangements of distinct isotype
In about 90% of the CBLs (103 of 113), either IGK orIGL rearrangements were present, indicating that ordered Ig light chain gene rearrangements ensured isotypic exclusion in these cases. Therefore, we conclude that monospecific Ig light chain expression is primarily determined by ordered rearrangement processes. Based on a computer simulation model of murine Ig light chain rearrangements, Mehr et al also suggested that allelic exclusion in B cells is maintained if recombination occurs in an ordered rather than a random process.36
Nevertheless, our data show that the ordered rearrangement process of Ig light chain genes is not absolute and that IGK andIGL rearrangements can coexist (10 of 113 cases). In such cases a different level of regulation of monospecific Ig light chain expression (ie, allelic and isotypic exclusion) should be expected. In 5 of these 10 CBLs, both an in-frame IGK and an in-frame IGL rearrangement were detected at the DNA level (Table 6). Four of these 5 cases could be analyzed by RT-PCR, and all 4 showed bitypic functional transcripts (ie, in-frame transcripts without stop codons because of somatic mutations). Therefore, there is no indication for regulation of isotypic exclusion at the level of transcription in these cases. Other possibilities include regulation at the level of translation, at the level of protein stability, or via preferential Ig light chain assembly with the Ig heavy chain. Although we did not find dual Igκ/Igλ chain expression in our CBL series (Figure 1), it has been described as occurring in normal and malignant human B lymphocytes.21-23 26
Summary of data of chronic B-cell leukemia with 2 complete immunoglobulin light chain rearrangements, as identified by reverse transcriptase–polymerase chain reaction analysis and sequencing
| . | IGK/IGK . | IGK/IGL . | IGL/IGL . | Total . |
|---|---|---|---|---|
| Patients (see Tables 3-5) | 12, 14-17 | 1-36-150, 5-10 | 46-151, 20-26, 28, 29, 32 | n = 26 |
| 2 functional rearrangements | 0/6 | 5/9 | 5/11 | 10/26 |
| Somatic mutations in cases with: | ||||
| 1 functional rearrangement | 2/6 | 1/4 | 2/6 | 5/16 |
| 2 functional rearrangements | − | 3/5 | 5/5 | 8/10 |
| Both functional rearrangements transcribed | − | 4/4 | 5/5 | 9/9 |
| Double Ig light chain protein expression | − | 0/5 | Not evaluable | 0/5 |
| . | IGK/IGK . | IGK/IGL . | IGL/IGL . | Total . |
|---|---|---|---|---|
| Patients (see Tables 3-5) | 12, 14-17 | 1-36-150, 5-10 | 46-151, 20-26, 28, 29, 32 | n = 26 |
| 2 functional rearrangements | 0/6 | 5/9 | 5/11 | 10/26 |
| Somatic mutations in cases with: | ||||
| 1 functional rearrangement | 2/6 | 1/4 | 2/6 | 5/16 |
| 2 functional rearrangements | − | 3/5 | 5/5 | 8/10 |
| Both functional rearrangements transcribed | − | 4/4 | 5/5 | 9/9 |
| Double Ig light chain protein expression | − | 0/5 | Not evaluable | 0/5 |
Ig indicates immunoglobulin; CBL, chronic B-cell leukemia.
Patient 3 was classified in the IGK/IGL category, because this Igκ+ CBL contained a functional IGLrearrangement.
Patient 4 (Igλ+ CBL) was classified in theIGL/IGL category, because both IGL rearrangements were identified, but the IGK rearrangement was not found.
Allelic exclusion in cases with biallelic IGK or biallelic IGL rearrangements
The mechanism of allelic exclusion was further studied using 24 CBLs with biallelic IGK or biallelic IGLrearrangements. The allelic exclusion of biallelic IGKrearrangements was solely regulated at the DNA level because only one functional rearrangement was present in all 6 evaluable cases (Table6). Theoretically, one third of the rearrangements on the secondIGK allele in Igκ+ CBLs could be in-frame. However, the murine experiments by Mostoslavsky et al showed that undermethylation of an IGK allele is required for and precedes a rearrangement.17 Moreover, they demonstrated that IGK gene demethylation takes place preferentially on only 1 allele in each cell, resulting in differential accessibility of the 2 IGK alleles for the recombinase system. If no productive rearrangement is obtained, a second rearrangement of the same allele involving an upstream Vκ and a downstream Jκ gene segment can take place.17Alternatively, the second allele is demethylated and rearranged. On top of the methylation-induced differential accessibility of IGKalleles, the feedback mechanism of down-regulation of the V(D)J recombinase system probably plays a role in the maintenance of allelic exclusion. This is in line with the ordered rearrangement model of Ig light chain genes and fits with the IGK gene data of our CBL series.
In 11 Igλ+ CBLs, 2 Vλ-Jλrearrangements could be identified. Six of them had 2 in-frame junctions (without stop codons), of which 5 were functional, and in one case a pseudo-Vλ segment was used. So, in 5 cases 2 Igλ protein chains may be expressed, but it was not possible to make a distinction between the expression of 1 or 2 Igλ chains, because there are no Vλ family–specific antibodies available. These 5 CBLs were all somatically mutated on either 1 allele (2 cases) or both alleles.
Possible mechanism explaining regulation of monospecific Ig light chain expression
The start of Ig light chain gene recombination seems to be strictly regulated by ordered accessibility of the IGK locus ensuring complete allelic exclusion at the DNA level. Igκ+ CBL with 2 functionally rearranged IGKgenes did not occur in our CBL series, suggesting that dual Igκ/Igκ-expressing B cells do not occur or are rare. However, whenIGL rearrangements were involved, 2 in-frame (functional) rearrangements and transcripts (IGK/IGL orIGL/IGL) were occasionally detected (Table 6), implying the possibility of dual Ig light chain expression (Igκ/Igλ or Igλ/Igλ).
Several mechanisms might operate to regulate ordered recombination, including demethylation of one allele, remodeling of chromatin structure, or selective accessibility of recombination machinery through differential presence of transcription factors (such as Rel/nuclear factor-κB or E12/E47 bHLH factors).37Demethylation indeed appeared to be necessary for rearrangement of Ig and TCR genes. In the mouse, monoallelic demethylation has been shown to occur at the IGK locus.17 In contrast toIGK genes, the ordered demethylation process might be less strict for the human IGL genes.
The occurrence of somatic mutations and subsequent receptor editing might have contributed to the presence of 2 functional Ig light chain rearrangements in our study. B-CLL can arise either from pre– or post–germinal center B cells.38 In the latter case the V regions carry somatic mutations (about 50% of cases).38Theoretically, the somatic mutation process can result in unfavorable mutations leading to receptor editing via secondary rearrangements and light chain replacement.39 40 It is not yet clear whether the secondary rearrangements for receptor editing follow an ordered pattern comparable to primary rearrangements. In cases with 2 functional rearrangements, one should expect somatic mutations in the in-frame rearrangement that is not expressed, whereas the expressed in-frame rearrangement can be mutated depending on whether the B cell underwent a second germinal center reaction. Indeed, 8 of 10 CBLs (80%) with 2 functional IGK/IGL orIGL/IGL rearrangements contained somatic mutations, in contrast to 5 of 16 cases (about 30%) of CBL with 2 complete Ig light chain gene rearrangements, one functional and one nonfunctional (Table 6). Nevertheless, this study indicates that isotypic and allelic exclusion is regulated by ordered rearrangement of Ig light chain genes in about 90% of cases. Even in cases with 2 functional Ig light chain transcripts, monospecific Ig expression can still be maintained, probably via mechanisms at the protein level, such as differential protein stability and preferential pairing of Ig chains.
Acknowledgment
The authors gratefully acknowledge Dr F. J. T. Staal for critical reading of the manuscript.
Supported by the Foundation “Vereniging Trustfonds Erasmus Universiteit Rotterdam” in The Netherlands.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
J. J. M. van Dongen, Dept of Immunology, Erasmus University Rotterdam, Dr Molewaterplein 50, 3015 GE Rotterdam, The Netherlands.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal