Abstract
Hepatitis C virus (HCV)–associated B cell lymphomas were previously shown to express a restricted repertoire of immunoglobulin VH and VL genes, VH1-69 and VκA27, respectively. Although this suggests a role for antigen selection in the pathogenesis of these lymphomas, the driving antigen involved in the clonal expansion has not been identified. B cell response to a viral antigen, the HCV envelope glycoprotein 2 (E2), was analyzed in an asymptomatic HCV-infected patient. Single B cells, immortalized as hybridomas and selected for binding E2, were analyzed for their V gene usage. Sequences of these V region genes demonstrated that each hybridoma expressed unique VH and VLgenes. Remarkably, these anti-E2 hybridomas preferentially used the VH1-69 gene. Analysis of replacement to silent mutation ratios indicated that the genes underwent somatic mutation and antigenic selection. In a separate report, human anti-E2 antibodies were also shown to express the same VH gene. These data strengthen the hypothesis that the HCV-associated lymphomas are derived from clonally expanded B cells stimulated by HCV.
Introduction
Hepatitis C virus (HCV) is estimated to have infected more than 100 million people globally.1The study of the natural history of this virus is curtailed because the virus does not infect small laboratory animals, nor can it be propagated in vitro. It is a major cause of chronic liver diseases in which 70% to 80% of infected patients become chronic carriers.2 The liver manifestations of HCV infection include chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma.3 Although HCV is a hepatotropic virus, the HCV genome and its replicative intermediates have also been detected in peripheral blood mononuclear cells and in lymphoid tissues of chronically infected patients.4-6 Furthermore, HCV has been recognized as the major causative agent of mixed cryoglobulinemia (MC)—70% to 100% of patients with MC are infected by HCV.7-10 MC is a disorder in which monoclonal or polyclonal B cell proliferation is triggered by HCV infection.11-14 It has been proposed that the benign lymphoproliferation associated with MC could lead to frank B cell neoplastic disorders. Indeed, a study from Italy in which 200 patients with MC were followed longitudinally showed that B cell non-Hodgkin lymphoma (NHL) developed in 14 patients.15 However, it should be noted that an association between HCV and B cell NHL has been reported only in certain geographic regions, and the cause of the varied incidence is not yet clear.
Immunochemically, MCs are classified as type II or type III, respectively, on the basis of the presence of monoclonal or polyclonal IgM with rheumatoid factors (RF). The monoclonal IgM was shown to be encoded by a restricted set of variable (V) region genes, specifically the VH1-69 (also known as 51p1) and the VκA27 (also known as κv325) germline genes.16-18 Interestingly, most patients with HCV-associated lymphomas express the same set of genes (VH1-69 and VκA27). Furthermore, analysis of these V region sequences revealed that they undergo somatic mutation, presumably during affinity maturation.19 It has, therefore, been proposed that these HCV-associated lymphomas may originate in B cells responding to a common antigen. Nevertheless, such a common antigen has not been identified.
We tested whether an immune response to a specific viral antigen is restricted in HCV infection. To this end we analyzed 10 human B cell hybridomas derived from the peripheral B cells of an asymptomatic HCV-infected patient. These hybridomas were selected by reactivity with the viral E2 glycoprotein.20 We demonstrate that the VH genes used by these anti-E2 B cells were highly restricted. Moreover, the same VH gene, VH1-69, seen in HCV-associated lymphomas and in MC is the one used in the anti-E2 immune response. An independent study that used a combinatorial Fab library approach and selected anti-E2 antibodies from an HCV-infected patient showed these antibodies to express the same gene bias.21 These results tie the HCV-associated lymphoproliferative disorders to the immune response to HCV antigens.
Materials and methods
Anti-HCV E2 human hybridomas
Peripheral B cells were isolated from an asymptomatic patient who had a high serum neutralization binding titer to the HCV 1b viral genotype. Peripheral blood B cells from this patient were activated by Epstein-Barr virus and then electrofused with heteromyeloma cells to produce human hybridomas as described.20 The hybridomas were screened for binding HCV E1/E2 proteins produced by recombinant baculovirus in insect cells. Ten hybridomas were selected, all of which produced an antibody that reacted with the HCV E2 envelope glycoprotein.20
RT-PCR and V region sequencing
Total cellular RNA was isolated using the RNeasy mini kit (QIAGEN, Valencia, CA). RNA (4 μg) was reverse transcribed, and V region genes were amplified using 5′ family-specific V leader primers and 3′ J region primers, as described previously.22Polymerase chain reaction (PCR) products were electrophoresed, size selected, and sequenced by an automated DNA sequencer (373; Applied Biosystems, Foster City, CA) using the ABI Prism Big Dye Terminator Kit (Perkin Elmer, Foster City, CA). The same primers were used for the original PCR reactions and for sequencing. To control for potential PCR error, all samples were evaluated through 2 independent rounds of PCR reaction and sequencing.
Analysis of mutations
Sequences were analyzed using MacVector and AssemblyLign (Oxford Molecular Group, Campbell, CA) and aligned with germline sequences using VBASE database and DNA plot on the Internet.23Somatic mutations were determined by comparison to germline genes with the highest homology (Table 1). The probability (P) of excess or scarcity of replacement (R) mutations in CDR and FR regions was calculated by a multinomial distribution model.24
Distribution of V gene mutations in anti-HCV E2 hybridomas
| Hybridomas . | VH Germline . | Homology (%) . | R:S mutations . | Scarcity of R mutation in FRs . | Excess of R mutation in CDRs . | |
|---|---|---|---|---|---|---|
| FRs . | CDRs . | |||||
| CBH4B | VH1-69 | 90.8 | 8:8 | 9:1 | P = .00300* | P = .00925* |
| CBH4D | VH1-69 | 93.3 | 8:3 | 3:5 | P = .08676 | P = .00011* |
| CBH4G | VH1-69 | 92.9 | 5:5 | 7:3 | P = .00176* | P = .02163* |
| CBH5 | VH1-69 | 88.3 | 9:11 | 11:2 | P = .00022* | P = .00247* |
| CBH7 | VH1-69 | 90.8 | 9:10 | 6:1 | P = .00925* | P = .00019* |
| CBH8E | VH1-69 | 96.4 | 3:3 | 5:2 | P = .00691* | P = .08563 |
| CBH11 | VH1-e | 91.5 | 8:5 | 4:7 | P = .00864* | P = .00002* |
| CBH2 | VH5-51 | 94.3 | 6:4 | 6:0 | P = .04465* | P = .04465* |
| CBH8C | VH4-59 | 92.8 | 1:7 | 9:3 | P = .00001* | P = .15227 |
| CBH17 | VH3-73 | 88.8 | 11:10 | 10:1 | P = .00592* | P = .00204* |
| Hybridomas . | VH Germline . | Homology (%) . | R:S mutations . | Scarcity of R mutation in FRs . | Excess of R mutation in CDRs . | |
|---|---|---|---|---|---|---|
| FRs . | CDRs . | |||||
| CBH4B | VH1-69 | 90.8 | 8:8 | 9:1 | P = .00300* | P = .00925* |
| CBH4D | VH1-69 | 93.3 | 8:3 | 3:5 | P = .08676 | P = .00011* |
| CBH4G | VH1-69 | 92.9 | 5:5 | 7:3 | P = .00176* | P = .02163* |
| CBH5 | VH1-69 | 88.3 | 9:11 | 11:2 | P = .00022* | P = .00247* |
| CBH7 | VH1-69 | 90.8 | 9:10 | 6:1 | P = .00925* | P = .00019* |
| CBH8E | VH1-69 | 96.4 | 3:3 | 5:2 | P = .00691* | P = .08563 |
| CBH11 | VH1-e | 91.5 | 8:5 | 4:7 | P = .00864* | P = .00002* |
| CBH2 | VH5-51 | 94.3 | 6:4 | 6:0 | P = .04465* | P = .04465* |
| CBH8C | VH4-59 | 92.8 | 1:7 | 9:3 | P = .00001* | P = .15227 |
| CBH17 | VH3-73 | 88.8 | 11:10 | 10:1 | P = .00592* | P = .00204* |
| Hybridomas . | VL Germline . | Homology (%) . | R:S mutations . | Scarcity of R mutation in FRs . | Excess of R mutation in CDRs . | |
|---|---|---|---|---|---|---|
| FRs . | CDRs . | |||||
| CBH4B | Vk3-A27 | 95.4 | 3:3 | 4:2 | P = .01085* | P = .07773 |
| CBH4D | Vλ2-2a2 | 95.6 | 6:3 | 3:0 | P = .31691 | P = .24510 |
| CBH4G | Vk1-A20 | 98.5 | 0:0 | 4:0 | P = .01350* | P = .00026* |
| CBH5 | Vk1-L12 | 94.3 | 1:7 | 5:2 | P = .00001* | P = .05436 |
| CBH7 | Vk1-O12 | 96.2 | 6:2 | 1:1 | P = .89335 | P = .97264 |
| CBH8E | Vk1-O12 | 97.3 | 0:4 | 3:0 | P = .01453* | P = .42831 |
| CBH11 | Vk1-L12 | 96.2 | 1:6 | 5:0 | P = .00017* | P = .02039* |
| CBH2 | Vk3-A27 | 96.2 | 1:2 | 5:2 | P = .00110* | P = .00848* |
| CBH8C | Vk3-L6 | 98.1 | 1:1 | 3:0 | P = .05049 | P = .01699* |
| CBH17 | Vλ3-3h | 90.9 | 10:4 | 8:2 | P = .05545 | P = .01976* |
| Hybridomas . | VL Germline . | Homology (%) . | R:S mutations . | Scarcity of R mutation in FRs . | Excess of R mutation in CDRs . | |
|---|---|---|---|---|---|---|
| FRs . | CDRs . | |||||
| CBH4B | Vk3-A27 | 95.4 | 3:3 | 4:2 | P = .01085* | P = .07773 |
| CBH4D | Vλ2-2a2 | 95.6 | 6:3 | 3:0 | P = .31691 | P = .24510 |
| CBH4G | Vk1-A20 | 98.5 | 0:0 | 4:0 | P = .01350* | P = .00026* |
| CBH5 | Vk1-L12 | 94.3 | 1:7 | 5:2 | P = .00001* | P = .05436 |
| CBH7 | Vk1-O12 | 96.2 | 6:2 | 1:1 | P = .89335 | P = .97264 |
| CBH8E | Vk1-O12 | 97.3 | 0:4 | 3:0 | P = .01453* | P = .42831 |
| CBH11 | Vk1-L12 | 96.2 | 1:6 | 5:0 | P = .00017* | P = .02039* |
| CBH2 | Vk3-A27 | 96.2 | 1:2 | 5:2 | P = .00110* | P = .00848* |
| CBH8C | Vk3-L6 | 98.1 | 1:1 | 3:0 | P = .05049 | P = .01699* |
| CBH17 | Vλ3-3h | 90.9 | 10:4 | 8:2 | P = .05545 | P = .01976* |
The probability (P) that excess or scarcity of replacement (R) mutations in CDRs and in FRs resulted only by chance was calculated using the multinominal distribution model.24
Significant.
Results
Anti-HCV E2 hybridomas use restricted VH genes
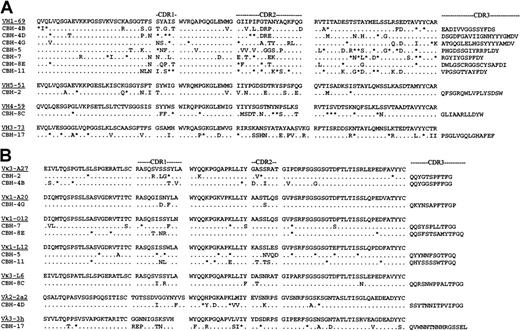
Individual B cells from a patient responding to HCV infection were isolated in the form of heterohybridomas. They were selected for reactivity with the viral envelope protein E2.20 Immunoglobulins secreted by 9 of these hybridomas recognized conformational, nondenatured epitopes within E2. The reactive E2 epitope of 5 immunoglobulins prevented binding of E2 to human CD81, the putative cellular receptor for HCV.25These hybridomas provide a unique opportunity to study a human B cell response to a specific HCV antigen. They enabled us to test whether these responding B cells were derived randomly from the V gene repertoire expressed in normal peripheral blood lymphocytes26-29 or whether they exhibited a biased V gene usage, as seen in the clonal populations in HCV-infected patients with B cell proliferative disorders.16,17,19 To this end, we sequenced V region genes expressed by the hybridomas. This analysis revealed that the VH gene usage was restricted (Figure1A). Six anti-HCV E2 hybridomas used a VH gene that matched best to germline VH1-69 (also known as 51p1). One additional VH1 hybridoma matched the VH1-e gene, which is highly homologous to VH1-69 because it has only one amino acid difference in FR3 (VH1-e Lys73 vs VH1-69 Glu73) and is considered to be VH1-69–related.30 All 7 hybridomas expressing these VH1 family genes were derived from different B cells because they had unique CDR3 regions. The preferential expression of VH1-69 in anti-HCV E2 hybridomas in 6 (and possibly 7) of 10 patients is exceptionally high. The VH genes encoding the remaining 3 heavy chains were most closely related to VH5-1, VH4-59, and VH3-73 (Figure1A). All VH genes showed numerous nucleotide differences from their corresponding germline genes (data not shown).
Deduced amino acid sequences of anti-HCV E2 hybridoma-region genes.
(A) VH genes. (B) VL genes. Amino acid sequences of the most homologous germline VH genes are shown above the cloned hybridoma V and VK region sequences. Amino acid replacement mutations are shown as uppercase letters, and silent mutations are indicated by asterisks.
Deduced amino acid sequences of anti-HCV E2 hybridoma-region genes.
(A) VH genes. (B) VL genes. Amino acid sequences of the most homologous germline VH genes are shown above the cloned hybridoma V and VK region sequences. Amino acid replacement mutations are shown as uppercase letters, and silent mutations are indicated by asterisks.
VL genes of anti-HCV E2 hybridomas
The isotype of the secreted light chain was first determined by enzyme-linked immunosorbent assay (data not shown) and confirmed by reverse transcription (RT)–PCR followed by sequencing (Figure 1B). The usage of VL genes was less restricted than that seen for the VH genes. Only 2 hybridomas used the VκA27 light chain, which has been associated with HCV lymphoproliferative disorders. The remaining 8 hybridomas used 6 different light chains. Interestingly, somatic mutations were observed in all VLsequences, but to a lesser extent than seen for the VHsequences. Overall, only one of the hybridomas, CBH-4B, used the canonical VH1-69/VκA27 combination seen in MC and in HCV-associated lymphomas. It is unclear why the VH, but not the light-chain V, genes used by these hybridomas resemble the biased V gene usage seen in B cell lymphoproliferative diseases. One likely explanation is that the selection of the hybridomas in vitro was based on reactivity with a recombinant protein, which might have recognized only a subset of B cells induced to proliferate in vivo by the virus.
Evidence of antigen selection
To address the role of antigen selection in the patient's immune response to the E2 protein, we analyzed the expressed V region genes for the distribution of replacement and silent mutations in CDR and FR regions (Table 1) using a multinomial distribution model.24 Calculated are the probabilities for obtaining replacement to silent mutations (R:S) in CDR and in FR regions by chance alone. A significant excess of R mutations was observed in CDR regions for 8 of 10 VH and 5 of 10 VLsequences, implying positive selection for antigen binding. A significant scarcity of R mutations was seen in FR regions in 9 of 10 VH sequences and in 6 of 10 VL sequences, indicating a negative selection pressure against change in the functional antibody framework structures. Thus, these hybridomas show strong evidence of antigenic selection.
Similar biased V gene usage by independent human anti-E2 antibodies
A recent study, aimed at isolating the protective antibodies against the HCV E2 protein, used a combinatorial library approach and RNA from another HCV-infected patient. Seven combinatorial Fab fragments, reacting with conformational epitopes of the E2 envelope glycoprotein of hepatitis C virus, were reported.21 We analyzed the V gene usage of these Fab fragments and found them to exhibit biased V gene usage. Three of them expressed the VH1-69 gene (accession numbers AJ236548, AJ236544,AJ236543), and a fourth one expressed the V1-e gene (AJ236542). Preferential usage was also found for the Vκ genes. The VκA27 gene was seen in 4 of the antibodies (AJ236555, AJ236552, AJ236551,AJ236549). Interestingly, 2 used the canonical VH1-69–VκA27 combination seen in MC and in lymphomas, and a third used the almost identical VH1-e–VκA27 combination.
Discussion
Approximately 1.7% of peripheral blood B cells express the VH1-69 gene,31 as expected for a random use of the total repertoire of functional VH gene regions. As discussed earlier, biased use of this gene has been seen in MC and in HCV-associated B cell lymphomas. Preferential use of this gene has also been seen in 10% to 20% of patients with CD5+ B cell chronic lymphocytic leukemia.32-34 In addition, biased use of VH1-69 has been demonstrated for salivary gland mucosa-associated lymphoid tissue (MALT) lymphomas in which 61% of the patients express the gene.35 Interestingly, MALT lymphomas that develop in the stomach do not show this bias. Taken together, these observations suggest immune stimulation and selection by an antigen that may be located only in the salivary gland for those lymphomas that arise in the salivary gland. The finding that the length of CDR3 is restricted in salivary gland MALT lymphomas, but not in other MALT lymphomas, strengthens this hypothesis.35
Restricted V gene usage combined with restricted CDR3 length has been seen in inbred mice strains responding to experimental vaccination protocols. B cells derived from such immunized mice and selected by reactivity with specific haptens such as 4-hydroxy-3-nitrophenylacetyl (NP),36 2-phenyloxazolone,37 and p-azophenylarsonate38 exhibit a V gene restriction bias. Fewer such studies have been reported in humans. However, studies by Carroll et al39 in volunteers who were vaccinated with the Haemophilus influenzae type b capsular polysaccharide (Hib PS) antigen show a restricted V gene response to this antigen. They reveal a markedly restricted immune response to the Hib PS antigen—approximately half the VH gene rearrangements are of the VH3b subfamily. Moreover, these restricted genes have extremely restricted CDR3 regions in which unrelated patients have identical V(D)J joints. The VL gene response to the Hib PS antigen was less restricted, as seen in the anti-E2 response in the current analysis, suggesting that in both cases the VH gene segments are the ones that play a more dominant role in antigen binding.
In summary, HCV-associated type II mixed cryoglobulinemia expresses immunoglobulin encoded mostly by germline VH1-69 and VκA27 genes.10,16,17 This same biased use of the VH1-69/VκA27 combination was found in HCV-associated lymphomas, indicating that they most likely represent the malignant counterpart of type II MC.15 Finally, our current study and the recent study by Allander et al21 support this hypothesis by demonstrating that human anti-HCV E2 monoclonal antibodies are preferentially encoded by the same V genes. These studies implicate the specific immune response to HCV E2 glycoprotein in the pathogenesis of B cell proliferative disorders and as a precursor to HCV-associated lymphomas. However, to prove directly that an HCV antigen plays a role in lymphomagenesis, it will be necessary to rescue immunoglobulins from HCV-associated lymphomas and to test their ability to bind HCV proteins.
Acknowledgments
We thank Dr Ronald Levy for his continuous interest in this project, Dr Izidore Lossos for guidance in the analysis of the distribution of somatic mutations, and Drs Ronald Levy, Izidore Lossos, and Elizabeth Quinn for reviewing the manuscript.
Supported by National Institutes of Health grants CA34233 (S.L.) and DA06596 and HL33811 (S.K.H.F.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Shoshana Levy, Department of Medicine, Division of Oncology, M-207, Stanford University Medical Center, Stanford, CA 94305-5115.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal