Abstract
A well-known complication of factor VIII replacement therapy in patients with hemophilia A is the development of inhibitory antibodies. Several studies have demonstrated the presence of a binding site for factor VIII inhibitors in the A3 domain. Six different human monoclonal single-chain variable domain antibody fragments (scFv) directed toward the A3-C1 domains of factor VIII have been isolated, using phage display technology. Sequence analysis revealed that the VH domains of 2 scFv were encoded by germline gene segments from the VH1 gene family and 4 by germline gene segments belonging to the VH3 gene family. Epitope mapping of the scFv was performed, using a series of hybrid factor VIII/factor V light chain fragments. This analysis revealed that 5 of 6 scFv were directed against a region encompassing amino acid sequence Q1778-D1840 in the A3 domain, a previously identified binding site for factor VIII inhibitors. Only 2 of 5 scFv directed against amino acid sequence Q1778-D1840 inhibited the procoagulant activity of factor VIII. Our results define the properties of human antibodies directed against region Q1778-D1840 in the A3 domain. Binding of one, noninhibitory scFv was independent of the region Q1778-D1840, suggesting the presence of an additional binding site for anti–factor VIII antibodies in the A3-C1 domains of factor VIII.
Introduction
Hemophilia A is an X-linked bleeding disorder that is associated with a functional absence of factor VIII. Upon initiation of factor VIII replacement therapy, approximately 25% of patients with severe hemophilia A develop antibodies that neutralize the procoagulant activity of factor VIII.1 On the basis of internal sequence homology, factor VIII can be defined by the domain structure A1-a1-A2-a2-B-a3-A3-C1-C2.2,3The majority of factor VIII inhibitors is directed toward epitopes within the A2, C2, and A3-C1 domains of factor VIII.4Within the A2 domain, the region R484-I508 has been identified as a major binding site for factor VIII inhibitors.5 In the C2 domain, amino acid residues E2181-V2243 and V2248-S2312 constitute binding sites for factor VIII inhibitors.6 7
The presence of an inhibitor binding site in the A3-C1 domains has been suggested by inhibitor neutralization experiments, using factor VIII light chain (a3-A3-C1-C2 domains) and isolated C2 domain.4,6 Epitope mapping with the use of in vitro synthesized factor VIII fragments defined the epitope in the A3 domain to amino acid residues Q1778-M1823.8 Furthermore, a synthetic peptide corresponding to residues K1804-V1819 competed for binding of factor VIII inhibitors to the light chain.9This region harbors amino acid residues E1811-K1818, which comprise a binding site for factor IXa.10 It was demonstrated that anti-A3 inhibitor immunoglobulin G (IgG) prevents factor IXa from binding to the factor VIII light chain.8,9 Recently, a region involved in the binding of factor VIII inhibitors has been localized to the acidic region a3, adjacent to the A3 domain.11 Replacement of the a3 region, comprising amino acid residues E1649-R1689 of human factor VIII, for the corresponding porcine sequence yielded a functional factor VIII molecule that was less antigenic to factor VIII inhibitors. The binding site for von Willebrand factor has been localized to the a3region, which is released following cleavage at position R1689 by thrombin.12-14 Inhibitory antibodies directed toward this region may prevent activation of factor VIII when complexed to von Willebrand factor.
Assembly of antibody genes occurs by random rearrangement of different gene segments.15 The primary variable region of the heavy chain (VH) is generated by random recombination of variable (VH), diversity (D), and joining (JH) gene segments that are present in multiple copies in the germline DNA. At the moment, 123 different VH gene segments have been identified, which can be classified into 7 different families (VH1 to VH7), based on nucleotide sequence homology.16,17 Two independent studies determined that only 3917 or 5116 of the VH gene segments are functional, whereas the remaining genes are mainly pseudogenes that are nonfunctional because of point mutations or truncations. For assembly of the primary variable light chain region (VL), a VL gene segment, either of κ or λ origin, is combined with a JL gene segment. Lack of the D gene segment in VL gene recombination results in a VL repertoire that displays limited diversity compared to the VH repertoire. As a consequence of the far greater diversity of the VH domain, it is suggested that the heavy chain provides the major contribution to antigen recognition and specificity. Besides combinatorial diversity, addition and deletion of nucleotides at the sites of V(D)J gene junction gives rise to diversity, particularly in the third complementarity-determining region (CDR3) of both VH and VL domains.
Recently, we have used phage display technology to isolate anti-A2 and anti-C2 antibodies from the immunoglobulin repertoires of patients with an inhibitor to factor VIII.18,19 Analysis of human anti-C2 antibodies revealed that their immunoglobulin heavy-chain variable (VH) domains were exclusively encoded by VH gene segments derived from the VH1 gene family. These findings suggest that only a subset of VHgene segments is used to generate human anti-C2 antibodies. Molecular analysis of anti-A2 antibodies revealed that a single VHdomain encoded by gene segment DP-10 (1-69) of the VH1 gene family is involved in assembly of a human antibody that binds to region R484-I508 in the A2 domain. Furthermore, an additional human antibody composed of a VH domain encoded by gene segment DP-47 (3-23) of the VH3 gene family, bound to residues D712-A736 in the acidic region a2, a previously unidentified binding site for anti–factor VIII antibodies.19
In the present study, phage display technology was used to further define the molecular characteristics of human antibodies reactive with the A3-C1 domains of factor VIII. The majority of the isolated antibodies was directed toward amino acid sequence Q1778-D1840 of factor VIII. Our results indicate that different residues in this region are involved in the binding of the various scFv. One of the isolated scFv did not react with this region, suggesting the presence of an additional binding site for anti–factor VIII antibodies in the A3-C1 domains of factor VIII.
Materials and methods
Materials
DNA modification enzymes were purchased from Life Technologies (Breda, The Netherlands) and New England Biolabs (Beverly, MA). The Baculovirus expression system (Pharmingen, San Diego, CA) was used to produce recombinant factor VIII fragments in insect cells as described previously.20 Insect-XPRESS medium was purchased from BioWhittaker (Alkmaar, The Netherlands). Oligonucleotides, protein G Sepharose-4FF, and protein A Sepharose CL-4B were purchased from Pharmacia-LKB (Woerden, The Netherlands). Plasma-derived factor VIII light chain was purified as described previously.21Thrombin-cleaved light chain was prepared as described previously.22 Monoclonal antibodies (mAbs) CLB-CAg A, 12, and 117 have been characterized previously8-10; mAbs ESH4 and ESH8 were purchased from American Diagnostica (Greenwich, CT).
Factor VIII assays
Factor VIII activity was measured by a one-stage clotting assay.23 Factor VIII inhibitor titers were measured using the Bethesda assay.24 Immunoprecipitation of metabolically labeled factor VIII fragments by anti–factor VIII IgG was performed essentially as described previously.20 Neutralization of factor VIII inhibitor activity by recombinant factor VIII fragments was performed as described.8
Construction of hybrid FVIII/FV light-chain hybrids
A series of hybrid factor VIII/factor V light-chain expression vectors were constructed by overlap extension mutagenesis, using vector pCLB-GP67-80K,20 encoding the light chain of factor VIII (amino acid residues E1649-Y2332), and factor V complementary DNA (cDNA) as templates. Three hybrid constructs were made in which amino acids residues R1803-K1818 (HV1803-1818), Q1778-H1821 (HV1778-1821), or K1804-D1840 (HV1804-1840) were replaced for the corresponding sequence of factor V (Figure 2A). Expression of metabolically labeled factor VIII light-chain hybrids in insect cells was performed as described.20
Phage display library construction and selection
In this study, peripheral blood mononuclear cells from a previously described inhibitor patient8 served as the source of RNA for the construction of a phage display library essentially as described previously.18 The patient's IgG4-specific VH gene repertoire was amplified, using appropriate VH gene family-based VH back primers and an IgG4 constant region primer as described.18Subsequently, each VH repertoire was cloned separately in the phagemid pHEN-1-VLrep,25 which contained an immunoglobulin light-chain variable (VL) gene repertoire of nonimmune origin.
Phage were selected for binding to the factor VIII light chain essentially as described previously.18 In this study, microtiter wells (Dynatech, Plockingen, Germany) were immobilized with antibody CLB-CAg 117, which is directed toward the C2 domain of factor VIII.8 Wells were blocked with Tris-buffered saline (TBS; 150 mM NaCl, 50 mM Tris, pH 7.4), 3% (wt/vol) human serum albumin (HSA) for 2 hours at 37°C. Phage in TBS, 3% HSA, and 0.5% (vol/vol) Tween-20 were preincubated for 2 hours at room temperature in CLB-CAg 117-coated wells to reduce nonspecific binding. Subsequently, CLB-CAg 117-coated wells were incubated for 2 hours at 37°C with 25 nM plasma-derived factor VIII light chain in 1 M NaCl, 50 mM Tris, pH 7.4, 2% HSA, and blocked with HSA as outlined above. For specific binding of phage directed toward factor VIII light chain, these wells were incubated for 2 hours at room temperature with nonbound phage transferred from the preincubations. After washing with TBS/0.1% (vol/vol), Tween-20, and TBS (both 20 times), bound phage were eluted with 100 mM triethylamine and rescued, using Escherichia coli TG1.26 The selection procedure was performed for a total of 3 rounds.
Screening and sequencing of selected clones
After the third round of selection, phage derived from 15 single infected colonies were tested for reactivity with factor VIII. The domain specificity was evaluated by testing reactivity of phage with factor VIII light chain and recombinant C2 fragment immobilized via antibody CLB-CAg 117.18 Bound phage was detected by anti-M13 antibody peroxidase conjugate (Pharmacia-LKB, Woerden, The Netherlands).27 Sequences encoding the VH and VL domains were determined using the BigDye Terminator sequencing kit on an Applied Biosystems 377XL automated DNA sequencer (Foster City, CA). Genes were aligned with a database of germline V genes as compiled in the V-BASE sequence directory.28
Characterization of scFv
For production of single-chain variable domain antibody fragments (scFv), V genes were subcloned into the vector pUC119-Sfi/Not-His6,29 which introduces a carboxy-terminal 6 × histidine tag in the scFv. Expression and purification of scFv was performed as described previously.30 Purified scFv were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and protein concentrations were measured spectrophotometrically at A280. Reactivity of scFv with hybrid factor VIII/factor V light-chain fragments and C2 domain was evaluated by immunoprecipitation analysis. Immunoprecipitation of metabolically labeled factor VIII fragments by scFv/Ni-NTA agarose (Qiagen, Hilden, Germany) complexes was performed as described previously.18
Reactivity of scFv with the region comprising amino acid residues E1649-R1689 of the factor VIII light chain was determined as follows. Factor VIII light chain and thrombin-cleaved factor VIII light chain (0.4 nM) were captured on antibody CLB-CAg 117–coated wells as described above. Wells were incubated with scFv (0-100 nM) in TBS/2% HSA and 0.1% (vol/vol) Tween-20 for 2 hours at room temperature. Bound scFv were detected using peroxidase-labeled antibody 9E10. Antibody 9E10 is directed toward the scFv's carboxy-terminal myc-tag.
Surface plasmon resonance
The kinetics of the interaction between scFv and factor VIII light chain were determined by surface plasmon resonance, using a BIAcore2000 biosensor system (Biacore AB, Uppsala, Sweden). ScFv were covalently coupled at the indicated densities to the dextran surface of an activated CM5-sensor chip, using the amine-coupling kit according to the manufacturer's instructions (Biacore AB). A control flow-channel was routinely activated and blocked in the absence of protein. Binding to coated channels was corrected for binding to noncoated channels (< 5% of binding to coated channels). Binding (association) of ligand was assessed in 200 mM NaCl, 2 mM CaCl2, 0.05% (v/v) Tween 20, and 20 mM HEPES, pH 7.4 at 25°C for 2 minutes at a flow rate of 20 μL/min. Dissociation was allowed for 2 minutes in the same buffer flow. Association (kon) and dissociation (koff) rate constants of data sets were determined using the BIAevaluation software. Binding data were corrected for bulk refractive index changes and fitted according to a one-site model. Equilibrium dissociation constants (Kd) were calculated askoff/kon.
Results
Characterization of anti–factor VIII antibodies in patient's plasma
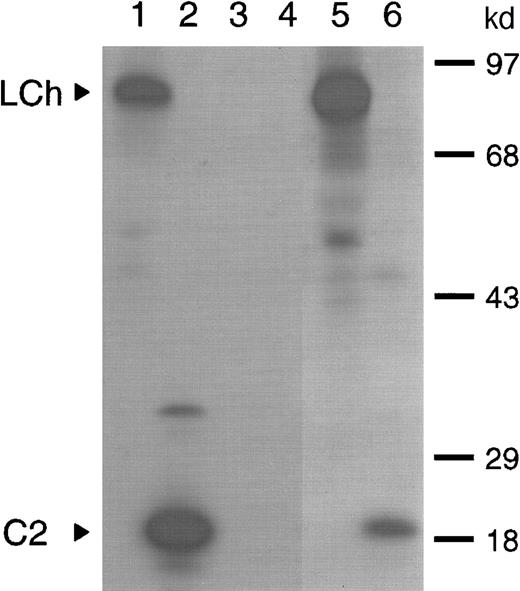
Previously, we have shown that amino acid residues Q1778-M1823 in the A3 domain constitute a binding site for factor VIII inhibitors. This initial characterization of factor VIII inhibitors was performed, using a plasma sample with an inhibitor titer of 40 BU/mL of a patient with severe hemophilia A.8 Following treatment of the same patient with factor VIII inhibitor bypassing agent (FEIBA), the inhibitor titer increased to 200 BU/mL. In this study, we first evaluated the domain specificity of anti–factor VIII antibodies in the latter plasma sample. Immunoprecipitation analysis showed reactivity of antibodies with the factor VIII light chain as well as the C2 domain (Figure 1; lanes 5 and 6). To determine to what extent anti–C2 domain antibodies contribute to the inhibitor titer, an inhibitor neutralization assay was performed. Addition of factor VIII light chain completely neutralized factor VIII inhibitory activity, whereas 30% neutralization was achieved on addition of C2 domain (data not shown). This suggests that the majority of factor VIII inhibitory antibodies is directed toward a region located outside the C2 domain. The patient's anti–factor VIII antibodies consisted predominantly of subclass IgG4 (data not shown). Therefore, a subclass-specific oligonucleotide primer was used to selectively amplify the patient's IgG4-specific VH gene repertoire. Recombination of the IgG4-enriched VH gene repertoire with a nonimmune VL gene repertoire in pHEN-1-VLrep yielded a phage library of 1.9 × 107 clones.
Characterization of antibodies present in plasma of a patient with inhibitors to factor VIII.
Binding of antibodies to metabolically labeled factor VIII fragments corresponding to the factor VIII light chain and the C2 domain was evaluated by immunoprecipitation. The samples were analyzed under reducing conditions on a 12.5% (wt/vol) sodium dodecyl sulfate–polyacrylamide gel. (lanes 1 and 2) Positive control (CLB-CAg 117); (lanes 3 and 4) negative control (normal plasma); (lanes 5 and 6) patient's plasma. Arrowheads indicate C2 domain (C2) and factor VIII light chain (LCh). Molecular weight markers in kd are indicated at the right.
Characterization of antibodies present in plasma of a patient with inhibitors to factor VIII.
Binding of antibodies to metabolically labeled factor VIII fragments corresponding to the factor VIII light chain and the C2 domain was evaluated by immunoprecipitation. The samples were analyzed under reducing conditions on a 12.5% (wt/vol) sodium dodecyl sulfate–polyacrylamide gel. (lanes 1 and 2) Positive control (CLB-CAg 117); (lanes 3 and 4) negative control (normal plasma); (lanes 5 and 6) patient's plasma. Arrowheads indicate C2 domain (C2) and factor VIII light chain (LCh). Molecular weight markers in kd are indicated at the right.
Isolation and sequence analysis of antibodies directed toward the A3 domain of factor VIII
After 3 rounds of selection, the domain specificity of phage derived from 15 single clones was analyzed. All analyzed clones reacted with the factor VIII light chain but not with the C2 domain (data not shown). Sequence analysis of these clones revealed the presence of 6 different VH genes. The VH genes of clones KM37 and KM41 were derived from germline gene segments DP-14 and DP-15, respectively (Table 1), both belonging to the VH1 gene family. The remaining 4 VH genes were derived from VH3 family gene segments; 2 from gene segment DP-49 (KM33 and KM38) and 2 from DP-77 (KM35 and KM36). The VH genes harbor 21 to 31 nucleotide substitutions compared to their nonmutated germline gene segments. This resulted in 9 to 18 amino acid replacements in the encoded VH domains. Deduced amino acid sequences of the VH domains are shown in Table2. Both VH genes of clones KM35 and KM36, which are highly homologous, used gene segment JH3b for VDJ rearrangement. JH4b was identified as the most homologous JH gene segment in clones KM38 and KM41 and JH6 in KM37. In the VH domain of KM33, no particular JH gene segment could be identified, possibly as a consequence of extensive somatic hypermutation and/or N-addition and deletion at the junction between D and JH gene segments. The presence of a particular D gene segment could only be established in clone KM37 in which gene segment D3-3 was rearranged. VL domains of clones KM35 and KM36 were encoded by germline gene segment DPL16, a member of the Vλ3 gene family. The other 4 VH domains paired with VL domains derived from different gene segments belonging to the VκI and VκIII gene families (Table 1).
Most homologous germline gene segments used in A3-C1 specific single-chain variable domain antibody fragments
| Clone . | VHdomain . | VL domain . | |||
|---|---|---|---|---|---|
| Germline . | Family . | Substitutions nucl/aa . | Germline . | Family . | |
| KM37 | DP-14 (1-18) | VH1 | 22/13 | DPK8 (L8) | VκI |
| KM41 | DP-15 (1-8) | VH1 | 21/14 | DPK22 (A27) | VκIII |
| KM33 | DP-49 (3-30) | VH3 | 21/10 | DPK3 (L11) | VκI |
| KM38 | DP-49 (3-30) | VH3 | 23/9 | L12a (L12) | VκI |
| KM35 | DP-77 (3-21) | VH3 | 31/18 | DPL16 (V2-13) | Vλ3 |
| KM36 | DP-77 (3-21) | VH3 | 26/15 | DPL16 (V2-13) | Vλ3 |
| Clone . | VHdomain . | VL domain . | |||
|---|---|---|---|---|---|
| Germline . | Family . | Substitutions nucl/aa . | Germline . | Family . | |
| KM37 | DP-14 (1-18) | VH1 | 22/13 | DPK8 (L8) | VκI |
| KM41 | DP-15 (1-8) | VH1 | 21/14 | DPK22 (A27) | VκIII |
| KM33 | DP-49 (3-30) | VH3 | 21/10 | DPK3 (L11) | VκI |
| KM38 | DP-49 (3-30) | VH3 | 23/9 | L12a (L12) | VκI |
| KM35 | DP-77 (3-21) | VH3 | 31/18 | DPL16 (V2-13) | Vλ3 |
| KM36 | DP-77 (3-21) | VH3 | 26/15 | DPL16 (V2-13) | Vλ3 |
VH and VL germline gene use and nomenclature according to V-BASE.28 The number of different nucleotides and amino acids compared to the nonmutated germlines are indicated.
Nucl indicates nucleotides; aa, amino acids.
Deduced protein sequences of isolated factor VIII A3-C1–specific single-chain variable domain antibody fragments
| Heavy chains . | |||||||
|---|---|---|---|---|---|---|---|
| . | FR1 . | CDR1 . | FR2 . | CDR2 . | FR3 . | CDR3 . | FR4 . |
| 1 | 1 1 | ||||||
| 1 2 3 | 4 | 5 6 | 7 8 9 | 0 | 0 1 | ||
| 123456789012345678901234567890 | 12345 | 67890123456789 | 012a3456789012345 | 67890123456789012abc345678901234 | 567890abcdefghijklm12 | 34567890123 | |
| DP-14 | QVQLVQSGAEVKKPGASVKVSCKASGYTFT | SYGIS | WVRQAPGQGLEWMG | WISAYNGNTNYAQKLQG | RVTMTTDTSTSTAYMELRSLRSDDTAVYYCAR | ||
| KM37 | ------------------E---------A | T---N | -------------- | -F-PHK-D------F-- | ---L------N--------N------------ | GKADQYYNLWRGSHSPNLLDN | WGQGTTVTVSS |
| DP-15 | QVQLVQSGAEVKKPGASVKVSCKASGYTFT | SYDIN | WVRQATGQGLEWMG | WMNPNSGNTGYAQKFQG | RVTMTRNTSISTAYMELSSLRSEDTAVYYCAR | ||
| KM41 | ----1--a-D------------T----I-- | ----D | -------------- | --------A-F----K- | -L-L--D--T-------RR-E----------- | STPHSYSGSGLPPTSDS.... | WGQGTLVTVSS |
| DP-49 | QVQLVESGGGVVQPGRSLRLSCAASGFTFS | SYGMH | WVRQAPGKGLEWVA | VISYDGSNKYYADSVKG | RFTISRDNSKNTLYLQMNSLRAEDTAVYYCAK | ||
| KM33 | e---------------------VD--L--- | ----- | -------A------ | ------ND--------- | --A-----A-----------TI---------- | DLIESNIAEAF.......... | WGQGTLVTVSS |
| KM38 | --n-r----------K-------------- | D-AI- | ------------L- | -------S--------- | -------------F-------V----L----- | DRHINISPPDS.......... | WGQGTLVTVSS |
| DP-77 | EVQLVESGGGLVKPGGSLRLSCAASGFTFS | SYSMN | WVRQAPGKGLEWVS | SISSSSSYIYYADSVKG | RFTISRDNAKNSLYLQMNSLRAEDTAVYYCAR | ||
| KM35 | ---------D--E---------T------R | R-DIH | ----T--------- | ----GGN--D------- | ---------N-VV-----------M---F--- | DGTIFGSAATWRAFDI..... | WGRGTMVTVSS |
| KM36 | -----------------------------R | R-DIH | ----T--------- | ----GGN--D------- | ---------N-VV-----------M---F--- | DGTIFGSAATWRAFDI..... | WGRGTMVTVSS |
| Heavy chains . | |||||||
|---|---|---|---|---|---|---|---|
| . | FR1 . | CDR1 . | FR2 . | CDR2 . | FR3 . | CDR3 . | FR4 . |
| 1 | 1 1 | ||||||
| 1 2 3 | 4 | 5 6 | 7 8 9 | 0 | 0 1 | ||
| 123456789012345678901234567890 | 12345 | 67890123456789 | 012a3456789012345 | 67890123456789012abc345678901234 | 567890abcdefghijklm12 | 34567890123 | |
| DP-14 | QVQLVQSGAEVKKPGASVKVSCKASGYTFT | SYGIS | WVRQAPGQGLEWMG | WISAYNGNTNYAQKLQG | RVTMTTDTSTSTAYMELRSLRSDDTAVYYCAR | ||
| KM37 | ------------------E---------A | T---N | -------------- | -F-PHK-D------F-- | ---L------N--------N------------ | GKADQYYNLWRGSHSPNLLDN | WGQGTTVTVSS |
| DP-15 | QVQLVQSGAEVKKPGASVKVSCKASGYTFT | SYDIN | WVRQATGQGLEWMG | WMNPNSGNTGYAQKFQG | RVTMTRNTSISTAYMELSSLRSEDTAVYYCAR | ||
| KM41 | ----1--a-D------------T----I-- | ----D | -------------- | --------A-F----K- | -L-L--D--T-------RR-E----------- | STPHSYSGSGLPPTSDS.... | WGQGTLVTVSS |
| DP-49 | QVQLVESGGGVVQPGRSLRLSCAASGFTFS | SYGMH | WVRQAPGKGLEWVA | VISYDGSNKYYADSVKG | RFTISRDNSKNTLYLQMNSLRAEDTAVYYCAK | ||
| KM33 | e---------------------VD--L--- | ----- | -------A------ | ------ND--------- | --A-----A-----------TI---------- | DLIESNIAEAF.......... | WGQGTLVTVSS |
| KM38 | --n-r----------K-------------- | D-AI- | ------------L- | -------S--------- | -------------F-------V----L----- | DRHINISPPDS.......... | WGQGTLVTVSS |
| DP-77 | EVQLVESGGGLVKPGGSLRLSCAASGFTFS | SYSMN | WVRQAPGKGLEWVS | SISSSSSYIYYADSVKG | RFTISRDNAKNSLYLQMNSLRAEDTAVYYCAR | ||
| KM35 | ---------D--E---------T------R | R-DIH | ----T--------- | ----GGN--D------- | ---------N-VV-----------M---F--- | DGTIFGSAATWRAFDI..... | WGRGTMVTVSS |
| KM36 | -----------------------------R | R-DIH | ----T--------- | ----GGN--D------- | ---------N-VV-----------M---F--- | DGTIFGSAATWRAFDI..... | WGRGTMVTVSS |
Sequence numbering is according to Kabat et al.31Sequences are available from GenBank under accession numbers: AF234247(VHKM33); AF234248 (VHKM35); AF234249 (VHKM36); AF234250 (VHKM37);AF234251 (VHKM38); AF234252 (VHKM41); AF234253 (VLKM33); AF234254(VLKM35); AF234255 (VLKM36); AF234256 (VLKM37); AF234257 (VLKM38);AF234258 (VLKM41).
scFv indicates single-chain variable domain antibody fragments; FR, framework region; CDR, complementarity-determining region. Dashes indicate sequence identity to germline. Lowercase indicates amino acid substitutions encoded by the PCR primer.
Biochemical characterization of A3-C1–specific scFv
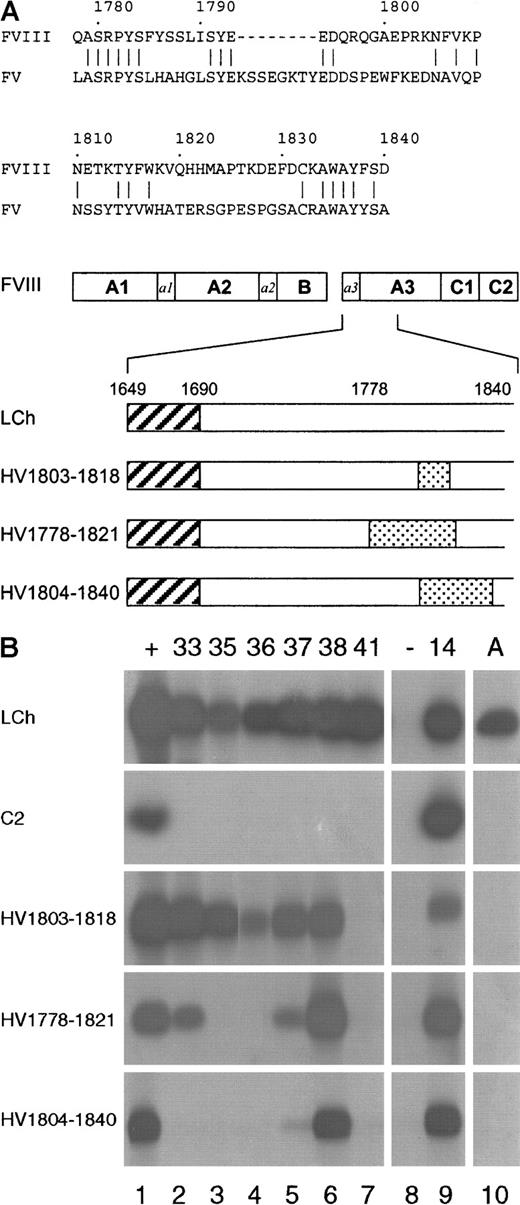
For the expression of scFv, the different VH and VL genes were subcloned in the expression vector pUC119-Sfi/Not-His6.29 To define the domain specificity of the isolated scFv, their reactivity was evaluated by immunoprecipitation analysis, using metabolically labeled factor VIII light chain and C2 domain. Expression of the factor VIII fragments was monitored by binding of scFv EL-14 (Figure2B; lane 9), a previously described anti-C2 domain scFv.18 All scFv, except for a control scFv that was directed toward the A2 domain (Figure 2B; lane 8), reacted with the factor VIII light chain. None of the isolated scFv bound to the C2 domain, indicating that all 6 patient-derived scFv are directed toward an epitope in the A3-C1 domains (Figure 2B).
Reactivity of isolated scFv with factor VIII/factor V light-chain hybrids.
(A) Below a sequence alignment of factor VIII region Q1778-D1840 with the corresponding sequence of factor V, a schematic representation of the hybrid factor VIII/factor V light-chain fragments used in this study is depicted. The regions R1803-K1818 (HV1803-1818), Q1778-H1821 (HV1778-1821), and K1804-D1840 (HV1804-1840) in the factor VIII light chain that have been replaced for the corresponding factor V sequence are indicated as dotted boxes. Hatched boxes correspond to the acidic region a3, composed of residues E1649-R1689. (B) Binding of scFv to recombinant factor VIII/factor V light-chain hybrids was assessed by immunoprecipitation. (Lane 1) positive (+) control (antibody CLB-CAg 117); (lanes 2-7) 33, 35, 36, 37, 38, and 41; scFv corresponding to clones KM33, KM35, KM36, KM37, KM38, and KM41; (lane 8) negative (−) control (scFv directed toward the A2 domain of factor VIII); (lane 9)14 positive control (scFv EL-14, directed toward the C2 domain of factor VIII); (lane 10) antibody (A) antibody CLB-CAg A. On the left, the used factor VIII fragments are indicated.
Reactivity of isolated scFv with factor VIII/factor V light-chain hybrids.
(A) Below a sequence alignment of factor VIII region Q1778-D1840 with the corresponding sequence of factor V, a schematic representation of the hybrid factor VIII/factor V light-chain fragments used in this study is depicted. The regions R1803-K1818 (HV1803-1818), Q1778-H1821 (HV1778-1821), and K1804-D1840 (HV1804-1840) in the factor VIII light chain that have been replaced for the corresponding factor V sequence are indicated as dotted boxes. Hatched boxes correspond to the acidic region a3, composed of residues E1649-R1689. (B) Binding of scFv to recombinant factor VIII/factor V light-chain hybrids was assessed by immunoprecipitation. (Lane 1) positive (+) control (antibody CLB-CAg 117); (lanes 2-7) 33, 35, 36, 37, 38, and 41; scFv corresponding to clones KM33, KM35, KM36, KM37, KM38, and KM41; (lane 8) negative (−) control (scFv directed toward the A2 domain of factor VIII); (lane 9)14 positive control (scFv EL-14, directed toward the C2 domain of factor VIII); (lane 10) antibody (A) antibody CLB-CAg A. On the left, the used factor VIII fragments are indicated.
The interaction of isolated scFv with the factor VIII light chain was explored in more detail by mutagenesis in the previously described binding site for anti-A3 antibodies.8,9 Three factor VIII/factor V light-chain hybrids were constructed that contained factor V replacements in the regions R1803-K1818 (HV1803-1818), Q1778-H1821 (HV1778-1821) or K1804-D1840 (HV1804-1840) (Figure 2A). Inspection of a 3-dimensional model of the triplicated A domains of factor VIII revealed that the majority of the amino acids within amino acid sequence Q1778-D1840 are located at the surface of the A3 domain.32 33
First, reactivity of antibody CLB-CAg A, whose binding site has been localized to residues K1804-K1818,10 with the different hybrids was evaluated. As expected, CLB-CAg A did not recognize any of the hybrid factor VIII/factor V fragments (Figure 2B, lane 10). In agreement with previous data, these findings indicate that the epitope of CLB-CAg A is localized within region R1803-K1818. Similar to CLB-CAg A, one of the isolated scFv, KM41, did not react with the different hybrid fragments, suggesting that the epitope of scFv KM41 is also contained within amino acid sequence R1803-K1818 (Figure 2B; lane 7).
No reactivity of scFv KM35 and KM36 with hybrids HV1778-1821 and HV1804-1840 was observed, whereas both scFv readily react with hybrid HV1803-1818 (Figure 2B; lanes 3 and 4). This points at an essential role for factor VIII residues surrounding sequence R1803-K1818 in the binding of scFv KM35 and KM36 to the light chain of factor VIII. ScFv KM33 and KM37 reacted with hybrid HV1803-1818, whereas binding of both scFv to HV1778-1821 was reduced (Figure 2B; lanes 2 and 5). Virtually no binding of scFv KM33 and KM37 to HV1804-1840 was observed, suggesting that residues within this region are essential for binding of these scFv to the A3 domain.
All tested factor VIII light-chain hybrids were bound to a similar extent by scFv KM38 (Figure 2B, lane 6). Therefore, its epitope must be located outside the region Q1778-D1840. Recently, a binding site for factor VIII inhibitors has been assigned to amino acid residues E1649-R1689 at the amino-terminus of the factor VIII light chain.11 Binding of KM38 to thrombin-cleaved factor VIII light chain was evaluated. Following thrombin cleavage at position R1689, the region E1649-R1689 is removed from the factor VIII light chain. In an enzyme-linked immunosorbent assay, scFv KM38 exhibited similar binding to factor VIII light chain and thrombin-cleaved factor VIII light chain (data not shown). These findings suggest that scFv KM38 recognizes an epitope elsewhere in the A3-C1 domains of factor VIII.
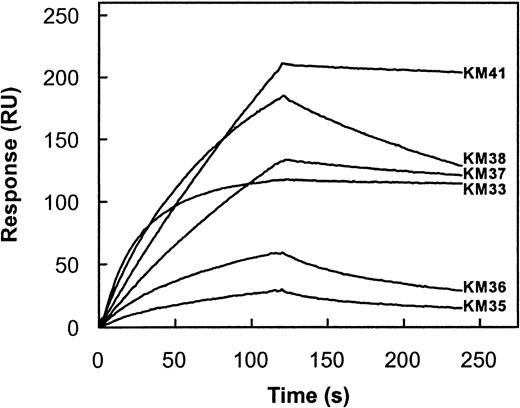
The capacity of each scFv to inhibit factor VIII procoagulant activity was evaluated in a Bethesda assay.24 Only scFv KM33 and KM41 inhibited factor VIII activity with inhibitor titers of 97 and 63 BU/mg, respectively. The other scFv did not inhibit factor VIII activity (titer < 5 BU/mg scFv). To determine whether the observed Bethesda titer of each scFv corresponds with its affinity for the factor VIII light chain, we measured the association rate constant (kon) and dissociation rate constant (koff) of the factor VIII light chain for the different scFv by surface plasmon resonance. Addition of factor VIII light chain to immobilized scFv KM41 resulted in a time-dependent increase in mass detected on the sensor chip that reflects binding of factor VIII light chain to immobilized scFv (Figure3). Fitting of this part of the curve yielded a kon of 1.8 × 105M−1s−1 for the interaction of factor VIII light chain with scFv KM41 (Table 3). Dissociation of bound factor VIII light chain from scFv KM41 was initiated at 2 minutes. Analysis of the obtained sensorgram revealed that factor VIII light chain dissociates with a slow rate from immobilized scFv KM41 (Figure 3). A koff of 2.9 × 10−4s−1 was obtained from the observed decay in binding (Table 3). On the basis of the experimentally determined values of kon and koff, a value of 1.6 nM was calculated for the Kd of scFv KM41 and factor VIII light chain (Table 3). The affinity of the other 5 scFv for factor VIII light chain was analyzed in a similar manner (Figure 3; Table 3). ScFv KM35 and KM36 bound with a 35-fold lower affinity to factor VIII light chain when compared to scFv KM41 (Figure 3). The konobserved for scFv KM38 is similar to that of scFv KM41, which indicates that factor VIII light chain readily binds to both scFv. The observed differences in affinity for the 2 scFv are due to an increased rate of dissociation of factor VIII light chain from scFv KM38 (Figure 3). Also, the reduced affinity of scFv KM37 compared to scFv KM41 can be attributed to an approximate 3-fold higher dissociation rate (Table 3). Factor VIII light chain associates with an approximate 10-fold higher rate to scFv KM33 when compared to scFv KM41, whereas the dissociation rate is similar for both scFv (Table 3). Consequently, the affinity of scFv KM33 is more than 10-fold higher than observed for scFv KM41 (Table 3). Overall, the affinity of the scFv isolated in this study ranges between 0.1 and 59 nM. Interestingly, scFv KM33 and KM41, having the highest affinity for factor VIII light chain, do inhibit the biological activity of factor VIII as determined in a Bethesda assay. These data indicate that the inhibitory capacity of the scFv is at least in part determined by their affinity for factor VIII.
Kinetic parameters for binding of factor VIII light chain to immobilized scFv.
Factor VIII light chain (20 nM) was incubated with immobilized scFv KM33 (3.3 fmol/mm2), KM35 (63.3 fmol/mm2), KM36 (67.5 fmol/mm2), KM37 (28.3 fmol/mm2), KM38 (32.3 fmol/mm2), and KM41 (25.0 fmol/mm2) in 200 mM NaCl, 2 mM CaCl2, 0.05% (v/v) Tween 20, and 20 mM HEPES, pH 7.4 at a flow rate of 20 μL/min for 2 minutes at 25°C. Dissociation was initiated on replacement of ligand solution by buffer. Response is indicated as resonance units (RU) and is corrected for nonspecific binding (< 5%). To obtain similar values of resonance units for all scFv, the amount of scFv KM33 immobilized was approximately 10-fold reduced compared to the other scFv. Similar values of kon and koffwere found when higher concentrations of scFv KM33 were immobilized.
Kinetic parameters for binding of factor VIII light chain to immobilized scFv.
Factor VIII light chain (20 nM) was incubated with immobilized scFv KM33 (3.3 fmol/mm2), KM35 (63.3 fmol/mm2), KM36 (67.5 fmol/mm2), KM37 (28.3 fmol/mm2), KM38 (32.3 fmol/mm2), and KM41 (25.0 fmol/mm2) in 200 mM NaCl, 2 mM CaCl2, 0.05% (v/v) Tween 20, and 20 mM HEPES, pH 7.4 at a flow rate of 20 μL/min for 2 minutes at 25°C. Dissociation was initiated on replacement of ligand solution by buffer. Response is indicated as resonance units (RU) and is corrected for nonspecific binding (< 5%). To obtain similar values of resonance units for all scFv, the amount of scFv KM33 immobilized was approximately 10-fold reduced compared to the other scFv. Similar values of kon and koffwere found when higher concentrations of scFv KM33 were immobilized.
Binding kinetics and affinities of isolated single-chain variable domain antibody fragments
| scFv . | kon(M−1s−1) . | koff(s−1) . | Kd (nM) . |
|---|---|---|---|
| KM33 | 1.8 × 106 | 1.9 × 10−4 | 0.1 |
| KM35 | 1.6 × 105 | 9.4 × 10−3 | 59 |
| KM36 | 1.7 × 105 | 9.5 × 10−3 | 55 |
| KM37 | 2.6 × 105 | 9.5 × 10−4 | 3.7 |
| KM38 | 3.8 × 105 | 3.5 × 10−3 | 9.0 |
| KM41 | 1.8 × 105 | 2.9 × 10−4 | 1.6 |
| scFv . | kon(M−1s−1) . | koff(s−1) . | Kd (nM) . |
|---|---|---|---|
| KM33 | 1.8 × 106 | 1.9 × 10−4 | 0.1 |
| KM35 | 1.6 × 105 | 9.4 × 10−3 | 59 |
| KM36 | 1.7 × 105 | 9.5 × 10−3 | 55 |
| KM37 | 2.6 × 105 | 9.5 × 10−4 | 3.7 |
| KM38 | 3.8 × 105 | 3.5 × 10−3 | 9.0 |
| KM41 | 1.8 × 105 | 2.9 × 10−4 | 1.6 |
Kd, kon, andkoff were determined by surface plasmon resonance as described in “Materials and methods.”
Discussion
A significant portion of plasmas of patients with inhibitors to factor VIII are known to contain antibodies that bind to the A3-C1 domains.4,6 In the present study, 6 different human antibodies directed toward the A3-C1 domains were isolated from the immunoglobulin repertoire of a single patient. Previously, we and others have shown that the VH domains of anti-C2 antibodies are preferentially encoded by germline gene segments derived from the VH1 gene family.18,34,35 The results of the present study suggest that anti–A3-C1 antibodies are composed of VH domains derived from multiple germline gene segments of the VH1 and VH3 family. Germline gene segments of the VH1 and VH3 family occur in, respectively, 15% and 55% of human peripheral IgG+ B cells.36 The individual germline gene segments DP-49 (3-30) DP-77 (3-21), and DP-14 (1-18) that encode anti–A3-C1 antibodies are present in 5% to 10% of the total immunoglobulin repertoire.36 In contrast, germline segment DP-15 (1-8) that has been used for assembly of scFv KM41 (Table 1) is present with a low frequency (< 0.5%) in the immunoglobulin repertoire. These observations suggest that, in contrast to, for example, human anti-Rh(D) antibodies, which are almost exclusively derived from members of the “VH3-33 superspecies”—closely related germline gene segments DP-49 (3-30/3-30.5), DP-46 (3-30.3), and DP-50 (3-33)—anti–A3-C1 antibodies are not encoded by a restricted set of VH gene segments.37 Further analysis of additional patients should confirm whether anti–A3-C1 antibodies are indeed recruited from B cells expressing immunoglobulins that incorporate frequently used VH gene segments.
Five of 6 scFv bound to amino acid sequences contained within the region Q1778-D1840 in the A3 domain. These findings confirm that this part of the A3 domain comprises a major binding site for factor VIII inhibitors.8,9 Within amino acid sequence Q1778-D1840, an interactive site for factor IXa has been localized to amino acid residues E1811-K1818.10 Binding of factor VIII inhibitors to this site in the A3 domain interferes with binding of factor IXa.8 9 Surprisingly, only 2 of 6 scFv described in this study effectively inhibited the procoagulant activity of factor VIII. In the following paragraphs, potential explanations for the different functional properties of the isolated scFv will be discussed.
Binding of the inhibitory scFv KM41 is dependent on residues R1803-K1818, which contain a binding site for factor IXa. Previously, we have shown that monoclonal antibody CLB-CAg A binds to a synthetic peptide comprising these residues.10 Replacement of residues R1803-K1818 by the corresponding sequence of factor V abolishes binding of CLB-CAg A to the factor VIII light chain (Figure2B). CLB-CAg A interferes with binding of factor IXa to the factor VIII light chain.21 In view of the similar epitope specificity of scFv KM41 and CLB-CAg A, it is likely that scFv KM41 competes with factor IXa for binding to the factor VIII light chain.
Also, scFv KM33 inhibits the procoagulant activity of factor VIII, although its epitope is located outside region R1803-K1818 (Figure 2B). Binding of scFv KM33 is primarily affected by replacements in the carboxy-terminal part of sequence Q1778-D1840. The epitope specificity of scFv KM33 is remarkably similar to that of scFv KM37, which does not inhibit factor VIII procoagulant activity. It should be noted that the VH domains of scFv KM33 and KM37 are derived from different VH gene segments, which may endow these scFv with different biochemical properties. Both scFv KM35 and KM36 do not inhibit factor VIII procoagulant activity, and binding of both scFv is dependent on residues contained within regions surrounding R1803-K1818. The VH domains of these scFv differ at only 3 amino acid positions, which may explain their similar epitope specificity.
The lack of inhibition of factor VIII activity by scFv KM35 and KM36 is most likely explained by their low affinity for the factor VIII light chain in comparison to scFv KM33 and KM41 (Figure 3). Because of their lower affinity for factor VIII, scFv KM35 and KM36 may not be able to efficiently compete for binding of factor IXa to the factor VIII light chain. The epitope specificity of the noninhibitory scFv KM37 overlaps with that of the inhibitory scFv KM33 (Figure 2B). The low affinity scFv KM37, compared to that of scFv KM33, for the factor VIII light chain may explain its inability to block the procoagulant activity of factor VIII. Taken together, it seems likely that high-affinity binding of scFv to factor VIII light chain is a prerequisite for factor VIII inhibition.
Inspection of the 3-dimensional homology model of the A domains of factor VIII reveals that, within region Q1778-D1840, amino acid residues Y1792-A1834 are exposed.32 The factor IXa binding site present within region R1803-K1818 is located at one site of this region, whereas regions Q1778-P1802 and V1819-D1840 are in close contact but oriented away from residues R1803-K1818.33 On the basis of the diameter of an antigen-binding site of 3.0 nm,38 region Q1778-D1840 can accommodate at least 2 separate binding sites for scFv. The first binding site may overlap with residues R1803-K1818, and antibodies directed against this site are likely to interfere with binding of factor IXa. The second binding site may be composed of residues derived from both regions Q1778-P1802 and V1819-D1840. This hypothesis is compatible with our findings on the reactivity of scFv with the hybrid factor VIII/factor V light-chain fragments. The inhibitory scFv KM41 and mAb CLB-CAg A are targeted to the factor IXa binding site present within region R1803-K1818. In contrast, binding of KM33, KM35, KM36, and KM37 to factor VIII is sensitive to replacement of amino acid sequences surrounding R1803-K1818. ScFv binding to this site may not necessarily interfere with factor VIII activity because most of these residues are not in close proximity to the factor IXa binding site at E1811-K1818. Because of the smaller size of scFv (30 kd) compared to that of complete antibodies (150 kd), we cannot exclude that a complete antibody molecule that binds to regions Q1778-P1802 and V1819-D1840 can interfere with factor VIII procoagulant activity. Also, complete antibody molecules may bind with a higher affinity to the factor VIII light chain compared to the corresponding scFv. Finally, it should be noted that the scFv described in this report contain VLdomains that are derived from a nonimmune source. Phage display does not preserve the original VH-VL pairing as found in anti–A3-C1 antibodies present in patient's plasma. Potentially, both affinity and epitope specificity may be modulated by substituting the VL chain of the scFv for a different, patient-derived light chain.
The epitope specificity of scFv KM38 clearly differs from that of the other scFv isolated in this study. Presumably, scFv KM38 corresponds to anti–factor VIII antibodies that recognize an additional epitope in the A3-C1 domains. ScFv KM38 is not directed against amino acid residues E1649-R1689, a recently described epitope for factor VIII inhibitors in the acidic region at the amino-terminus of the factor VIII light chain.11 Human antibodies directed against this region are often accompanied by inhibitory antibodies directed against other parts on factor VIII and are not present in plasma of all inhibitor patients. Most likely, antibodies with this specificity are not sufficiently represented in the repertoire of the patient described in this study to allow for their isolation by phage display. Competition experiments revealed that the epitope of scFv KM38 overlaps with that of antibody CLB-CAg 12, a noninhibitory mAb directed against the A3-C1 domains of factor VIII (data not shown). Only a single scFv that bound outside region Q1778-D1840 has been found in this study, suggesting that human antibodies corresponding to scFv KM38 occur with a low frequency in the immunoglobulin repertoire of inhibitor patients.
Acknowledgments
We thank C. van der Zwaan for providing factor V cDNA, P. H. N. Celie for providing purified factor VIII light chain, and G. van Stempvoort for the construction of HV1803-1818. We are also grateful to W. G. van Aken, R. C. Aalberse, and P. J. Lenting for critical reading of the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jan Voorberg, Department of Plasma Proteins, CLB, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail:j_voorberg@clb.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal