Aberrant hypermethylation of tumor suppressor genes plays an important role in the development of many tumors. Recently identified new DNA methyltransferase (DNMT) genes, DNMT3Aand DNMT3B, code for de novo methyltransferases. To determine the roles of DNMT3A, DNMT3B, as well as DNMT1, in the development of leukemia, competitive polymerase chain reaction (PCR) assays were performed and the expression levels of DNMTs were measured in normal hematopoiesis, 33 cases of acute myelogenous leukemia (AML), and 17 cases of chronic myelogenous leukemia (CML). All genes were constitutively expressed, although at different levels, in T lymphocytes, monocytes, neutrophils, and normal bone marrow cells. Interestingly, DNMT3B was expressed at high levels in CD34+ bone marrow cells but down-regulated in differentiated cells. In AML, 5.3-, 4.4-, and 11.7-fold mean increases were seen in the levels of DNMT1, 3A, and3B, respectively, compared with the control bone marrow cells. Although CML cells in the chronic phase did not show significant changes, cells in the acute phase showed 3.2-, 4.5-, and 3.4-fold mean increases in the levels of DNMT1, 3A, and3B, respectively. Using methylation-specific PCR, it was observed that the p15INAK4B gene, a cell cycle regulator, was methylated in 24 of 33 (72%) cases of AML. Furthermore, AML cells with methylatedp15INAK4B tended to express higher levels ofDNMT1 and 3B. In conclusion, DNMTswere substantially overexpressed in leukemia cells in a leukemia type- and stage-specific manner. Up-regulated DNMTs may contribute to the pathogenesis of leukemia by inducing aberrant regional hypermethylation.
Introduction
DNA methylation plays an important role in tissue- and stage-specific gene regulation,1,2 genomic imprinting,3,4 and X-chromosome inactivation,5 and has been shown to be essential for normal mammalian development.6 Recent studies have revealed that both global DNA hypomethylation and regional hypermethylation occur in tumorigenesis.7-9 Such aberrant DNA methylation is observed in a nonrandom, tumor type-specific manner.10 In particular, certain types of tumors show regional hypermethylation of CpG islands associated with the promoter regions of tumor suppressor genes, such asRB,11,VHL,12,p16INAK4A,13 andhMLH1.14 Furthermore, the regional hypermethylation is often associated with the inactivation of the tumor suppressor genes.15 These data suggest that this epigenetic process has a pathogenetic role in the clonal evolution of cancer.9
In hematologic malignancies, aberrant DNA hypermethylation is thought to have relevance to leukemogenesis.16 For example, during the progression of chronic myelogenous leukemia (CML), theABL1 promoter of the BCR-ABL fusion gene becomes significantly hypermethylated.17,18 Also, aberrant hypermethylation of the p15INAK4B tumor suppressor gene is associated with its inactivation in at least half of the patients with acute lymphoblastic leukemia (ALL) and acute myelogenous leukemia (AML).19,20 Furthermore, hypermethylation of p15INAK4B is observed concomitant with the disease progression in myelodysplastic syndrome (MDS).21 In addition to these tumor-related genes, a number of other genes are concurrently hypermethylated in AML,22 suggesting that there might be a dysregulation in the normal DNA methylation mechanism, by which the leukemic cells become predisposed to hypermethylation.
Until recently, only one mammalian DNA methyltransferase,DNMT1, had been known, which has a higher maintenance DNA methylase activity rather than a de novo methylase activity in vitro.23 Recently, new mammalian DNA methyltransferase genes, DNMT3A and DNMT3B, have been cloned.24-26 Both the mouse Dnmt3a and3b (the orthologs of human DNMTs) enzymes were shown to methylate hemimethylated and unmethylated DNA with equal efficiencies in vitro.24 In transgenic Drosophila melanogaster, Dnmt3a clearly exhibited de novo methylase activity, whereas Dnmt1 did not.27Furthermore, a simultaneous inactivation of both Dnmt3a andDnmt3b blocked the de novo methylation activity in embryonic stem cells and embryos,28 suggesting that these enzymes are the long sought de novo methyltransferases.
To understand the mechanisms underlying the aberrant, tumor type-specific hypermethylation, it is important to know the role of each DNA methyltransferase in the pathogenetic process. In colon cancer, increased DNMT1 expression has been demonstrated when compared with normal mucosa.29 In both colon and lung cancers, DNMT1 activity increases progressively, along with the advancement of their tumor stage.30,31 In hematologic malignancies, overexpression of DNMT1 was shown in 12 leukemia samples including AML, ALL, and MDS.32 As toDNMT3A and 3B, Robertson and coworkers have recently shown that both genes, as well as DNMT1, are up-regulated in some malignancies, such as bladder, colon, kidney, and pancreas tumors, though at different levels.25 Xie and colleagues have also reported the increased expression of all 3DNMTs in several tumor cell lines.26
We have obtained DNMT3A and 3B complementary DNAs (cDNAs) independently from the other laboratories by database search. Then we have set out to study the roles of these enzymes in the pathogenesis of hematologic malignancies. In the present study, we report the expression levels of DNMT3A, DNMT3B,and DNMT1 in normal hematopoiesis, AML, and CML, studied by a competitive polymerase chain reaction (PCR) assay. Furthermore, in AML cases, we have investigated whether the expression levels ofDNMTs are correlated with aberrant hypermethylation of thep15INAK4B tumor suppressor gene.
Materials and methods
Cloning of the human DNMT3A and 3B cDNAs
We performed a TBLASTN search33 of the dbEST database using modified mouse Dnmt1 motif IV sequences as queries and identified 3 matching mouse EST sequences (AA177277, AA116694, and AA1550177) and 4 matching human EST sequences (W76111, N88352,AA361360, and AA216697). Two of the human EST clones deposited by the I.M.A.G.E. Consortium were obtained from Research Genetics and sequenced in their entirety. These sequences were identical to the corresponding part of the DNMT3A and DNMT3B cDNAs discovered by Robertson and colleagues25 and Xie and coworkers.26 PCR primers (Table1) were prepared to amplify part of the catalytic domain of each DNMT3 cDNA. They were designed not to amplify genomic fragments, considering the exon-intron organization.
Oligonucleotide primers
| . | Oligonucleotide sequence . | Nucleotide position* (exon number) . | Target size (bp) . | Competitor size (bp) . |
|---|---|---|---|---|
| 5′ GAPDH primer | 5′-AAGGCTGAGAACGGGAAGCTTGTCATCAAT-3′ | 180-210 (exon 4) | 500 | 710 |
| 3′ GAPDH primer | 5′-TTCCCGTTCAGCTCAGGGATGACCTTGCCC-3′ | 651-680 (exon 8) | ||
| 5′ PCNA primer | 5′-TCCATCCTCAAGAAGGTGTTGGAG-3′ | 27-50 (exon 1) | 664 | 782 |
| 3′ PCNA primer | 5′-CAGACATACTGAGTGTCACCGTTG-3′ | 668-691 (exon 5) | ||
| 5′ DNMT1 primer | 5′-ACCGCTTCTACTTCCTCGAGGCCTA-3′ | 3242-3266 (exon 28) | 335 | 460 |
| 3′ DNMT1 primer | 5′-GTTGCAGTCCTCTGTGAACACTGTGG-3′ | 3551-3576 (exon 30) | ||
| 5′ DNMT3A primer | 5′-CACACAGAAGCATATCCAGGAGTG-3′ | 2070-2093† | 551 | 668 |
| 3′ DNMT3A primer | 5′-AGTGGACTGGGAAACCAAATACCC-3′ | 2597-2620 | ||
| 5′ DNMT3B primer | 5′-AATGTGAATCCAGCCAGGAAAGGC-3′ | 1942-1965 (exon 17) | 190 | 331 |
| 3′ DNMT3B primer | 5′-ACTGGATTACACTCCAGGAACCGT-3′ | 2109-2132 (exons 18/19) | ||
| 5′ p15-M primer‡ | 5′-GCGTTCGTATTTTGCGGTT-3′ | 148 | ||
| 3′ p15-M primer | 5′-CGTACAATAACCGAACGACCGA-3′ | |||
| 5′ p15-U primer1-153 | 5′-TGTGATGTGTTTGTATTTTGTGGTT-3′ | 154 | ||
| 3′ p15-U primer | 5′-CCATACAATAACCAAACAACCAA-3′ |
| . | Oligonucleotide sequence . | Nucleotide position* (exon number) . | Target size (bp) . | Competitor size (bp) . |
|---|---|---|---|---|
| 5′ GAPDH primer | 5′-AAGGCTGAGAACGGGAAGCTTGTCATCAAT-3′ | 180-210 (exon 4) | 500 | 710 |
| 3′ GAPDH primer | 5′-TTCCCGTTCAGCTCAGGGATGACCTTGCCC-3′ | 651-680 (exon 8) | ||
| 5′ PCNA primer | 5′-TCCATCCTCAAGAAGGTGTTGGAG-3′ | 27-50 (exon 1) | 664 | 782 |
| 3′ PCNA primer | 5′-CAGACATACTGAGTGTCACCGTTG-3′ | 668-691 (exon 5) | ||
| 5′ DNMT1 primer | 5′-ACCGCTTCTACTTCCTCGAGGCCTA-3′ | 3242-3266 (exon 28) | 335 | 460 |
| 3′ DNMT1 primer | 5′-GTTGCAGTCCTCTGTGAACACTGTGG-3′ | 3551-3576 (exon 30) | ||
| 5′ DNMT3A primer | 5′-CACACAGAAGCATATCCAGGAGTG-3′ | 2070-2093† | 551 | 668 |
| 3′ DNMT3A primer | 5′-AGTGGACTGGGAAACCAAATACCC-3′ | 2597-2620 | ||
| 5′ DNMT3B primer | 5′-AATGTGAATCCAGCCAGGAAAGGC-3′ | 1942-1965 (exon 17) | 190 | 331 |
| 3′ DNMT3B primer | 5′-ACTGGATTACACTCCAGGAACCGT-3′ | 2109-2132 (exons 18/19) | ||
| 5′ p15-M primer‡ | 5′-GCGTTCGTATTTTGCGGTT-3′ | 148 | ||
| 3′ p15-M primer | 5′-CGTACAATAACCGAACGACCGA-3′ | |||
| 5′ p15-U primer1-153 | 5′-TGTGATGTGTTTGTATTTTGTGGTT-3′ | 154 | ||
| 3′ p15-U primer | 5′-CCATACAATAACCAAACAACCAA-3′ |
The 5′ and 3′ primers of a pair for standard and competitive RT-PCR were designed for sequences in different exons so that they amplify only cDNA and not genomic DNA. Target size refers to the size of the PCR products from the gene transcripts and competitor size refers to the size of the PCR products from the competitor DNA. For methylation-specific PCR, primers designed by Herman and coworkers39 were used.
The nucleotide sequence positions are relative to the ATG start.
The exon-intron organization has not been determined.
M primer, a primer specific for methylated DNA.
U primer, a primer specific for unmethylated DNA.
Clinical samples and cell lines
Heparinized peripheral blood and bone marrow cells were collected from patients with AML and CML, and from healthy donors after obtaining their informed consent. Immediately after harvest, mononuclear cells (MNCs) were isolated by sedimentation on Ficoll-Hypaque gradients. More than 90% of the MNC populations from AML and acute phase CML were leukemic blasts. Granulocyte fractions were collected from normal peripheral blood with Polymorphperp (Nycomed Pharma, Oslo, Norway). Plastic-adherent cells from normal MNCs were used as the monocyte fraction. Peripheral lymphocytes were collected as nonadherent cells from normal MNCs, and T lymphocytes were enriched by negative selection, using Dynabeads M-450 CD19 (Dynal, Oslo, Norway). Purified T lymphocytes were stimulated with 2.5 μg/mL phytohemagglutinin (PHA; Sigma Chemical Co, St Louis, MO) for 72 hours and used as the activated T cells. CD34+ cells were isolated from normal bone marrow cells by positive selection, using Dynabeads M-450 CD34 (Dynal),34 and a flow cytometric analysis confirmed that more than 95% of the cells were CD34+. In addition to these cells, an Epstein-Barr virus (EBV)-transformed B-lymphocyte cell line (LCL), and 3 human leukemic cell lines (HL60, K562, and KU812) maintained under the standard conditions were used in this study.
In vitro assay for hematopoietic progenitors
Clonogenic progenitor assays were performed using the methylcellulose culture system.35 Five hundred CD34+ cells were cultured in 1 mL Iscove modified Dulbecco medium (GIBCO, Grand Island, NY) supplemented with 30% fetal calf serum (ICN Biochemicals, Osaka, Japan), 50 ng recombinant human interleukin-3 (rhIL-3), 50 ng recombinant human stem cell factor (rhSCF), 10 ng rhIL-6, 10 ng recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF), 10 ng recombinant human granulocyte CSF (rhG-CSF), 3 U recombinant human erythropoietin (all cytokines were from Kirin Brewery Co, Tokyo, Japan), 5 × 10−5 M 2-mercaptoethanol, and 0.88% methylcellulose in 35-mm Nunc 171099 culture dishes (Nunc Inc, Naperville, IL) at 37°C under a humidified atmosphere with 5% CO2. After 14 days of culture, individual colonies were picked up using finely drawn-out Pasteur pipettes and processed for the reverse transcriptase-PCR (RT-PCR) analysis.
Isolation of RNA and RT-PCR analysis
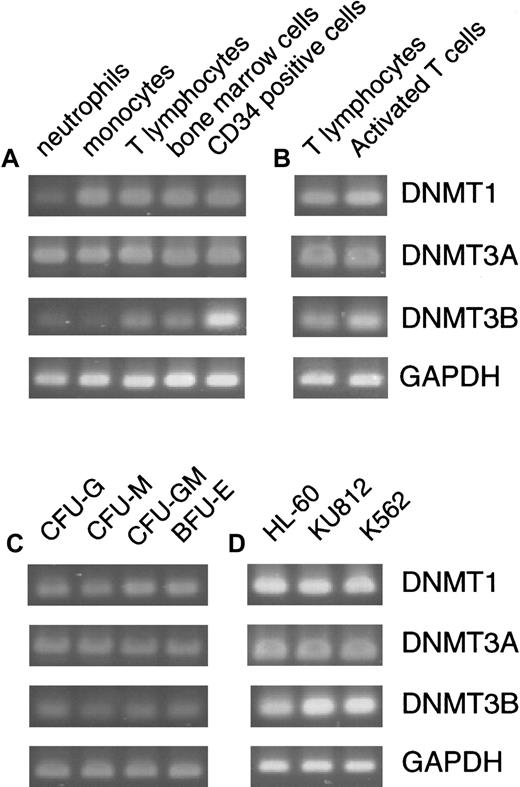
Total RNA was extracted from various cell samples by acid the guanidine/phenol/chloroform method.36 To isolate RNA from the individually picked up colonies, MS2 phage RNA (Boehringer Mannheim, Mannheim, Germany) was added as a carrier. First-strand cDNA was synthesized from 2 μg total RNA in a 50-μL reaction mixture containing random hexamers as primers by using a cDNA synthesis kit (Stratagene, La Jolla, CA). One μL cDNA mixture was brought into 25 μL 1 × PCR buffer, 10 mM Tris-HCl (pH 9.5), 50 mM KCl, 0.1% Triton X-100), 0.2 mM of each dNTP, 1.5 mM MgCl2, 10 pmol of each primer, and 2.0 U of Taq-DNA polymerase (Promega, Madison, WI). The primers used are listed in Table1. PCR was performed in a Perkin Elmer GeneAmp PCR System 9600. Each PCR cycle consisted of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 1 minute. The PCR cycle numbers were 26 for GAPDH, 30 for DNMT1 and3A, and 35 for DNMT3B (Figure1). Under the conditions used, the cDNAs were exponentially amplified, and thus a semiquantitative estimation of the products was possible (data not shown).
Expression of
DNMTs in normal hematopoiesis and leukemia cell lines. Expression of DNMTs was assayed by a standard RT-PCR method in normal peripheral cells and bone marrow cells (A), resting and PHA-activated T cells (B), various colonies formed in hematopoietic progenitor assays (C), and 3 leukemia cell lines (D). With the cells other than the hematopoietic progenitor colonies, PCR amplification was performed for 26 cycles for GAPDH (a housekeeping control), 30 cycles for DNMT1 and3A, and 35 cycles for DNMT3B (A,B,D). To amplify transcripts from the picked hematopoietic colonies, PCR was performed for 35 cycles for DNMT1, 3A, andGAPDH, and 40 cycles for DNMT3B.
Expression of
DNMTs in normal hematopoiesis and leukemia cell lines. Expression of DNMTs was assayed by a standard RT-PCR method in normal peripheral cells and bone marrow cells (A), resting and PHA-activated T cells (B), various colonies formed in hematopoietic progenitor assays (C), and 3 leukemia cell lines (D). With the cells other than the hematopoietic progenitor colonies, PCR amplification was performed for 26 cycles for GAPDH (a housekeeping control), 30 cycles for DNMT1 and3A, and 35 cycles for DNMT3B (A,B,D). To amplify transcripts from the picked hematopoietic colonies, PCR was performed for 35 cycles for DNMT1, 3A, andGAPDH, and 40 cycles for DNMT3B.
Competitive PCR for quantitative analysis
To develop a competitive PCR assay37 for each gene, a competitor plasmid containing an extra DNA fragment in the target sequence was prepared. The GAPDH competitor plasmid was prepared by inserting a 210-bp SacII fragment from λ DNA into the unique SacII site of a 500-bp GAPDH cDNA fragment cloned in a plasmid vector pCR2.1 (Invitrogen, Gronigen, The Netherlands). The PCNA competitor was prepared by cloning a 118-bp HaeIII fragment from φX174 DNA into theStuI site of a 664-bp PCNA cDNA fragment in pCR2.1. The DNMT1 competitor was prepared by inserting a 125-bp HindIII fragment from λ DNA into theHindIII site of a 335-bp DNMT1 cDNA fragment in a modified pBluescriptSK+ vector, from which the HindIII site within the multicloning site had been deleted. The DNMT3Acompetitor was prepared by inserting a 117-bp BstPI fragment from λ DNA into the BstPI site of a 551-bpDNMT3A cDNA fragment in pCR2.1. The DNMT3Bcompetitor was prepared by cloning a 141-bp TthHB8I fragment from pBR322 DNA into the NspV site of a 190-bpDNMT3B cDNA fragment in pCR2.1. All the above cDNA fragments were amplified from RNA of HL60 cells by RT-PCR.
The copy number of each competitor plasmid was determined and 3- or 4- fold consecutive dilutions were prepared. PCR was performed under the conditions described above. Series of PCR products were electrophoresed on a 1.5% agarose gel, stained with Vistra green (Amersham, Heidelberg, Germany), exposed to a FluorImager (Molecular Dynamics, Sunnyvale, CA) and quantitated using ImageQuant Software version 4.1 (Molecular Dynamics).
To calculate the initial amount of target transcripts, a range of dilutions in which the ratio of the products from the competitor to the products from the target was between 0.2 and 5.0 were taken. Then, log(products from competitor/products from target) was plotted against log(initial molecular amount of added competitor). A regression analysis revealed that this provided a near-linear curve in each case. The initial amount of target DNA was then determined from the equivalence point of the curve, where log(products from competitor/products from target) = 0. Expression levels of eachDNMT were displayed as relative values by setting the mean of the levels in normal bone samples as 1, after normalizing the levels of DNMT by those of GAPDH or PCNA in the same sample.
Bisulfite modification and methylation-specific PCR
One μg genomic DNA was denatured in 50 μL 0.2 M NaOH at 37°C for 10 minutes and then added with 30 μL 10 mM hydroquinone (Sigma, St Louis, MO) and 520 μL 3 M sodium bisulfite (pH 5.0) (Sigma). Samples were incubated at 50°C for 16 hours under mineral oil. Modified DNA was purified using the Wizard purification resin and the Vacuum Manifold (Promega) and eluted into 50 μL water. After addition of 5.6 μL of 3 M NaOH (final 0.3 M), samples were let stand at room temperature for 5 minutes for final desulphonation. After ethanol precipitation, samples were dissolved in 50 μL water. Bisulfite treatment of DNA converts all unmethylated, but not methylated, cytosines to uracils.38
Based on the sequence differences resulting from this modification, methylation-specific PCR (MSP) was performed for thep15INA4B gene with the primer sets designed by Herman and colleagues (Table 1).39 PCR was performed in a 50-μL reaction mixture containing 100 ng bisulfite modified DNA, 1 × PCR reaction buffer (Promega), 0.2 mM of each dNTP, 1.5 mM MgCl2, 20 pmol of each primer, and 5% dimethylsulfoxide. Reactions were initiated by a hot start procedure: an initial denaturation at 95°C for 5 minutes, a further incubation at 98°C for 30 seconds, and then an addition of 2.0 U Taq-DNA polymerase (Promega). This was followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. A final extension was done at 72°C for 10 minutes. Products were loaded on a 2% agarose gel, stained with ethidium bromide, and observed under UV illumination.
Statisticalanalysis
To compare the DNMT levels between different sample categories, the Kruskal-Wallis test with the Bonferroni method for multiple comparisons was used. The strength of the association between the expression levels of each DNMT in different sample categories was calculated by the Spearman rank-correlation coefficient. The Mann-Whitney U test was used to compare the expression levels of DNMTs between 2 sample groups with or withoutp15INAK4B methylation.
Results
Expression of DNMTs in normal hematopoiesis and leukemia cell lines
We first examined whether the 3 DNMT genes were expressed in normal hematopoiesis. By a standard RT-PCR method, transcripts from all DNMTs were detected in peripheral neutrophils, monocytes, T lymphocytes, total bone marrow cells, CD34+ bone marrow cells, and various colonies derived from the CD34+ cells (Figure 1A-C). However, DNMT3Bwas expressed at levels lower than those of the other 2 genes, because 5 or more additional PCR cycles were required to obtain band signals comparable to those of DNMT1 and 3A. DNMT3B expression was especially low in differentiated cell populations such as neutrophils, monocytes (Figure 1A), and colonies of both the myeloid and erythroid lineages (Figure 1C). In contrast, total bone marrow cells and CD34+ cells expressedDNMT3B at somewhat higher levels (Figure 1A). We also examined 3 established leukemia cell lines HL60, KU812, and K562, by a standard PCR method and found that all 3 genes are respectively expressed at levels higher than those in normal hematopoiesis (Figure1D). This suggests that overexpression of DNMTs might have some relevance to leukemic transformation (discussed later).
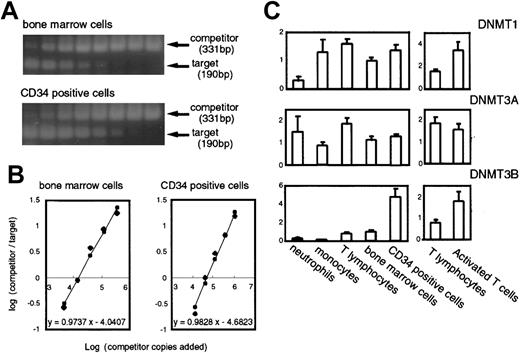
To obtain more accurate information on DNMT expression in normal hematopoiesis, we adopted the competitive PCR method using a synthetic mimic DNA as an internal control. Competitive PCR is an effective method to address an expression level of a certain gene37, however, it has been reported that there could be some pitfalls for making an absolute estimation of targets.40 41 In this study, we demonstrated the results as fold-increase or decrease relative to the mean value of the same gene in control bone marrow cells after the normalization of the level of expression of each DNMT in a particular sample byGAPDH (a housekeeping control) or PCNA (a cell proliferation marker) in that sample (Figures2 and 3). Examples showing actual PCR-amplified bands from both competitors and targets (Figure 2A) and graphs used to measure the expression levels of the transcripts (Figure 2B) are shown.
Expression levels of
DNMTs in normal hematopoietic cells measured by competitive PCR. (A) Examples of the competitive PCR assay forDNMT3B. Phosphorimages of 2 agarose gels stained with Vistra green are shown. The lanes contained the products amplified from a constant amount of the cDNA mixture, and the competitor DNA prepared in successive 3-fold dilutions. (B) Graphs show the regression analysis of the results obtained in panel A. The initial amount of target DNA was estimated by the point where log(products from competitor/products from target) = 0. (C) Levels of the DNMT transcripts in normal hematopoietic cells. The number of samples are neutrophils, n = 3; monocytes, n = 3; peripheral T cells, n = 3; PHA-activated T cells, n = 3; bone marrow cells, n = 6; CD34+ cells, n = 5. The levels of DNMTs are displayed as relative values calculated such that the mean of the normal bone marrow cells would equal a value of 1 after correcting the variations by the levels ofGAPDH.
Expression levels of
DNMTs in normal hematopoietic cells measured by competitive PCR. (A) Examples of the competitive PCR assay forDNMT3B. Phosphorimages of 2 agarose gels stained with Vistra green are shown. The lanes contained the products amplified from a constant amount of the cDNA mixture, and the competitor DNA prepared in successive 3-fold dilutions. (B) Graphs show the regression analysis of the results obtained in panel A. The initial amount of target DNA was estimated by the point where log(products from competitor/products from target) = 0. (C) Levels of the DNMT transcripts in normal hematopoietic cells. The number of samples are neutrophils, n = 3; monocytes, n = 3; peripheral T cells, n = 3; PHA-activated T cells, n = 3; bone marrow cells, n = 6; CD34+ cells, n = 5. The levels of DNMTs are displayed as relative values calculated such that the mean of the normal bone marrow cells would equal a value of 1 after correcting the variations by the levels ofGAPDH.
Expression levels of
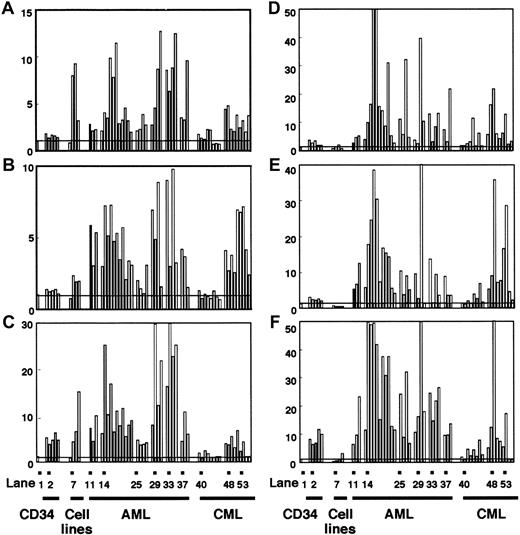
DNMTs in normal cells and individual leukemia cases measured by competitive PCR. Expression levels of DNMT1(A,D), 3A (B,E), and 3B (C,F) in CD34+ cells, leukemia cell lines, AML, and CML were assayed by competitive PCR. The levels of DNMTs were displayed as in Figure 2, but normalization was done by using the levels of eitherGAPDH (A-C) or PCNA (D-F). Lane 1 shows the mean value of the levels in normal bone marrow cells; lanes 2-6, CD34+ cells; lane 7, EBV-transformed B-cell line; lane 8, HL60; lane 9, KU812; lane 10, K562; lane 11, AML (FAB subtype M0); lanes 12 and 13, AML (M1); lanes 14-24, AML (M2); lanes 25-28, AML (M3); lanes 29-31, AML (M4); lane 32, AML (M5); lanes 33 and 34, AML (M6); lanes 35 and 36, AML (M7); lanes 37-39, AML (mixed lineage); lanes 40-47, CML (chronic phase); lanes 48-52, CML (myeloid crisis); lanes 53-56, CML (lymphoid crisis).
Expression levels of
DNMTs in normal cells and individual leukemia cases measured by competitive PCR. Expression levels of DNMT1(A,D), 3A (B,E), and 3B (C,F) in CD34+ cells, leukemia cell lines, AML, and CML were assayed by competitive PCR. The levels of DNMTs were displayed as in Figure 2, but normalization was done by using the levels of eitherGAPDH (A-C) or PCNA (D-F). Lane 1 shows the mean value of the levels in normal bone marrow cells; lanes 2-6, CD34+ cells; lane 7, EBV-transformed B-cell line; lane 8, HL60; lane 9, KU812; lane 10, K562; lane 11, AML (FAB subtype M0); lanes 12 and 13, AML (M1); lanes 14-24, AML (M2); lanes 25-28, AML (M3); lanes 29-31, AML (M4); lane 32, AML (M5); lanes 33 and 34, AML (M6); lanes 35 and 36, AML (M7); lanes 37-39, AML (mixed lineage); lanes 40-47, CML (chronic phase); lanes 48-52, CML (myeloid crisis); lanes 53-56, CML (lymphoid crisis).
As shown in Figure 2C, expression of DNMT1 was rather uniform in various cell types except neutrophils, where its expression was relatively weak. DNMT3A was expressed at higher levels in T lymphocytes and neutrophils than in other cells although the differences were not so distinct (Figure 2C). It was characteristic, however, that CD34+ cells expressed DNMT3B at a level about 5-fold higher than that in normal bone marrow cells (Figures 2C and 1A). Furthermore, in contrast to the CD34+cells, differentiated cells such as neutrophils and monocytes showed very low levels of DNMT3B expression (Figure 2C), indicating that this gene is dramatically down-regulated on hematopoietic cell differentiation, at least in some lineages. In addition, on activation with PHA, T lymphocytes showed a 2-fold increase in expression ofDNMT1 and 3B (Figures 2C and 1B). Thus, all 3DNMTs were expressed at detectable levels in normal hematopoietic cells, and their expression levels varied among cell types and differentiation conditions.
DNMTs were overexpressed in AML
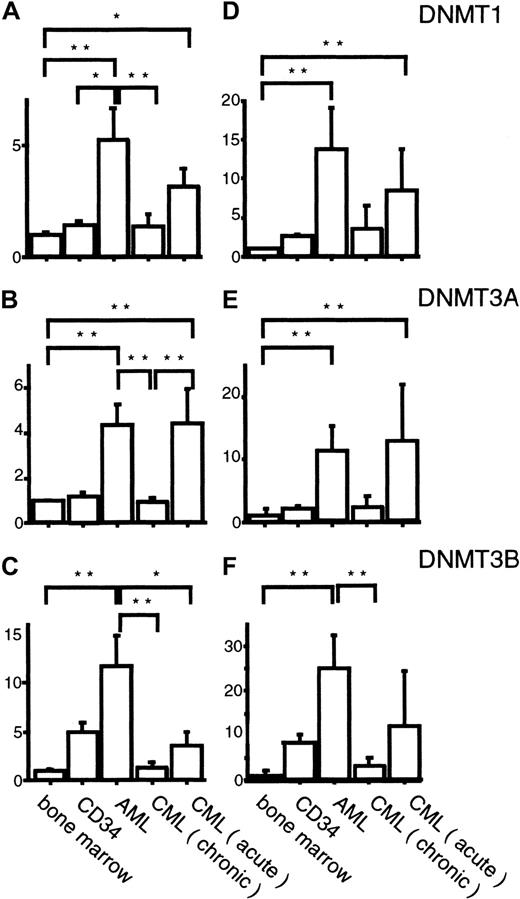
To know the possible role of DNMTs in leukemogenesis, we analyzed their expression levels in 33 AML cases by competitive PCR. When normalized by the GAPDH levels, we observed 5.3-, 4.4-, and 11.7-fold mean increases in DNMT1, 3A, and3B levels, respectively, compared with their levels in normal bone marrow cells (Figures 3A-C and 4A-C), although the exact level varied from case to case (Figure 3A-C). These increases in the mean expression levels of DNMTs were statistically significant (Figure4A-C). It is noteworthy that among the 3 DNMT genes,DNMT3B showed the largest fold increase (11.7-fold; Figure4C). However, when the mean DNMT3B level in AML cells was compared with that in CD34+ cells, only 2.4-fold mean increase was observed (Figure 4C). It is also interesting that all 4 M3 AML patients expressed lower levels of all DNMTs compared with the other subtypes (Figure 3A-C) although the number of cases studied was rather small.
Summary of the expression levels of
DNMTs in AML and CML measured by competitive PCR.Expression levels of DNMTs, corrected by the levels ofGAPDH (A-C) or PCNA (D-F) transcripts are shown. The Kruskal-Wallis test with the Bonferroni method for multiple comparison was used to compare the levels of each DNMT in different sample categories. *, ** indicates statistical significance (*, P < .05; **, P < .01).
Summary of the expression levels of
DNMTs in AML and CML measured by competitive PCR.Expression levels of DNMTs, corrected by the levels ofGAPDH (A-C) or PCNA (D-F) transcripts are shown. The Kruskal-Wallis test with the Bonferroni method for multiple comparison was used to compare the levels of each DNMT in different sample categories. *, ** indicates statistical significance (*, P < .05; **, P < .01).
Several reports pointed out that, in colon cancer, DNMTsappear to be overexpressed when the levels are normalized by those of a housekeeping gene but not overexpressed when corrected by those of cell proliferation marker genes.42 43 To exclude the possibility that the observed overexpression was due to accelerated cell proliferation of the AML cells, we corrected the levels ofDNMTs by those of a cell proliferation markerPCNA (Figures 3D-F and 4D-F). As a result, the overexpression of DNMTs in 3 leukemia cell lines was clearly canceled (Figure 3D-F). However, the overexpression of allDNMTs in AML was not affected, or became even more evident, by this treatment (Figures 3D-F and 4D-F).
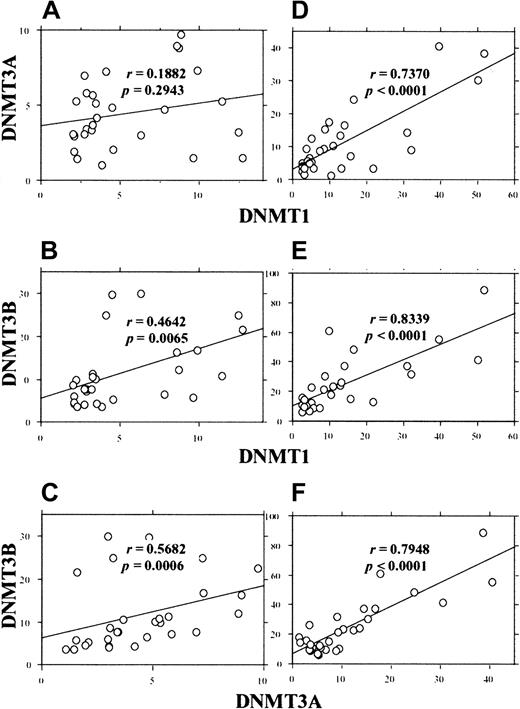
Because the extent of overexpression varied among the AML cases, we next investigated whether the 3 DNMT genes are overexpressed coordinately or independently. As shown in Figure5, analysis of the expression levels in AML cases showed coordinate overexpression of DNMTs. When the values obtained by GAPDH correction were used, moderate correlations between DNMT1 and 3B(r = 0.464, P = .0065) and betweenDNMT3A and 3B (r = 0.568,P = .0006) were observed (Figure 5B,C). Furthermore, when the values by PCNA correction were used, we observed strong correlations between DNMT1 and 3A(r = 0.737, P < .0001), DNMT1 and3B (r = 0.8339, P < .0001), andDNMT3A and 3B (r = 0.794,P < .0001) (Figure 5D-F).
Correlation between the levels of different
DNMTs in individual AML cases. The levels ofDNMTs were corrected by GAPDH (A-C) orPCNA (D-F) and the correlation coefficients were calculated by the Spearman rank-correlation method.
Correlation between the levels of different
DNMTs in individual AML cases. The levels ofDNMTs were corrected by GAPDH (A-C) orPCNA (D-F) and the correlation coefficients were calculated by the Spearman rank-correlation method.
Expression of DNMTs in CML
Chronic myelogenous leukemia is distinct from AML with its phasic clinical course that reflects the clonal evolution of leukemia cells. We investigated the expression levels of DNMTs in CML cells in each clinical phase. In contrast to AML, CML cells in the chronic phase expressed DNMTs at levels almost equal to those in normal bone marrow cells (Figures 3A-C and 4A-C). CML cells in the acute phase showed 3.2-, 4.5-, and 3.4-fold mean increases in the expression levels of DNMT1, 3A, and3B, respectively, compared with their levels in normal bone marrow cells (Figures 3A-C and 4A-C). As in AML, the overexpression was not abolished even when the expression levels were corrected byPCNA (Figures 3D-F and 4D-F). It is of interest that the overexpression of DNMT3B was not evident in acute phase CML when the mean expression level was compared to that in CD34+ cells (Figures 3C and 4C).
Methylation status of p15INAK4Band its correlation with DNMT overexpression in AML
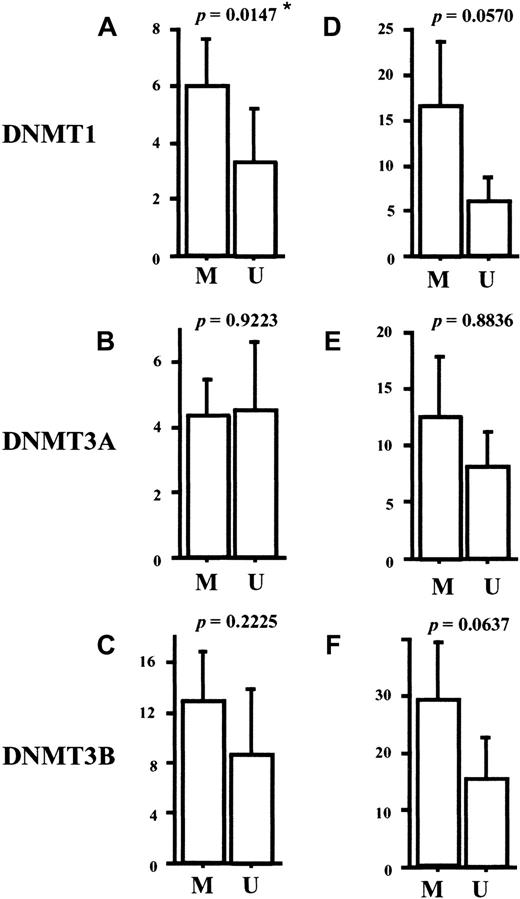
It has been reported that a tumor suppressor gene,p15INA4B, is frequently methylated and silenced in AML.19,20 Under the physiologic condition, expression of p15INA4B is induced by transforming growth factor-β (TGF-β)44 and its protein product inhibits cell cycle progression.45 To explore the possible role of the DNMT overexpression in p15INA4Bmethylation, we investigated the methylation status of the promoter region of this gene in our AML samples. By using methylation-specific PCR (Figure 6 represents the results),39 we observed thatp15INAK4B was methylated in 24 (72%) of the 33 cases of AML (Table 2). Interestingly, the AML cases with methylated p15INAK4B had a tendency to express higher levels of DNMT1, 3B, and potentially 3A, compared to the cases with unmethylatedp15INAK4B (Figure7). Although a statistically significant result was obtained only for DNMT1 (P = .0147), our findings suggests that the overexpressed DNMTs play a role in aberrant regional hypermethylation observed in AML.
Methylation-specific PCR for the p15INAK4Bpromoter in normal bone marrow cells and leukemia samples.
Three of the 4 AML cases gave the methylation-specific PCR band, whereas normal bone marrow cells and CML cases showed only the unmethylated DNA-specific band. M-primer indicates primers specific for methylated DNA; U-primers, primers specific for unmethylated DNA.
Methylation-specific PCR for the p15INAK4Bpromoter in normal bone marrow cells and leukemia samples.
Three of the 4 AML cases gave the methylation-specific PCR band, whereas normal bone marrow cells and CML cases showed only the unmethylated DNA-specific band. M-primer indicates primers specific for methylated DNA; U-primers, primers specific for unmethylated DNA.
Summary of the methylation status of p15INAK4Bin AML
| FAB type . | No. of patients . | No. of methylated cases (%) . |
|---|---|---|
| M0 | 1 | 0 (0) |
| M1 | 2 | 1 (50) |
| M2 | 11 | 8 (72) |
| M3 | 4 | 3 (75) |
| M4 | 3 | 3 (100) |
| M5 | 1 | 1 (100) |
| M6 | 2 | 2 (100) |
| M7 | 2 | 1 (50) |
| Mix | 3 | 2 (66) |
| Total | 29 | 21 (72) |
| FAB type . | No. of patients . | No. of methylated cases (%) . |
|---|---|---|
| M0 | 1 | 0 (0) |
| M1 | 2 | 1 (50) |
| M2 | 11 | 8 (72) |
| M3 | 4 | 3 (75) |
| M4 | 3 | 3 (100) |
| M5 | 1 | 1 (100) |
| M6 | 2 | 2 (100) |
| M7 | 2 | 1 (50) |
| Mix | 3 | 2 (66) |
| Total | 29 | 21 (72) |
Comparisons of the expression levels of
DNMTs between the AML groups with or withoutp15INAK4B methylation. The levels of DNMTs were corrected by GAPDH (A-C) orPCNA (D-F). The level of DNMT1 corrected byGAPDH is significantly higher in the group with methylatedp15INAK4B than in the group with unmethylatedp15INAK4B (A) (P = .0147). Comparisons were made by the Mann-Whitney U test. M indicates methylated cases; U, unmethylated cases.
Comparisons of the expression levels of
DNMTs between the AML groups with or withoutp15INAK4B methylation. The levels of DNMTs were corrected by GAPDH (A-C) orPCNA (D-F). The level of DNMT1 corrected byGAPDH is significantly higher in the group with methylatedp15INAK4B than in the group with unmethylatedp15INAK4B (A) (P = .0147). Comparisons were made by the Mann-Whitney U test. M indicates methylated cases; U, unmethylated cases.
Discussion
In the present paper, we have described the expression levels of the 3 DNMTs in normal hematopoiesis and leukemia cells. We first found that all DNMTs are expressed at detectable levels in normal hematopoietic cells. However, there are some gene-specific features such as the down-regulation ofDNMT3B on differentiation of hematopoietic progenitor cells and the induction of both DNMT1 and 3B on activation of peripheral T cells. These findings are of interest from the viewpoint of hematopoietic cell differentiation because it has been reported that some hematopoietic cell-specific genes, such as those coding for myeloperoxidase,46 globin,47c-fms,48 and G-CSF receptor,49 are regulated by methylation in a lineage- and differentiation-dependent manner. In addition, it is known that differentiation of naive T cells into Th1 or Th2 cells is accompanied by changes in methylation at the genes coding for cytokines such as interferon-γ, IL-4, and IL-13.50Furthermore, the importance of DNMT3B in the hematopoietic system has been also shown in ICF (immunodeficiency, centromeric instability, facial anomalies) syndrome. ICF syndrome is a rare autosomal recessive disorder characterized by variable immunologic defects, such as a decreased number of lymphocytes and variable reduction in serum immunoglobulin levels,51,52 and recently, mutations were identified in DNMT3B in some ICF patients.28,53 54 Although how the defects inDNMT3B cause immunologic disorders remains unknown, it has been proposed that a loss of the proper methylation pattern may impair stage-specific gene regulation, resulting in interference with lymphocyte maturation. Therefore, the expression of DNMTs in hematopoietic cells is thought to have important roles for the differentiation process of blood cells.
We next found that all DNMTs are substantially overexpressed in most cases of AML when compared to normal bone marrow cells. Also, all DNMTs are expressed at higher levels in AML cells than in CD34+ cells, although this is statistically significant only for DNMT1. In addition, this overexpression observed in AML is not due to the accelerated proliferation of the leukemic cells because the data are basically unaffected even after corrected by the levels of a cell proliferation marker, PCNA. Furthermore, DNMTs were coordinately, not independently, up-regulated in AML cells. These results suggest that AML cells may possess higher maintenance and de novo methyltransferase activities than normal hematopoietic cells and these methyltransferases may contribute to the leukemogenesis by inducing aberrant methylation. Among the FAB subtypes, however, M3 (4 cases) showed only a minimal increase in the expression levels of DNMTs. Although this observation has to be confirmed in other M3 cases, it suggests that the possible roles of DNMTs in leukemogenesis vary among subtypes of AML.
In contrast to AML, CML shows phase-dependent expression ofDNMTs. In the chronic phase, levels of DNMTs are not significantly different from those in normal bone marrow cells. However, CML cells in the acute phase expressed higher levels ofDNMTs than normal bone marrow cells. DNMT1 and3A, but not 3B, are also expressed at higher levels in acute phase CML cells than in CD34+ cells. Thus, the expression pattern of DNMTs in CML was different from that of AML, suggesting that there might be different roles ofDNMTs for leukemogensis. The near-normal expression ofDNMTs in chronic phase is compatible with the fact that the increased cell population in this phase is seemingly mature myeloid cells with essentially normal morphology and function. These results also suggest that DNMTs are not involved in the onset or maintenance of the chronic phase but could be associated with blast crisis. Because the ABL1 promoter nested within theBCR-ABL fusion gene has been demonstrated to be methylated with disease progression,17 18 it is conceivable that the overexpressed DNMTs methylate the ABL1 promoter region, and this in turn leads to clonal evolution. In the future study, it is important to know whether DNMTs are overexpressed prior to blastic crisis in progenitor cells of CML.
The overexpressed DNMTs could be involved in the development of leukemia by inducing hypermethylation of tumor suppressor genes. We focused on p15INAK4b because this gene frequently becomes inactivated with disease progression in acute leukemia and MDS by hypermethylation of its 5′ CpG island.19-21 The protein product ofp15INAK4B is a cell cycle regulator induced by TGF-β44 and involved in the inhibition of G1phase progression.45 It has been reported that a loss of sensitivity of leukemia cells to growth inhibition by TGF-β is correlated with the inactivation ofp15INAK4B.19 Consistent with the previous reports, this gene was methylated in 23 of the 33 (72%) cases with AML. Furthermore, we observed that the AML cases with hypermethylated p15INAK4B had a tendency to show higher levels of DNMT1 and 3B although the correlation was statistically significant only for DNMT1. These results suggest that up-regulated DNMTs could have roles for p15INAK4B methylation, although further studies will be required to test this possibility.
We studied the expression levels of DNMTs to gain a clue to their roles in aberrant regional hypermethylation in leukemia. AlthoughDNMTs were substantially overexpressed in leukemia cells in a leukemia type- and stage-specific manner, the mechanism responsible for aberrant methylation remains to be solved. As for DNMT1, it has been proposed that a loss of p21 function in cancer facilitates the formation of DNA-DNMT1-PCNA complexes, resulting in aberrant methylation of CpG island.55,56 However, the mechanisms of recently identified de novo methyltransferases, DNMT3A and3B, for aberrant methylation are largely unknown. In the study of D. melanogaster, induced expression of bothDnmt1 and Dnmt3a cooperated to establish and maintain methylation, resulting in increased methylation.27 As we have also observed coordinate overexpression of DNMTs in AML, there is a possibility thatDNMTs act cooperatively in leukemia for the establishment of aberrant hypermethylation. It should also be noted thatDNMT3B has at least 4 alternatively spliced transcripts, which are expressed in a tissue-specific manner.25 Because 2 of the splice variants lack the highly conserved sequence motifs in the methyltransferase catalytic domain, it will be important to know which splice variants are overexpressed in leukemia.
In conclusion, the DNMT genes were expressed constitutively in normal hematopoiesis and were overexpressed in some types of leukemia. The overexpressed DNMTs with maintenance or de novo methyltransferase activity may contribute to the pathogenesis of leukemia by inducing aberrant regional hypermethylation. Further studies will be needed to clarify the precise pathogenetic roles of DNMTs in clonal evolution of leukemia and to develop therapeutic alternatives designed to suppress abnormal hypermethylation.
The authors are indebted to Dr Naoko Kinukawa (Department of Medical Informatics, Faculty of Medicine, Kyushu University) for statistical analysis of the data.
First Department of Internal Medicine, Faculty of Medicine, Kyushu University, Fukuoka, Japan; Division of Disease Genes, Institute of Genetic Information, Kyushu University, Fukuoka, Japan; and Division of Human Genetics, Department of Integrated Genetics, National Institute of Genetics, and Department of Genetics, Graduate University for Advanced Studies, Mishima, Shizuoka, Japan.
Submitted June 15, 2000; accepted November 6, 2000.
Supported in part by grants from the Ministry of Health and Welfare of Japan (Research on Human Genome and Therapy), the Ministry of Education, Science, Sports and Culture of Japan, and Uehara Memorial Foundation of Japan. S.M. is supported by a fellowship from the Japan Society for Promotion of Sciences. T.C. was a research resident supported by Japan Human Sciences Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Shin-ichi Mizuno, First Department of Internal Medicine, Faculty of Medicine, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka, 812-8582, Japan; e-mail:smizuno@gen.kyushu-u.ac.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal