A large east Texas family with autosomal dominant inheritance of a novel bleeding disorder has been identified. The disorder is characterized clinically by easy bruising, life-threatening bleeding with trauma or surgery, and menorrhagia in affected women. Laboratory studies demonstrated prolongation of the prothrombin time and activated partial thromboplastin time in affected individuals. Paradoxically, assays of known coagulation factors are all within normal limits. To determine the molecular basis of this disease, a candidate gene linkage analysis in this kindred was done. Initially it was hypothesized that the cause of the disease in this family could be an antithrombin III (AT3) mutation that resulted in a constitutively active AT3 in the absence of heparin binding. Linkage studies using DNA from the family and an intragenic polymorphic marker within the AT3 gene showed that the disease mapped to this locus. The coding region and intron/exon junctions of AT3were sequenced using the proband's DNA, but this analysis failed to identify a mutation. Additional family members were recruited for the study, and 16 polymorphic markers around the AT3 gene were analyzed. Using 2 recombinants, the critical interval for the defective gene was narrowed to approximately 1.5 Mb, centromeric toAT3. The factor V (FV) gene was mapped into the disease interval and sequenced; there were no mutations found. Elucidation of the genetic defect causing the bleeding disorder in this family may reveal a novel protein involved in the coagulation cascade.
Introduction
Inherited bleeding disorders have a variety of etiologies including impaired function of platelets, blood vessels, blood coagulation pathways, the anticoagulant pathways, or the fibrinolytic system. The clinical manifestations and laboratory findings of this phenotype vary greatly between and within families. The severity of the underlying protein defect often determines the clinical presentation of affected patients, and bleeding may vary from spontaneous hemorrhage to significant bleed loss following surgery or physical trauma. Hemophilia A, hemophilia B, and von Willebrand disease (vWD) comprise more than 80% of all inherited bleeding disorders.1-4
A large kindred from east Texas has been identified with a novel bleeding disorder that is inherited in an autosomal dominant manner. The bleeding disorder is moderate in severity and characterized by a prolonged prothrombin time (PT) and activated partial thromboplastin time (aPTT) and normal levels of all known coagulation factors. The disorder has been mapped to a 1.47-Mb region at chromosome 1q23, and known candidate genes at this location have been excluded as the cause of the disease in this family. Identification of the defective gene causing this disorder may identify a previously unrecognized protein involved in the coagulation cascade and further expand our knowledge about the proteins involved in this system.
Patients, materials, and methods
Clinical description
We studied a Texas family of 4 generations with 46 members (Figure 1). The proband (III:10) was a 35-year-old man with a bleeding diathesis since childhood characterized by bruising, epistaxis, bleeding from gums, and significant bleeding after minor trauma, but no hemarthrosis or spontaneous hematomas. The proband required 2 blood transfusions as a child: at the age of 2 years, after sustaining a laceration from a fall, and at 12 years, following a tooth extraction. The PT was 18.4 seconds (normal range, 11.1-13.1 seconds), and the aPTT was 48.7 seconds (normal range, 25-34 seconds) (Table 1). There was a modest variability of these assays from day to day. The following studies were all normal: thrombin time, functional fibrinogen level, von Willebrand antigen and ristocetin cofactor, bleeding time, and platelet number and aggregation. The following factor activity levels were normal: II, V, VII, VIII, IX, X, XI, and XII. Factor V and X levels were the same, with serial dilutions up to 1:16, and the dilution curves were parallel to the standard curve. The protein C activity and resistance ratio was normal. Mixing studies were performed to detect a circulating inhibitor (Table2).
Pedigree of the family with the inherited bleeding disorder.
Closed symbol indicates affected; open symbol, unaffected; and open symbol with ?, disease status unknown. The PT and aPTT are indicated after the generation and family number; ND indicates not determined. Genetic haplotypes are indicated for all individuals who were genotyped. The filled black bar indicates the haplotype cosegregating with the disease. Recombinations are indicated by open bars.
Pedigree of the family with the inherited bleeding disorder.
Closed symbol indicates affected; open symbol, unaffected; and open symbol with ?, disease status unknown. The PT and aPTT are indicated after the generation and family number; ND indicates not determined. Genetic haplotypes are indicated for all individuals who were genotyped. The filled black bar indicates the haplotype cosegregating with the disease. Recombinations are indicated by open bars.
Laboratory evaluation of the proband
| Laboratory test . | Proband data . | Normal range . |
|---|---|---|
| PT, seconds | 18.4 | 11.1-13.1 |
| aPTT, seconds | 48.7 | 25.0-34.0 |
| Fibrinogen, (μmol/L) | 6.1 | 4.9-12.1 |
| (mg/dL) | (208) | (170-410) |
| Factor II, % | 94 | 83-117 |
| Factor V, % | 111 | 50-150 |
| Factor VII, % | 70 | 65-135 |
| Factor VIII, % | 59 | 55-145 |
| Factor IX, % | 74 | 60-140 |
| Factor X, % | 81 | 45-156 |
| Factor XI, % | 76 | 75-135 |
| Factor XII, % | 82 | 50-150 |
| Thrombin time, seconds | 20.1 | 18.0-22.0 |
| Antithrombin III function, % | 90 | 80-180 |
| Antithrombin III antigen, mg/L | 260 | 210-350 |
| (mg/dL) | (26) | (21-35) |
| vWF, ristocetin cofactor | 54 | 45-140 |
| Bleeding time, minutes | 6.0 | 3.0-8.0 |
| Platelet number and aggregation | Normal | — |
| RVVT, seconds | 26.6 | 25-30 |
| Protein C, % | 68 | 55-140 |
| Activated protein C resistance, ratio | 2.8 | 2.2-4.0 |
| Laboratory test . | Proband data . | Normal range . |
|---|---|---|
| PT, seconds | 18.4 | 11.1-13.1 |
| aPTT, seconds | 48.7 | 25.0-34.0 |
| Fibrinogen, (μmol/L) | 6.1 | 4.9-12.1 |
| (mg/dL) | (208) | (170-410) |
| Factor II, % | 94 | 83-117 |
| Factor V, % | 111 | 50-150 |
| Factor VII, % | 70 | 65-135 |
| Factor VIII, % | 59 | 55-145 |
| Factor IX, % | 74 | 60-140 |
| Factor X, % | 81 | 45-156 |
| Factor XI, % | 76 | 75-135 |
| Factor XII, % | 82 | 50-150 |
| Thrombin time, seconds | 20.1 | 18.0-22.0 |
| Antithrombin III function, % | 90 | 80-180 |
| Antithrombin III antigen, mg/L | 260 | 210-350 |
| (mg/dL) | (26) | (21-35) |
| vWF, ristocetin cofactor | 54 | 45-140 |
| Bleeding time, minutes | 6.0 | 3.0-8.0 |
| Platelet number and aggregation | Normal | — |
| RVVT, seconds | 26.6 | 25-30 |
| Protein C, % | 68 | 55-140 |
| Activated protein C resistance, ratio | 2.8 | 2.2-4.0 |
PT indicates prothrombin time; aPTT, activated partial thromboplastin time, AT3, antithrombin III; vWF, von Willebrand factor; RVVT, Russell viper venom time.
Various concentrations of patient's plasma mixed with control plasma and the clotting times determined immediately and one hour later
| Control plasma, % . | Patient plasma, % . | aPTT (immediate), seconds . | aPTT, (1 h), seconds . |
|---|---|---|---|
| 100 | 0 | 28.7 | 30.7 |
| 80 | 20 | 28.2 | 31.1 |
| 60 | 40 | 30.1 | 34.6 |
| 50 | 50 | 29.7 | 33.3 |
| 40 | 60 | 32.7 | 35.5 |
| 20 | 80 | 33.1 | 39.7 |
| 0 | 100 | 42.1 | 47.3 |
| Control plasma, % . | Patient plasma, % . | aPTT (immediate), seconds . | aPTT, (1 h), seconds . |
|---|---|---|---|
| 100 | 0 | 28.7 | 30.7 |
| 80 | 20 | 28.2 | 31.1 |
| 60 | 40 | 30.1 | 34.6 |
| 50 | 50 | 29.7 | 33.3 |
| 40 | 60 | 32.7 | 35.5 |
| 20 | 80 | 33.1 | 39.7 |
| 0 | 100 | 42.1 | 47.3 |
The assays were done at 37°C.
aPTT indicates activated partial thromboplastin time.
The proband's sister (III:18) also had bleeding problems including easy bruising and menorrahagia. As a child she had excessive bleeding with shedding of her baby teeth, which required packing with gauze. At the age of 7 years, 3 days following a tonsillectomy, she had an onset of bleeding that required an infusion of 2 units of blood and 2 units of plasma. Excessive bleeding also occurred with the removal of an ingrown toenail and a tooth extraction, but no transfusions were required. At 15 years, she was operated on for possible appendicitis and was found to have a right hemorrhagic ovarian cyst. She received a blood transfusion postoperatively. Her PT was prolonged at 13.7 seconds (normal range, 9.5-12 seconds), and her aPTT was prolonged at 51.1 seconds (normal range, 29-39 seconds), but all factor assays, thrombin time, functional fibrinogen level, von Willebrand antigen and ristocetin cofactor, bleeding time, and platelet number and aggregation were normal. At 18 years she underwent labor and delivery without bleeding complications. At 19 years she was again hospitalized for ovarian cystic hemorrhage, but did not require a transfusion.
The proband's brother (III:12) had epistaxis, bleeding gums during tooth brushing, and easy bruising, but no other problems. His PT and aPTT were prolonged (Figure 1). Another sister of the proband (III:20) had recurrent episodes of epistaxis and bleeding that began during her childhood. She had an appendectomy and gave birth to her first child without bleeding complications. Delivery of her second child was complicated by excessive bleeding, and she required a blood transfusion.
Other members of the family were clinically assessed, and the PT and aPTT were determined (Figure 1). For the purpose of this study, affected members were defined by the presence of clinical manifestations of the bleeding disorder and either a mild or moderately prolonged PT and/or aPTT. The clinical features of the bleeding disorder include severe postsurgical bleeding and easy bruising, and affected women also experienced menorrhagia. Informed consent to draw blood for the hematological studies and DNA isolation was obtained from 29 of these family members, 16 affected individuals, and 13 unaffected individuals. Nine individuals contributed DNA (buccal cells) but did not consent to have their blood drawn to confirm prolongation of either the PT or aPTT. The genotypes of these individuals are included in the pedigree, but the phenotypes were scored as unknown for the statistical analysis. There is no known consanguinity in the family.
Biochemical test for coagulation
The screening coagulation assays were performed with an aPTT reagent, Platelin (Organon Teknika, Durham, NC) and a PT reagent, Thromboplastin C+ (Dade Behring, Miami, FL) in an automatic coagulation analyzer, Electra 1600 C (Dade, Deerfield, IL). The prothrombin times were also performed using another thromboplastin reagent, Simplastin (General Diagnostic, Morris Plains, NJ), which demonstrated similar prolongation. The affected family members also had prolonged coagulation tests in other hospitals using other assay systems. Assays for coagulation factors XII, XI, IX, and VIII were measured using a PTT reagent (Platelin), and factors VII, X, and V and prothrombin assays were measured using the PT reagent Thromboplastin C+. Inhibitor screens were performed with a PTT reagent (Reactif APTT, Sigma Chemical, St Louis, MO) in a fibrometer (BBL Fibrosystem, Becton Dickinson, San Jose, CA). Various concentrations of plasma from the proband were incubated with plasma from control subjects (Instrumentation Laboratory, Lexington, MA), and each aPTT was determined following incubation for 1 hour at 37°C. RVVT was performed as described previously.5
Genotyping
Blood or buccal cell samples were collected from all consenting individuals, and DNA was prepared using standard methods. The intragenic AT3 marker and microsatellite markers from chromosome 1 were selected using the most recent Massachusetts Institute of Technology (MIT) STS map,6 the GDB database,7 the CHLC database,8 and previous physical maps of this region.9-12 The forward primer of each polymerase chain reaction (PCR) primer pair (Research Genetics, Huntsville, AL) was end-radiolabeled with phosphorous-33 γ–adenosine 5′-triphosphate (33P-γATP) using polynucleotide kinase. The amplifications were performed in a final volume of 25 μL containing 50 ng DNA, 50 ng of each primer, 10 ng end-labeled primer, 0.25 μM each dinucleoside 5′-triphosphate (dNTP), 1.5 mM magnesium dichloride (MgCl2), 1 times Taq buffer, and 0.5 U Taq polymerase (Gibco-BRL Life Technologies, Rockville, MD). The products were electrophoresed through 7% denaturing polyacrylamide gels using M13mp18 C & T sequence ladders as sizing markers. The products were visualized by autoradiography.13
PCR amplification and sequencing
Primers were designed to amplify each individual exon of the AT3 and the FV genes along with each of the 5′ and 3′ flanking intronic sequences based on the genomic sequence of these genes.14-18 Amplification conditions included 100 ng genomic DNA, 100 ng of each primer, 20 μM of each dNTP, 1 × Taq buffer, 1.5 mM MgCl2, and 1.5 U Taq polymerase. PCR products were purified by excision from a 1.7% agarose gel and purified using QIAquick spin columns (Qiagen, Valencia, CA). The fragments were sequenced with an ABI 377 DNA sequencer (Department of Microbiology Sequencing Core Facility, University of Texas–Houston Medical School).
Linkage analysis
Two-point pair-wise linkage analysis was performed using the MLINK program of the LINKAGE package and the version FASTLINK 2.2.19-22 Multipoint analysis was performed between the disease locus and 7 markers mapping to the subtelomeric region of chromosome 1q using the LINKMAP program. The disease allele frequency was set at 0.0001. Haplotypes were reconstructed from the genotype data with the GENEHUNTER program.23
Construction of the BAC map for the critical interval
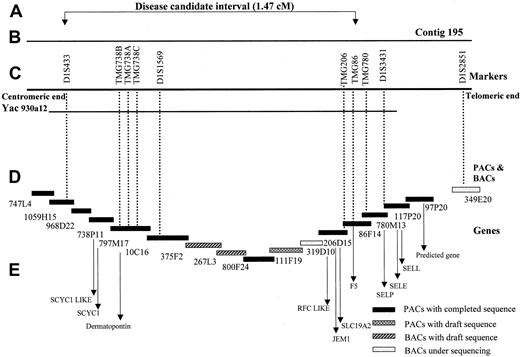
The tiling path of critical interval containing the disease gene lies near the SLC19A2 and open-angle glaucoma genes.10-12,24-26 Polymorphic markers were used to narrow the critical region. The PACs and BACs for contig 195, which completely covers the critical interval, were sequenced by the Sanger Centre.27 Full analysis of specific clones is available. The order of the PACs and BACs within the contig was confirmed using the human gene map FPC database.29 The gaps in the contig were filled by sequenced BACs from other genome centers.30 31 Only the PACs and BACs with completed sequences were used to identify and arrange the known and predicted genes. Based on the assembled contigs and sequenced BACs, the critical region is calculated to be 1.47 Mb.
Results
Linkage studies and sequencing of theAT3 gene
Initially we hypothesized that a mutant antithrombin III which was constitutively active in the absence of heparin binding could account for the bleeding disorder and the laboratory results of affected individuals in this family. Linkage to AT3 was initially tested using an intragenic microsatellite marker and DNA samples from individuals in the first 2 generations of the family (Figure1).9 32 Evidence of significant linkage was obtained with the AT3 marker, giving a 2-point lod (logarithm plus odds) score of Zmax = 4.47, with zero recombination (θ = 0).
AT3 was sequenced because of the evidence of linkage of the disease to an intragenic marker within AT3. Intron-based, exon-specific primers were designed to amplify each of the 6 exons ofAT3 individually, along with flanking intronic sequence. The amplified DNA fragments were sequenced in both the sense and antisense directions. Two previously reported polymorphisms in exon 4 of the gene were found in the proband (7596G > A and 7626G > A), but neither change resulted in an amino acid change. No mutations were identified in the gene.
Protein studies of antithrombin III were also completed to verify that there were no cryptic mutations which we had failed to identify. Western analysis using plasma from an affected individual and a healthy control showed normal migration of antithrombin III. Antigen levels and antithrombin III were normal in plasma from 2 affected individuals.
Multipoint linkage and haplotype analysis
The chromosome region 1q23-25 was further examined using an additional 16 markers covering a 19-cM region.10,11 In addition, DNA was obtained from the third generation of the family for linkage studies. PT and aPTT results were completed on children in the third generation of the family who were of an age to give assent for drawing blood. The markers were ordered based on previously published physical maps of the region.10-12,24 25 The lod scores for the markers are indicated in Table3. The highest lod score, a θ = 0, was obtained using an expressed polymorphism identified in theFV gene (see below), which gave a lod score of 7.22.
Lod scores for bleeding disorder family with chromosome 1q markers
| Marker . | Recombination fraction, θ . | |||||||
|---|---|---|---|---|---|---|---|---|
| 0.01 . | 0.05 . | 0.10 . | 0.20 . | 0.30 . | 0.40 . | θmax . | Zmax . | |
| DIS431 | 2.82 | 3.81 | 3.87 | 3.30 | 2.33 | 1.12 | 0.10 | 3.87 |
| DIS2799 | 4.82 | 5.09 | 4.83 | 3.90 | 2.70 | 1.29 | 0.05 | 5.90 |
| DIS433 | 1.51 | 1.92 | 1.81 | 1.22 | 0.49 | −0.05 | 0.05 | 1.92 |
| F5 | 7.11 | 6.64 | 6.03 | 4.71 | 3.21 | 1.54 | 0.00 | 7.22 |
| DIS452 | −0.35 | 0.29 | 0.51 | 0.60 | 0.51 | 0.31 | 0.20 | 0.60 |
| DIS2851 | 4.69 | 4.96 | 4.71 | 3.80 | 2.62 | 1.24 | 0.05 | 4.96 |
| DIS2815 | 4.82 | 5.09 | 4.83 | 3.90 | 2.70 | 1.29 | 0.05 | 5.09 |
| AT3 | 3.90 | 4.25 | 4.09 | 3.38 | 2.40 | 1.19 | 0.05 | 4.25 |
| DIS1589 | 3.75 | 4.06 | 3.85 | 3.06 | 2.04 | 0.89 | 0.05 | 4.06 |
| DIS242 | 4.60 | 4.88 | 4.63 | 3.74 | 2.58 | 1.22 | 0.05 | 4.88 |
| DIS2691 | 3.64 | 3.99 | 3.83 | 3.12 | 2.15 | 0.99 | 0.05 | 3.99 |
| DIS416 | 6.49 | 6.06 | 5.50 | 4.28 | 2.90 | 1.37 | 0.00 | 6.49 |
| DIS2786 | 1.56 | 2.02 | 2.06 | 1.75 | 1.23 | 0.57 | 0.05 | 2.02 |
| DIS212 | 4.70 | 4.97 | 4.72 | 3.81 | 2.63 | 1.25 | 0.05 | 4.97 |
| DIS2883 | 4.03 | 4.39 | 4.22 | 3.46 | 2.41 | 1.14 | 0.05 | 4.39 |
| DIS2619 | 3.26 | 3.62 | 3.48 | 2.83 | 1.93 | 0.90 | 0.05 | 3.62 |
| DIS2848 | −1.88 | 0.58 | 1.34 | 1.57 | 1.20 | 0.58 | 0.20 | 1.57 |
| DIS222 | 0.28 | 0.25 | 0.21 | 0.12 | 0.04 | 0.01 | 0.00 | 0.29 |
| Marker . | Recombination fraction, θ . | |||||||
|---|---|---|---|---|---|---|---|---|
| 0.01 . | 0.05 . | 0.10 . | 0.20 . | 0.30 . | 0.40 . | θmax . | Zmax . | |
| DIS431 | 2.82 | 3.81 | 3.87 | 3.30 | 2.33 | 1.12 | 0.10 | 3.87 |
| DIS2799 | 4.82 | 5.09 | 4.83 | 3.90 | 2.70 | 1.29 | 0.05 | 5.90 |
| DIS433 | 1.51 | 1.92 | 1.81 | 1.22 | 0.49 | −0.05 | 0.05 | 1.92 |
| F5 | 7.11 | 6.64 | 6.03 | 4.71 | 3.21 | 1.54 | 0.00 | 7.22 |
| DIS452 | −0.35 | 0.29 | 0.51 | 0.60 | 0.51 | 0.31 | 0.20 | 0.60 |
| DIS2851 | 4.69 | 4.96 | 4.71 | 3.80 | 2.62 | 1.24 | 0.05 | 4.96 |
| DIS2815 | 4.82 | 5.09 | 4.83 | 3.90 | 2.70 | 1.29 | 0.05 | 5.09 |
| AT3 | 3.90 | 4.25 | 4.09 | 3.38 | 2.40 | 1.19 | 0.05 | 4.25 |
| DIS1589 | 3.75 | 4.06 | 3.85 | 3.06 | 2.04 | 0.89 | 0.05 | 4.06 |
| DIS242 | 4.60 | 4.88 | 4.63 | 3.74 | 2.58 | 1.22 | 0.05 | 4.88 |
| DIS2691 | 3.64 | 3.99 | 3.83 | 3.12 | 2.15 | 0.99 | 0.05 | 3.99 |
| DIS416 | 6.49 | 6.06 | 5.50 | 4.28 | 2.90 | 1.37 | 0.00 | 6.49 |
| DIS2786 | 1.56 | 2.02 | 2.06 | 1.75 | 1.23 | 0.57 | 0.05 | 2.02 |
| DIS212 | 4.70 | 4.97 | 4.72 | 3.81 | 2.63 | 1.25 | 0.05 | 4.97 |
| DIS2883 | 4.03 | 4.39 | 4.22 | 3.46 | 2.41 | 1.14 | 0.05 | 4.39 |
| DIS2619 | 3.26 | 3.62 | 3.48 | 2.83 | 1.93 | 0.90 | 0.05 | 3.62 |
| DIS2848 | −1.88 | 0.58 | 1.34 | 1.57 | 1.20 | 0.58 | 0.20 | 1.57 |
| DIS222 | 0.28 | 0.25 | 0.21 | 0.12 | 0.04 | 0.01 | 0.00 | 0.29 |
The haplotype segregating with the bleeding disorder in this family is indicated in Figure 1 by the filled black bars under the affected individuals. Eight recombinants, indicated by open bars, were detected on both normal and disease-linked chromosomes. The most informative recombinants occurred in individuals III:2 and IV:11. To further refine the disease interval, we analyzed some additional markers from PAC contig 195.26 Individual IV:11 showed a recombination in marker TMG86, assigning the critical region for the disease gene between D1S433 and TMG86. The BAC clones of this chromosomal region have been sequenced, and the contig assembled from the sequenced BACs is shown in Figure 2. Based on overlapping sequenced BACs, the size of the critical interval is calculated to be 1.47 Mb without any gaps in the sequence.
Genomic organization of the bleeding disorder candidate region.
(A) The critical region for the disease gene between the markers D1S433 and TMG86 spanning an interval of 1.47 cM. (B) The region covered by the PAC contig 195 from the Sanger Center. (C) The markers used for genotyping (D), PACs, and BACs included in the contig 195, representing the tiling path. The contig contains mainly Sanger Centre clones, with minor gaps filled by BACs from other centers (267L3, Whitehead Institute/MIT Genome Center, Cambridge, MA, and 319D10, Washington University Genome Sequencing Center, St Louis, MO). Black boxes, PACs with completed and fully analyzed sequences; patterned boxes, PACs and BACs with working draft sequences; and grey boxes, BACs in the initial stages of sequencing. (E) The candidate genes in the critical region.
Genomic organization of the bleeding disorder candidate region.
(A) The critical region for the disease gene between the markers D1S433 and TMG86 spanning an interval of 1.47 cM. (B) The region covered by the PAC contig 195 from the Sanger Center. (C) The markers used for genotyping (D), PACs, and BACs included in the contig 195, representing the tiling path. The contig contains mainly Sanger Centre clones, with minor gaps filled by BACs from other centers (267L3, Whitehead Institute/MIT Genome Center, Cambridge, MA, and 319D10, Washington University Genome Sequencing Center, St Louis, MO). Black boxes, PACs with completed and fully analyzed sequences; patterned boxes, PACs and BACs with working draft sequences; and grey boxes, BACs in the initial stages of sequencing. (E) The candidate genes in the critical region.
Mutational screen of the FV gene
The gene for FV mapped within the critical interval. Although FV activity assays were normal, the gene was sequenced directly using DNA from the proband to exclude FV as the defective gene. The sequence analysis of the FV gene identified a total of 7 nucleotide variations (Table4). Six of the nucleotide variations were previously reported polymorphisms. However, a novel 2440A > G nucleotide substitution was detected in the first nucleotide of codon 756 in exon 13, resulting in the substitution of serine (S) by glycine. Direct sequencing of exon 13 of theFV gene revealed that the S756 alteration was present in DNA from the affected individuals but not in the unaffected individuals. The S756 alteration was not found in the DNA from 62 unrelated controls.
Nucleotide substitutions detected in the FVgene of the proband
| Codon . | Nucleotide . | Amino acid . |
|---|---|---|
| 708 | ATT → ATC | I |
| 717 | AAC → AAT | N |
| 735 | GAA → GAG | E |
| 756 | AGT → GGT | S → G |
| 830 | AGA → AAA | R → K |
| 837 | CGT → CAT | R → H |
| 1 198 | CTC → CTT | L |
| Codon . | Nucleotide . | Amino acid . |
|---|---|---|
| 708 | ATT → ATC | I |
| 717 | AAC → AAT | N |
| 735 | GAA → GAG | E |
| 756 | AGT → GGT | S → G |
| 830 | AGA → AAA | R → K |
| 837 | CGT → CAT | R → H |
| 1 198 | CTC → CTT | L |
Discussion
We have mapped the locus for a novel inherited bleeding disorder in a large east Texas family to an interval of 1.47 Mb flanked by D1S433 and TMG86 on chromosome 1q23. The bleeding diathesis in this family is characterized by moderate severity, with significant bleeding following trauma. Bleeding after surgery or trauma is profuse, and some affected individuals in the family have required blood transfusions after these events. No petechiae are present, the bleeding symptoms are characteristic of disorders of secondary hemostasis, and the bleeding times are normal. This disorder is inherited in an autosomal dominant manner without evidence of decreased penetrance. Despite the moderate bleeding and prolongation of the PT and aPTT, all known coagulation factor assays are within normal limits. The mixing studies were inconclusive; these studies did not clearly indicate either the presence of an inhibitor or the lack of a factor. Based on this constellation of findings, we proposed that the bleeding disorder in this family is due to either a slow-acting inhibitor or a deficiency state.
The autosomal dominant inheritance of the disorder lead us to postulate that a known inhibitor of coagulation (eg, antithrombin III) was constitutively active to cause the disorder in the family, rather than a deficiency of a known factor. We initially chose AT3as a candidate locus based on the hypothesis that the bleeding diathesis observed in this family could be due to an antithrombin III that was constitutively active in the absence of heparin binding. However, sequencing of AT3 and the functional and structural studies of the antithrombin III protein failed to reveal any defect in this protein. The confirmation that AT3 was not the defective gene was determined with the evaluation of the fourth generation of family members, which revealed a recombination betweenAT3 and the presence of the disease (individual IV:11, Figure 1).
The evidence of significant linkage to the AT3 gene indicated that the defective gene causing the bleeding disorder in this family was closely linked to the AT3 gene on the long arm of chromosome 1. The lod score of 4.25 at a θ = 0.05 indicated that the linkage was not due to chance alone; this lod score indicates that there are more than 10 000 to 1 odds that the defective gene is closely linked to the AT3 gene. Further analysis of other markers in this region of chromosome 1 confirmed the linkage of the disease to this chromosomal location.
Although FV activity assays were within the normal range in the plasma of the affected individuals, we sequenced the FV gene because it mapped into the disease interval and is in the common pathway of the coagulation cascade. Direct sequencing of the gene using DNA from the proband revealed 6 previously reported expressed polymorphisms, all within exon 13.33 In addition, a novel alteration was found in exon 13 of the FV gene that resulted in the substitution of a glycine for a serine at amino acid 756. This alteration was found to segregate with the disease phenotype and was not found in 124 chromosomes from unrelated control individuals. Exon 13 of the FV gene codes for the B region or connecting region, which is cleaved from the active protein and is not necessary for procoagulant activity. Exon 13 is a highly polymorphic region of the gene; the finding of 6 sequence variations among this region in the proband illustrates the loose sequence constraints of this exon.17,18,34,35 The lack of sequence constraint in this region of the molecule is also reflected by the fact that there is only a 59% sequence homology between the human and bovine exon 13, whereas other exons are 85% identical.34 35 The FV clotting activity was tested and was within normal limits in 2 of the affected family members. Based on these facts, we conclude that theFV S756 alteration is a private polymorphism in this family, although we cannot exclude the possibility that this alteration could have some unexpected effect on protein function. We hypothesize that another gene, which is closely linked to FV and within a 1.47-Mb interval, is responsible for the bleeding disorder in this family.
Three groups have published YAC contigs of 1q23 that were constructed to identify the defective gene causing open-angle glaucoma.10-12,36 More recently, a PAC contig has been assembled for this region for the identification of theSLC19A2 gene that causes thiamine-responsive megloblastic anemia.26 The interval containing the defective gene causing the novel bleeding disorder is contained within YAC 930a12 and PAC 195. Construction of overlapping BAC sequences for this interval indicates that the critical interval is 1.47 Mb. This interval contains 5 known genes (including FV) and 2 predicted genes that have homology to known genes. In addition, electronic gene mapping of the genomic sequence predicts 65 genes in the interval. Known genes includeFV, dematopontin (proteoglycan cell-binding protein),SCYC1 (encodes lymphotactin, the sole member of the C family of chemokines), JEM-1 (leucine-zipper transcription factor), and SLC19A2 (thiamine transporter, which is the defective gene in thiamine-responsive megaloblastic anemia syndrome). The 2 predicted genes have homology to chemokines (SCYC1-like) and folate carrier proteins (RFC-like). Genes encoding the selectin proteins, P selectin, L selectin, and E selectin, map outside the interval.37 38 The prolongation of both the PT and aPTT suggests that the defective protein causing the bleeding disorder in this family is either a soluble protein in plasma (a secreted protein or soluble fragment of a membrane protein) or a protein that modifies a soluble protein. Therefore, dematopontin and SCYC1 are possible candidates for the defective gene causing this bleeding disorder.
In summary, we have identified a family with a novel bleeding disorder that is inherited in an autosomal dominant manner. Linkage analysis demonstrated that the defective gene causing the disorder in this family maps to a 1.47-Mb region at 1q23. Discernment of the defective gene may result in the identification of a previously unrecognized protein involved in the coagulation system. Alternatively, the defective gene product may not normally participate in the coagulation cascade, but it may disrupt the coagulation cascade when mutated, a so-called “gain-of-function” mutation. In either case, identification of the defective gene will advance current knowledge about the proteins involved in the coagulation cascade and thrombosis.
We would like to thank the family members for participating in this research, Norma Adams for the preparation of the manuscript, Khalid Hanafy and Hua Chen for technical assistance, Prateek Gupta for assistance in collecting samples, and Madeleine Jewell and Samantha Jordan for excellent nursing assistance.
Supported by grant 011618-096 (D.M.M.) from the Advance Technology Program, the Texas Higher Education Coordinating Board, and National Institutes of Health grant M01-RR02558 to the University Clinical Research Center, the University of Texas–Houston Medical School.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dianna M. Milewicz, Department of Internal Medicine, University of Texas–Houston Medical School, 6431 Fannin, MSB 1.614, Houston, TX 77030; e-mail: Dianna.M.Milewicz@uth.tmc.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal