Abstract
Diamond-Blackfan anemia (DBA) is a rare congenital hypoplastic anemia that usually presents early in infancy and is inherited in 10% to 20% of cases. Linkage analysis has shown that DBA in many of both dominant and recessive DBA families mapped to chromosome 19q13.2 leading to the cloning of a gene on chromosome 19q13.2 that encodes a ribosomal protein, RPS19. However, subsequently, mutations of theRPS19 gene have only been identified in 25% of all patients with DBA. This study analyzed 14 multiplex DBA families, 9 of which had 19q13.2 haplotypes inconsistent with 19q linkage. A genome-wide search for linked loci suggested the presence of a second DBA locus in a 26.4-centimorgan (cM) interval on human chromosome 8p. Subsequently, 24 additional DBA families were ascertained and all 38 families were analyzed with additional polymorphic markers on chromosome 8p. In total, 18 of 38 families were consistent with linkage to chromosome 8p with a maximal LOD score with heterogeneity of 3.55 at D8S277 assuming 90% penetrance. The results indicate the existence of a second DBA gene in the 26.4-cM telomeric region of human chromosome 8p23.3-p22, most likely within an 8.1-cM interval flanked by D8S518 and D8S1825. Seven families were inconsistent with linkage to 8p or 19q and did not reveal mutations in the RPS19 gene, suggesting further genetic heterogeneity.
Introduction
Diamond-Blackfan anemia (DBA) is a rare congenital hypoplastic anemia that usually presents early in infancy. The disease is characterized by a moderate to severe normochromic, often macrocytic anemia, reticulocytopenia, and selective erythroblastopenia in the bone marrow.1,2 Although most cases are apparently sporadic, 10% to 20% show clear patterns of inheritance, most commonly autosomal dominant.1 DBA is markedly heterogeneous clinically, and within affected families anemia may be mild or absent in some individuals, with only subtle indications of the erythroid abnormality such as increased mean corpuscular volume (MCV) or erythrocyte adenosine deaminase activity (eADA) or both.3More than 40% of patients have congenital abnormalities, particularly of the upper limb and craniofacial regions.1,4-6 Recently, linkage analysis of 29 European families showed that DBA in 26 of both dominant and recessive families mapped to chromosome 19q.7Identification of a patient with a translocation led to the cloning of a gene on chromosome 19q13.2 that encodes a ribosomal protein, RPS19.8,9 However, mutations of the RPS19 gene have only been identified in 25% of cases,8,10 suggesting a greater degree of genetic heterogeneity than expected from the initial linkage studies.7 11 In this report we provide evidence for the existence of a second DBA gene on chromosome 8p, and also for further genetic heterogeneity.
Patients, materials, and methods
Patients
Thirty-eight families with DBA from the United States, Poland, Germany, Japan, Australia, France, the Czech Republic, and the United Kingdom formed the basis of the study. Initially we ascertained DNA samples from 14 families (P1-19; Figure1). We analyzed them for linkage to chromosome 19q and submitted them for a genome-wide screen. Subsequently these families, together with 24 additional families ascertained later (P20-49; Figure 1), were tested for linkage to candidate regions on chromosome 8. Informed consent was obtained under protocols in effect at Children's Hospital, Boston, Mount Sinai Medical Center, New York, or each country of origin.
Chromosomes 19q and 8p haplotypes.
Structure and affected status of 38 multiplex DBA pedigrees (P). Solid symbols represent affected individuals, question mark symbols (?) represent possibly affected family members, clear symbols represent unaffected individuals. Pedigree numbers are coded to indicate that data are consistent with linkage to 8p (rectangle), 19q (oval), 8p and 19q (rectangle + oval), or inconsistent with linkage to both 8p and 19q (bold underlined) or inconclusive (parentheses), assuming 100% penetrance. The most likely haplotypes for markers D19S197, D19S408, LIPE, D19S178 and D8S518, D19S277, D8S1819, D8S439, D8S1825, and D8S1130 were created by the Cyrillic program (version 2.1).
Chromosomes 19q and 8p haplotypes.
Structure and affected status of 38 multiplex DBA pedigrees (P). Solid symbols represent affected individuals, question mark symbols (?) represent possibly affected family members, clear symbols represent unaffected individuals. Pedigree numbers are coded to indicate that data are consistent with linkage to 8p (rectangle), 19q (oval), 8p and 19q (rectangle + oval), or inconsistent with linkage to both 8p and 19q (bold underlined) or inconclusive (parentheses), assuming 100% penetrance. The most likely haplotypes for markers D19S197, D19S408, LIPE, D19S178 and D8S518, D19S277, D8S1819, D8S439, D8S1825, and D8S1130 were created by the Cyrillic program (version 2.1).
The affection status of each family member was determined by the primary hematologist in the country of origin and confirmed by our review of the clinical and laboratory data. Diagnosis was based on accepted criteria for DBA, namely, normochromic anemia in infancy or early childhood, reticulocytopenia, normal leukocyte and platelet counts, and decreased to absent bone marrow erythroid precursors. One family (P18; Figure 1) had 2 affected members with typical DBA phenotype, but normal bone marrow erythropoiesis, as described in 5% of cases.1 All members of the DBA families were examined for congenital malformations and macrocytosis; eADA and hemoglobin F (HbF) were determined in most but not all families.
Diamond-Blackfan anemia is markedly heterogeneous with respect to the presence or absence of congenital abnormalities, the response to treatment, and evolution of the disease over time. Moreover, within families containing individuals with typical DBA, silent or mild expressions of the disease such as mild anemia or increased MCV or eADA or both have been reported in apparently unaffected family members3; indeed, in some families the RPS19mutation cosegregates with affected individuals and with other family members who are macrocytic or in whom eADA is elevated more than 3 SDs above the mean value.10 On clinical grounds we distinguished 2 different types of kindreds among all the multiplex DBA families. The majority of families comprised 2 or more affected individuals who presented with classical DBA with anemia, reticulocytopenia, and erythroblastopenia in the bone marrow. Coexistence of mild and severe forms of anemia in 3 families (P16, P23, and P34; Figure 1) was observed. The individuals who presented with mild anemia in these pedigrees were coded as affected in the linkage study, because they were offspring/siblings and parents of severely affected patients. Moreover, in one pedigree (P23; Figure 1), 5 affected individuals, including the member with mild anemia, presented with increased MCV (macrocytosis) and elevated eADA. This family has been described elsewhere.12 A second group of 12 pedigrees (P14, 15, 17, 18, 22, 30, 37, 39, 43, 45, 48, 49; Figure 1) comprised classically affected probands and one or 2 individuals who presented with normal or slightly decreased hemoglobin level but with unexplained macrocytosis or highly elevated eADA or both, more then 3 SD above the mean value. These individuals were designated as genetically affected with variable expressivity of the phenotype.3 A number of affected individuals in both groups presented with various congenital malformations such as webbed neck, thumb abnormalities, congenital heart defect, urogenital malformations, and short stature. In one patient mild mental retardation was present. Family members were designated as unaffected if they had normal red blood cell counts, no congenital malformations, and did not have an unexplained elevated eADA or MCV. Thirty-two families manifested autosomal dominant inheritance; 6 pedigrees (P21, 32, 38, 44, 45, 47; Figure 1) with 2 severely affected children had parents with no clinical or laboratory signs of anemia and could represent either autosomal dominant inheritance with incomplete penetrance, gonadal mosaicism for a dominant mutation in one of the parents, or autosomal recessive inheritance. The paternity in these families was confirmed by extended haplotype analysis. The affection status of unaffected individuals in these families was coded as unknown.
DNA genotyping
DNA was isolated from whole blood by standard methods.13 A 10-centimorgan (cM) resolution genome-wide screen was carried out on DNA from 14 DBA families (28 potentially informative meioses) by the National Heart, Lung and Blood Institute Mammalian Genotype Service in Marshfield, Wisconsin, using their screening set 9.14 Polymerase chain reaction (PCR) and gel analysis of polymorphic markers from within chromosome 19q13.2 and chromosome 8p were performed with 32P-labeled deoxycytosine triphosphate (dCTP), as described.13 Primer sequences and optimal conditions for PCR were obtained from Genome Data Base (GDB) (http://gdbwww.gdb.org/) and Généthon.15
RPS19 gene sequencing
The genomic DNA (gDNA) samples from affected individuals from all 38 studied families were amplified by PCR. Five PCR fragments spanning the 6 exons and 5′ UTR were amplified using 150- to 200-ng template DNA in 50-μL reactions with Taq DNA polymerase (Qiagen, Valencia, CA). PCR products were purified using a High Pure PCR Product Purification Kit (Boehringer Mannheim, Indianapolis, IN). The PCR primers and additional internal primers were used for fluorescent automated DNA sequencing, performed using Applied Biosystem 373 or 377 DNA sequencer and ABI Big Dye Terminator sequencing kits (Perkin Elmer, Foster City, CA). The DNA sequence was analyzed using Sequencher program software (GeneCodes, version 3.1).
Statistical analysis
The GENEHUNTER program (version 1.1) was used for parametric 2- and multipoint LOD score calculation16 with the assumption of autosomal dominant pattern of inheritance. Varied penetrance values were used (0.7, 0.8, 0.85, 0.9, and 0.95), with an assumed disease-gene frequency of 0.000001 (that is, disease incidence 2/1 000 000 births) and no sex difference with reference to disease frequency. Changing the disease gene frequency by several orders of magnitude had minimal influence on the results. The allele frequencies for each polymorphism, locus order, and the genetic and physical distances between them were obtained from GDB, NCBI (http://www.ncbi.nlm.nih.gov/) and Généthon.
Results
Chromosome 19q linkage
The PCR primers were used to amplify polymorphic markers on chromosome 19q13.2 (D19S197, LIPE, D19S408, D19S178) known to be linked to the DBA phenotype.7RPS19 is located between D19S197 and LIPE.8 Haplotype data for 5 of 14 initially studied families (Table 1) were consistent with linkage to chromosome 19q13.2 (P5, 7, 8, 10, 14; Figure1). Haplotype analysis performed in the other 9 families (P1, 2, 4, 11, 15, 16, 17, 18, 19; Figure 1, Table 1) were not consistent with linkage to this region.
Results of 19q and 8p haplotype analysis andRPS19 gene sequencing in the 38 DBA pedigrees
| . | UPN (P) . | 19q haplotype . | 8p haplotype . | RPS19 gene mutation . | DBA phenotype in the pedigree . | |||
|---|---|---|---|---|---|---|---|---|
| A . | MCV . | eADA . | CA . | |||||
| A | 1 | − | + | − | SA | H | ↑ | Webbed neck, duplicated thumb, congenital heart defect |
| 2 | − | + | − | SA | N | ND | Double ureter | |
| 4 | − | + | − | SA | N | ND | None | |
| 5 | + | + | − | SA | N | ND | Short stature | |
| 7 | + | − | − | SA | N | ND | Webbed neck, mental retardation | |
| 8 | + | − | − | SA | N | ND | None | |
| 10 | + | − | − | SA | H | ND | High arched palate | |
| 11 | − | + | − | SA | H | ↑ | Abnormal toes | |
| 14 | + | + | exon 5, T392G | SA | N | ↑ | None | |
| 15 | − | − | − | SA | H | ↑ | Extra lobe of kidney | |
| 16 | − | − | − | SA, MA | H | ↑ | Abnormal thumb | |
| 17 | − | + | − | SA | H | N | None | |
| 18 | − | + | − | SA | H | ND | None | |
| 19 | − | − | − | SA | N | ND | None | |
| B | 20 | + | − | − | SA | H | ND | None |
| 21 | No vertical transmission | − | SA | N | ↑ | None | ||
| 22 | − | − | − | SA | H | ↑ | None | |
| 23 | + | − | exon 4, G185A | SA, MA | H | ↑ | Short stature | |
| 25 | + | − | − | SA | H | ND | Minor myelomeningocoele | |
| 26 | − | + | − | SA | N | ↑ | None | |
| 27 | − | − | − | SA | N | ND | None | |
| 28 | + | + | − | SA | H | ↑ | None | |
| 30 | − | + | − | SA | N | ↑ | None | |
| 32 | No vertical transmission | − | MA | N | ND | Short stature, mental retardation | ||
| 34 | − | + | − | SA, MA | H | ND | Hypertelorism, blue sclera | |
| 37 | − | + | − | SA | H | ↑ | Double kidney | |
| 38 | No vertical transmission | exon 3, G167A | SA | N | ↑ | None | ||
| 39 | − | + | − | SA | N | ↑ | None | |
| 40 | + | + | − | SA | H | ↑ | None | |
| 41 | Inconclusive | − | − | SA | H | ↑ | Triphalyngeal thumbs | |
| 42 | − | − | − | SA | H | ↑ | DBA facies | |
| 43 | − | + | − | SA | H | ↑ | Growth retardation, abnormal toes, bifid thumbs, pes cavus, facies | |
| 44 | No vertical transmission | − | SA | H | ↑ | DBA facies, growth retardation | ||
| 45 | Inconclusive-unaffected grandparents | − | MA | H | ↑ | None | ||
| 46 | − | + | − | SA | H | ↑ | None | |
| 47 | No vertical transmission | − | SA | H | ↑ | None | ||
| 48 | − | + | − | SA | N | ↑ | High arched palate, DBA facies, absent radial pulse | |
| 49 | − | − | − | SA | N | ↑ | None | |
| . | UPN (P) . | 19q haplotype . | 8p haplotype . | RPS19 gene mutation . | DBA phenotype in the pedigree . | |||
|---|---|---|---|---|---|---|---|---|
| A . | MCV . | eADA . | CA . | |||||
| A | 1 | − | + | − | SA | H | ↑ | Webbed neck, duplicated thumb, congenital heart defect |
| 2 | − | + | − | SA | N | ND | Double ureter | |
| 4 | − | + | − | SA | N | ND | None | |
| 5 | + | + | − | SA | N | ND | Short stature | |
| 7 | + | − | − | SA | N | ND | Webbed neck, mental retardation | |
| 8 | + | − | − | SA | N | ND | None | |
| 10 | + | − | − | SA | H | ND | High arched palate | |
| 11 | − | + | − | SA | H | ↑ | Abnormal toes | |
| 14 | + | + | exon 5, T392G | SA | N | ↑ | None | |
| 15 | − | − | − | SA | H | ↑ | Extra lobe of kidney | |
| 16 | − | − | − | SA, MA | H | ↑ | Abnormal thumb | |
| 17 | − | + | − | SA | H | N | None | |
| 18 | − | + | − | SA | H | ND | None | |
| 19 | − | − | − | SA | N | ND | None | |
| B | 20 | + | − | − | SA | H | ND | None |
| 21 | No vertical transmission | − | SA | N | ↑ | None | ||
| 22 | − | − | − | SA | H | ↑ | None | |
| 23 | + | − | exon 4, G185A | SA, MA | H | ↑ | Short stature | |
| 25 | + | − | − | SA | H | ND | Minor myelomeningocoele | |
| 26 | − | + | − | SA | N | ↑ | None | |
| 27 | − | − | − | SA | N | ND | None | |
| 28 | + | + | − | SA | H | ↑ | None | |
| 30 | − | + | − | SA | N | ↑ | None | |
| 32 | No vertical transmission | − | MA | N | ND | Short stature, mental retardation | ||
| 34 | − | + | − | SA, MA | H | ND | Hypertelorism, blue sclera | |
| 37 | − | + | − | SA | H | ↑ | Double kidney | |
| 38 | No vertical transmission | exon 3, G167A | SA | N | ↑ | None | ||
| 39 | − | + | − | SA | N | ↑ | None | |
| 40 | + | + | − | SA | H | ↑ | None | |
| 41 | Inconclusive | − | − | SA | H | ↑ | Triphalyngeal thumbs | |
| 42 | − | − | − | SA | H | ↑ | DBA facies | |
| 43 | − | + | − | SA | H | ↑ | Growth retardation, abnormal toes, bifid thumbs, pes cavus, facies | |
| 44 | No vertical transmission | − | SA | H | ↑ | DBA facies, growth retardation | ||
| 45 | Inconclusive-unaffected grandparents | − | MA | H | ↑ | None | ||
| 46 | − | + | − | SA | H | ↑ | None | |
| 47 | No vertical transmission | − | SA | H | ↑ | None | ||
| 48 | − | + | − | SA | N | ↑ | High arched palate, DBA facies, absent radial pulse | |
| 49 | − | − | − | SA | N | ↑ | None | |
Part A contains 14 initially ascertained pedigrees; part B comprises 24 pedigrees ascertained and studied subsequently. UPN (P) indicates unique pedigree number (P); +/−, consistent/inconsistent with linkage to haplotype; A, anemia; SA, severe anemia; MA, mild anemia; H, high MCV; N, in normal range; ↑, eADA > 3 SD; CA, congenital anomalies.
Genomewide screen and linkage to chromosome 8p
Because these data indicated that the locus on chromosome 19q did not explain many of our cases, a genome-wide search was performed for linked loci in all 14 pedigrees. Haplotype analysis revealed a subgroup of 8 pedigrees consistent with linkage to chromosome 8p (P1, 2, 4, 5, 11, 14, 17, 18; Figure 1); 6 pedigrees were not consistent with linkage to this region (P7, 8, 10, 15, 16, 19; Figure 1). Two of 8 families consistent with linkage to chromosome 8p were also consistent with linkage to chromosome 19q (P5, 14; Figure 1). The genome-wide search showed the highest number of pedigrees consistent with linkage to chromosome 8p, suggesting the possibility of a second DBAgene in that region. However, the result was inconclusive because of the relatively small size and number of families studied.
To address this issue, we ascertained 24 additional pedigrees (P20-49; Figure 1), and, together with the first subset of 14 families, thus analyzed 38 pedigrees in total. Eighteen of 38 families were consistent with linkage to 8p, 14 families were inconsistent, and 6 families did not show vertical transmission of the disease; it is thus difficult to assess linkage to this region in these 6 families (Table 1). They may represent autosomal dominant inheritance with reduced penetrance, mosaicism, germline mutation in one of the parents, or an autosomal recessive pattern of inheritance.
RPS19 gene mutations
Twelve of 38 pedigrees were consistent with linkage to chromosome 19q, 19 were inconsistent with linkage to this region, and results of linkage analysis of 7 pedigrees were inconclusive (Figure 1, Table 1). Subsequent sequence analysis of the RPS19 gene in affected individuals from all 38 families revealed different missense mutations in 3 families. One of them is a novel mutation located in exon 5 at codon 131 (T392G; Leu → Arg) in pedigree P14. Two other mutations, detected in exon 3 at codon 56 (G167A; Arg → Gln) in P38, and in exon 4 at codon 62 (G185A; Arg → Gln) in affected individuals from pedigree P23 (Figure 1, Table 1), were previously described.10 17 No other mutations were found in the 6 exons or their flanking regions in the other 35 pedigrees.
Multipoint linkage analysis with heterogeneity
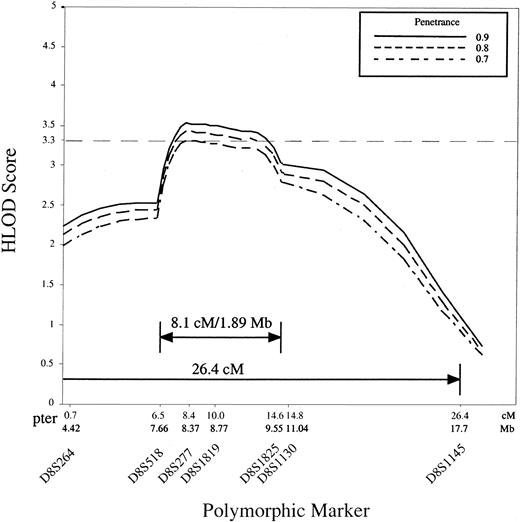
Because the high degree of genetic heterogeneity and small family size precluded unambiguous identification of the subset of families with the second DBA locus we used the GENEHUNTER program to analyze all studied families except the 3 with mutations in exons 3, 4, and 5 of the RPS19 gene, giving a total of 35 families. The parametric multipoint LOD score with heterogeneity (HLOD score), varied between 3.33 and 3.59 at penetrance 0.7 to 0.95 for marker D8S277, with the proportion (α) of families linked to the locus equal to 0.56 to 0.48 (Table 2, Figure2). In other words we estimate from our data that approximately 50% of families are consistent with linkage to the region of 8p with the highest HLOD score. The HLOD score decreases to 0.64 and 0.735 for locus D8S1145 at penetrances 0.7 to 0.95 (Table2, Figure 2), suggesting strongly that the gene resides telomeric to this marker. These results provide evidence for a second DBAgene in the 26.4-cM interval of human chromosome 8p23.3-p22.
GENEHUNTER analysis of 35 DBA pedigrees
| Chromosome 8p; HLOD scores of 35 DBA pedigrees at θ = 0.0 . | |||||||
|---|---|---|---|---|---|---|---|
| . | . | . | Penetrance . | ||||
| Locus (from pter) . | Genetic distance (cM) . | Physical distance (Mb) . | 70% . | 80% . | 85% . | 90% . | 95% . |
| HLOD (α) . | HLOD (α) . | HLOD (α) . | HLOD (α) . | HLOD (α) . | |||
| D8S264 | 0.7 | 4.42 | 2.00 (0.47) | 2.12 (0.46) | 2.20 (0.44) | 2.24 (0.43) | 2.29 (0.41) |
| D8S518 | 6.5 | 7.66 | 2.34 (0.48) | 2.44 (0.46) | 2.48 (0.45) | 2.53 (0.43) | 2.57 (0.41) |
| D8S277 | 8.4 | 8.37 | 3.33 (0.56) | 3.45 (0.53) | 3.50 (0.52) | 3.55 (0.50) | 3.59 (0.48) |
| D8S1819 | 10 | 8.77 | 3.29 (0.55) | 3.41 (0.53) | 3.47 (0.51) | 3.52 (0.50) | 3.57 (0.47) |
| D8S439 | 8.77 | 3.23 (0.55) | 3.34 (0.52) | 3.40 (0.51) | 3.45 (0.49) | 3.50 (0.47) | |
| D8S1825 | 14.6 | 9.55 | 2.81 (0.52) | 2.93 (0.50) | 2.98 (0.50) | 3.04 (0.47) | 3.1 (0.45) |
| D8S1130 | 14.8 | 11.04 | 2.79 (0.52) | 2.91 (0.50) | 2.97 (0.49) | 3.03 (0.47) | 3.1 (0.45) |
| D8S1145 | 26.4 | 17.7 | 0.64 (0.36) | 0.68 (0.35) | 0.71 (0.35) | 0.73 (0.34) | 0.75 (0.33) |
| Chromosome 8p; HLOD scores of 35 DBA pedigrees at θ = 0.0 . | |||||||
|---|---|---|---|---|---|---|---|
| . | . | . | Penetrance . | ||||
| Locus (from pter) . | Genetic distance (cM) . | Physical distance (Mb) . | 70% . | 80% . | 85% . | 90% . | 95% . |
| HLOD (α) . | HLOD (α) . | HLOD (α) . | HLOD (α) . | HLOD (α) . | |||
| D8S264 | 0.7 | 4.42 | 2.00 (0.47) | 2.12 (0.46) | 2.20 (0.44) | 2.24 (0.43) | 2.29 (0.41) |
| D8S518 | 6.5 | 7.66 | 2.34 (0.48) | 2.44 (0.46) | 2.48 (0.45) | 2.53 (0.43) | 2.57 (0.41) |
| D8S277 | 8.4 | 8.37 | 3.33 (0.56) | 3.45 (0.53) | 3.50 (0.52) | 3.55 (0.50) | 3.59 (0.48) |
| D8S1819 | 10 | 8.77 | 3.29 (0.55) | 3.41 (0.53) | 3.47 (0.51) | 3.52 (0.50) | 3.57 (0.47) |
| D8S439 | 8.77 | 3.23 (0.55) | 3.34 (0.52) | 3.40 (0.51) | 3.45 (0.49) | 3.50 (0.47) | |
| D8S1825 | 14.6 | 9.55 | 2.81 (0.52) | 2.93 (0.50) | 2.98 (0.50) | 3.04 (0.47) | 3.1 (0.45) |
| D8S1130 | 14.8 | 11.04 | 2.79 (0.52) | 2.91 (0.50) | 2.97 (0.49) | 3.03 (0.47) | 3.1 (0.45) |
| D8S1145 | 26.4 | 17.7 | 0.64 (0.36) | 0.68 (0.35) | 0.71 (0.35) | 0.73 (0.34) | 0.75 (0.33) |
Multipoint plot of HLOD scores against genetic distance (centimorgans).
The analysis of 35 DBA families is shown, and the GENEHUNTER HLOD scores were calculated for the markers bounded by D8S264 (telomeric) and D8S1145 (centromeric). The data show significant linkage of a DBA locus to D8S277 at penetrance 0.7, 0.8, and 0.9, and to D8S1819 at penetrance 0.8 and 0.9.
Multipoint plot of HLOD scores against genetic distance (centimorgans).
The analysis of 35 DBA families is shown, and the GENEHUNTER HLOD scores were calculated for the markers bounded by D8S264 (telomeric) and D8S1145 (centromeric). The data show significant linkage of a DBA locus to D8S277 at penetrance 0.7, 0.8, and 0.9, and to D8S1819 at penetrance 0.8 and 0.9.
Evidence for non-19q non-8p DBA
In total, haplotype analysis did not reveal linkage of DBA to chromosome 8p in 14 of the 38 families (P7, 8, 10, 15, 16, 19, 20, 22, 23, 25, 27, 41, 42, 49; Figure 1, Table 1). Seven of these families also showed no evidence for linkage to chromosome 19q (P15, 16, 19, 22, 27, 42, 49); all of them were negative forRPS19 mutations (Table 1) by sequencing. Thus, data on 7 pedigrees support the existence of a non-19q non-8p gene (or genes). In 3 of these families haplotype segregation provided strong evidence for non-19q non-8p DBA gene location (P22, 27, 49) in that discordance of 19q and 8p haplotypes was found in affected family members.
Discussion
Our data suggest that a second gene for DBA resides in the telomeric region of human chromosome 8p23.3-p22. Statistical analysis revealed significant HLOD scores for the telomeric region of the chromosome 8p markers flanked by D8S1145 at penetrance 0.7 and higher. Estimation of the disease gene penetrance in DBA is difficult because of the high degree of clinical heterogeneity in this disease; however, using the single ascertainment approach18 in pedigrees that show vertical disease transmission we calculated that the disease penetrance is approximately 0.9. This result supports the significance of the HLOD score calculated at a penetrance of 0.9 (Figure2).
Additionally we found recombinations in affected individuals in 5 of the 8p-linked families (P1, 4, 11, 17, 43) within that region. Recombinations in pedigrees P17 and P43 occurred with marker D8S518, in families P1 and P4 with D8S1145, and in pedigree P11 with marker D8S1825, suggesting that the potential critical region is flanked by the polymorphic markers D8S518 and D8S1825. These data suggested that the second DBA gene is possibly located within an 8.1-cM interval that contains approximately 1.89 Mb DNA and maps to chromosome 8p23.2-p23.1 (GDB html as above).
In total, haplotype analysis identified 18 families that were consistent with linkage to an 8p locus. Two of these families (P17, 18) include both severely affected members and individuals with only an increased MCV (> 3 SD above mean value) but no history of anemia. Unexplained macrocytosis in DBA kindreds in which clinically affected members also have an increased MCV suggests strongly that macrocytic individuals carry a DBA gene mutation and have variable expressivity of the phenotype. Variable expressivity was described in 2 families each with 2 clinically affected half-siblings and an unaffected common parent, who was therefore an obligatory gene carrier; the parent in each family had no symptoms of anemia, but did have an elevated MCV and HbF.19,20 Moreover, Willig et al reported a family with an RPS19 mutation in individuals with classical DBA and in a clinically unaffected father who presented only with macrocytosis.10
In addition to the chromosome 8p- and 19q-linked families, we found 7 pedigrees inconsistent with linkage to either 8p or 19q, and negative for RPS19 gene mutations (P15, 16, 19, 22, 27, 42, 49). In 4 of these, (P15, 16, 19, 42), possible explanations for this finding could be either genetic heterogeneity or lack of penetrance of the disorder in healthy siblings. However, in 3 families (P22, 27, 49) the segregation of 2 different haplotypes of chromosome 19q and 8p in affected family members provided strong evidence for a non-19q non-8p locus.
We found that the 19q families, the 8p families, and the non-8p non-19q families all contained pedigrees with the classical severe DBA phenotype as well as families with severe and mild anemia. Moreover, some individuals from each group had increased MCV or elevated eADA or both, that is, variable expressivity of the phenotype. Thus, with the proviso that insufficient families have been studied, our data do not show any correlations between clinical phenotype and genotype at this time.
Examination of candidate genes that map to the critical interval reveal no ribosomal protein genes known to map to 8p23.3-p22 region. Seventy-five of the 80 known mammalian ribosomal protein genes have been mapped,21 so it is possible that one of the 5 unmapped genes will localize to chromosome 8p. The region does not contain any obvious candidates, but one gene, GATA4, warrants discussion. GATA4 is expressed during embryogenesis in extraembryonic visceral endoderm and appears to play an indirect role in the development of the primitive hemangioblasts that give rise to both endothelial and hematopoietic cells within blood islands of the yolk sac22-24; it is also thought to be important in cardiac development. Targeted disruption of GATA4 in mice is embryonic lethal,23,24 butGATA4−/− embryonic stem cells can contribute to erythropoiesis in chimeric embryoid bodies.25,26 Thus a defect in GATA4 might be expected to be extrinsic to the erythroid precursor. Although this is inconsistent with the data in DBA that suggest a defect intrinsic to erythroid cells,27 28GATA4 will be considered a candidate to be theDBA2 gene in future studies.
In conclusion, we provide strong evidence for genetic heterogeneity of DBA. Our data indicate the existence of a second DBA gene on chromosome 8p23.3-p22, most likely within an 8.1-cM interval that contains approximately 1.89 Mb DNA and maps to chromosome 8p23.2-p23.1. Our results also indicate further genetic heterogeneity; that is, there is likely at least one more DBA gene in addition to those on chromosomes 19q and 8p. However, the high degree of genetic heterogeneity makes it difficult to generate statistical proof concerning the latter point. Confirmation of an 8p DBA locus will need to await the results of follow-up studies in additional patient populations as well as mutation studies of candidate genes in the region. The genetic data raise the possibility that proteins encoded by the different DBA genes interact in a common and novel pathway that affects erythropoiesis and other specific aspects of development.
Supported by grants from the Daniella Maria Arturi Foundation, the Diamond Blackfan Anemia Foundation, the Association Française contre les Myopathies, Généthon, National Institutes of Health (NIH) AR44345, DK 26263, Direction de la Recherche Clinique (CRC 95183) and the Max Reinhardt Charitable Trust. H.G. was supported by F05 TW05395 from the Fogarty International Center, NIH, and A.H.B. by NIH KO2 award AR02026.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hanna Gazda, Pediatric Oncology, Rm M615, Dana Farber Cancer Institute, 44 Binney St, Boston, MA 02115.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal