Abstract
Interleukin-12 (IL-12) plays an important role in the production of interferon gamma (IFN-γ) and is essential for protection against intracellular pathogens such as Mycobacterium andSalmonella. A 31-year-old man had disseminatedMycobacterium avium complex (MAC) infection. The production of IFN-γ by peripheral blood mononuclear cells stimulated with phytohemagglutinin (PHA-PBMCs) was found severely impaired (40.7 pg/mL compared with 833 ± 289 pg/mL for the patient's and healthy subjects' (n = 3) PHA- PBMCs, respectively), and the patient's PHA-PBMCs completely lacked surface IL-12 receptor β1 (IL-12Rβ1) chain. The IL-12Rβ1 gene transcript in his PHA-PBMCs had an R213W substitution in each allele. Family history showed that both parents were heterozygotes in the R213W substitution. Transfection of a human embryonal kidney cell line 293 (HEKC293) with wild-type IL-12Rβ1wt gene led to cell surface IL-12Rβ1 expression; however, no expression was seen in HEKC293 transfected with the mutated IL-12Rβ1R213W gene. TheIL-12Rβ1 gene transcript, but no IL-12Rβ1 protein, was detected in PHA-PBMCs and T cells, suggesting a post-translational event(s), most likely a shortened turnover of the protein. The R213W substitution was not detected in the cells of 32 healthy persons or of 25 patients with tuberculosis or MAC infection. Six amino acid substitutions (Q214R, M365T, G378R, H438Y, A525T, and G594E) were identified, but the incidences of such substitutions were not significantly different between the groups. The Q214R substitution is reportedly linked to IL-12Rβ1 deficiency; however, the study showed that 19 and 10 of 57 Japanese and 6 and 4 of 33 healthy white persons were heterozygous and homozygous for Arg-214, respectively, suggesting that the Q214R substitution represents a polymorphism and is not related to IL-12Rβ1 deficiency but that the R213W substitution is responsible for IL-12Rβ1 deficiency.
Introduction
Interleukin-12 (IL-12) is a heterodimeric cytokine composed of 2 disulfide-bound glycoprotein subunits, p35 and p40.1 It is secreted from macrophages and dendritic cells and exerts effects on T cells and natural killer cells,2which, in response to IL-12, produce interferon gamma (IFN-γ) that functions to activate macrophages. This cascade is critical to host defense against intracellular pathogens such as Mycobacteriaand Salmonella. Indeed, an inherited deficiency of the IFN-γR gene presents susceptibility to even attenuated mycobacteria such as bacille Calmette-Guérin and nontuberculous mycobacterial infections.3 4
High-affinity receptors for IL-12 (IL-12R) are of heterodimer, composed of β1 and β2 subunits. IL-12Rβ1 chain is a type 1 transmembrane glycoprotein with a molecular size of approximately 100 kd and a cytoplasmic region shorter than that of IL-12Rβ2 chain. Both β1 and β2 chains are essential to confer high affinity to IL-12 on the receptor.2 The IL-12Rβ2 chain has a long intracytoplasmic region, which is necessary for IL-12 signaling through the Jak/Stat pathway.5,6 Type 1 helper T-cell (Th1) clones express mRNA for IL-12Rβ2 chain, whereas Th2 clones do not, and the IL-12Rβ2 chain is expressed on human naive T cells during differentiation to Th1 but not to Th2.7 8 Therefore, the IL-12Rβ2 chain is thought to be associated with Th1 development.
Inherited IL-12Rβ1 deficiency has been identified recently in 7 patients with bacille Calmette-Guérin and nontuberculous mycobacterial infections.9 10 IL-12Rβ1 chain deficiency causes an illness characterized by a selective susceptibility to poorly pathogenic mycobacteria, resembling IFN-γ receptor 1 (IFN-γR1) deficiency, attesting to the importance of IL-12R in the host defense against intracellular pathogens. In patients with IL-12Rβ1 chain deficiency, which differs from complete IFN-γR1 deficiency, the formation of granulomatous lesion is observed, and its clinical course is mild.
In this report, we describe a patient with IL-12Rβ1 deficiency with disseminated Mycobacterium avium complex (MAC) infection. A missense mutation in the IL-12Rβ1 chain was identified in this patient and apparently caused rapid proteolysis of IL-12Rβ1, resulting in null phenotype of the IL-12Rβ1 chain. We believe that this is the first report of a patient with IL-12Rβ1 deficiency caused by a missense mutation.
Patients, materials, and methods
Case description
A 31-year-old man was referred to our hospital because of persistent generalized lymphadenopathy. The patient was born of a consanguineous marriage between cousins. On examination, enlarged elastic lymph nodes were palpated in the neck and the axillary and inguinal regions. Computed abdominal tomography revealed swollen para-aortic lymph nodes as well. A lymph node biopsy from the neck showed histiocytic granuloma. The gastric juice specimens (stained with Ziehl-Neelsen basic fuchsin dyes) contained atypicalMycobacterium (Gaffky 5), and the same atypicalMycobacterium was identified in biopsied neck lymph node cells. The diagnosis was disseminated MAC infection. The patient was seronegative for human immunodeficiency virus type 1 (HIV-1); antibody-dependent cell cytotoxicity activity of his peripheral blood mononuclear cells (PBMCs) scored 80% (normal range, 41%-72%), and natural killer cell activity was 49% (18%-40%) assayed as previously described.11,12 His serum immunoglobulin levels were within normal ranges. Clarithromycin (400 mg/d), rifampicin (450 mg/d), and ciprofloxacin hydrochloride (600 mg/d) were administered for 18 months when these drugs were discontinued because lymph node swelling subsided. After 18 months, the lymph nodes in his neck enlarged again, antibiotics were resumed, and subcutaneous injection of 5 × 105 U recombinant IFN-γ 3 times a week was begun.13
Cells, cell lines, and method of transfection
Peripheral blood mononuclear cells from the patient and healthy donors were isolated by density gradient centrifugation using Ficoll Paque (Pharmacia Biotech, Uppsala, Sweden) and cultured with 5 μg/mL phytohemagglutinin (PHA-P; Sigma, St Louis, MO) for 3 days. PHA-activated lymphoblasts from the patient were co-cultured with lethally irradiated (120 Gy) human T-cell leukemia virus type 1 (HTLV-1)–producing MT-2 cells and were propagated in the presence of IL-2 as previously described.14 From this co-culture, TS-1HTLV-1, an HTLV-1–transformed T-cell line, was established. To construct a vector carrying the IL-12Rβ1–chain gene, wild-type and mutated IL-12Rβ1–chain genes were cloned into an expression vector, pCEP4 (Invitrogen, Carlsbad, CA). A human embryonal kidney cell line, HEKC293, was transfected with such expression vectors using Superfect transfect reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Stable transfectants were selected by incubation in Dulbecco minimal essential medium-containing 0.5 mg/mL hygromycin B (Sigma).
Flow cytometric analysis
Flow cytometric analysis was performed using an Epics XL-MCL (Beckman Coulter, Fullerton, CA) as previously described.15 In brief, cells (1 × 106) were incubated with 1 μg anti–IL-12Rβ1 monoclonal antibody (TOS mAb; Pharmingen, San Diego, CA) or anti–IFN-γR1 (Genzyme, Cambridge, MA) or a control IgG-1 monoclonal antibody (0.5β16) followed by 50 μL of a 1:100 dilution of fluorescein isothiocyanate (FITC) goat-antimouse IgG (Cosmo Bio, Tokyo, Japan).
Reverse transcription coupled with polymerase chain reaction
Total RNA was isolated using TRIzol reagent (Gibco BRL, Gaithersburg, MD) according to the manufacturer's instructions and reverse-transcribed into cDNA using the Superscript II preamplification system (Gibco BRL) with oligo dT. cDNA of the human IL-12Rβ1 (GenBank accession number U03187) and β2 (GenBank accession number U64198) chain genes were then amplified using primers as follows: IL-12Rβ1, sense 5′-TGAACCTCGCAGGTGGCAGA-3′ (nucleotides 7-26); IL-12Rβ1 antisense, 5′-TCGGGCGAGTCACTCACCCT-3′ (nucleotides 2070-2089); IL-12Rβ2 sense, 5′-GGCGACACGTGGAAGAATAC-3′ (nucleotides 594-613); and IL-12Rβ2 antisense, 5′-AGAGATGACAGCTGCTGGAG-3′ (nucleotides 3303-3322). The polymerase chain reaction (PCR) reaction mixture contained 1.5 mM MgCl2, 0.2 μM each primer, 0.1 mM each dNTP, 2 U LA Taq polymerase (Takara, Kyoto, Japan), and 3 μL reverse transcriptase (RT) reaction mixture. To increase the specificity of PCR amplification, the hot-start method was performed with AmpliWax PCR Gem 50 (PerkinElmer, Norwalk, CT). Conditions of PCR amplification were as follows: 95°C for 3 minutes, 35 cycles of 95°C for 60 seconds, 64°C for 60 seconds, 72°C for 120 seconds, and a final extension at 72°C for 4 minutes. PCR reaction was performed with a Robocycler Gradient 40 (Stratagene, San Diego, CA).
Cloning of RT-PCR products and sequencing
PCR products were cloned into pGEM-T Easy vectors (Promega, Madison, WI), and their sequences were determined using Big Dye Terminator (Applied Biosystems, Foster City, CA) with ABI 377 autosequencer (PerkinElmer Applied Biosystems). Sequencing primers used were as follows: for IL-12Rβ1, 5′-TGAACCTCGCAGGTGGCAGA-3′ (sense, nucleotides 7-26), 5′-TCGGGCGAGTCACTCACCCT-3′ (antisense, nucleotides 2070-2089); 5′-GATAACCAGGTTGGTGCTGA-3′ (sense, nucleotides 551-570), 5′-CGCAGTCGCCCAACTTCCAT-3′ (antisense, nucleotides 604-623); 5′-GCCTACAACGTGGCTGTCAT-3′ (sense, nucleotides 1001-1020), 5′-ATGCAATACGTCATGCTCTG-3′ (antisense, nucleotides 1151-1170); 5′-CACCTGTCCCGGCGTCCTAA-3′ (sense, nucleotides 1180-1499), 5′-CTGTTTGCTGTCTTCATCTC-3′ (antisense, nucleotides 1521-1540); for IL-12Rβ2, 5′-GGCGACACGTGGAAGAATAC-3′ (sense, nucleotides 594-613), 5′-AGAGATGACAGCTGCTGGAG-3′ (antisense, nucleotides 3303-3322); 5′-GTCTGCAAACTGGCCTGTAT-3′ (sense, nucleotides 938-957), 5′-TAAGTGGGTGTCTCGTCCTC-3′ (antisense, nucleotides 1080-1099); 5′-GCAGGCTCTGGAATATGGTT-3′ (sense, nucleotides 1431-1450), 5′-GTGCTCTCAATGATTCACTC-3′ (antisense, nucleotides 1564-1583); 5′-GAGGGCATGGACAACATTCT-3′ (sense, nucleotides 1934-1953), 5′-ATTGTAGGGTCGACTCCGTA-3′ (antisense, nucleotides 2061-2080); 5′-CGAGTGACATATGTCCTGTG-3′ (sense, nucleotides 2411-2430), 5′-GCTGGAAGTAATGCGTTGAG-3′ (antisense, nucleotides 2563-2582); and 5′-AGCTGAGAGCAGACAACTGG-3′ (sense, nucleotides 2911-2930).
Northern blot analysis
Expression of the IL-12Rβ1 chain–encoding gene was examined using Northern blot analysis as previously described.17 In brief, total RNA (10 μg each) purified from various cell populations was subjected to 1% agarose–formaldehyde gel electrophoresis, blotted onto a nylon transfer membrane (Hybond N+; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom), and hybridized with a [32P]dCTP-labeled whole IL-12Rβ1 cDNA probe in a hybridization solution (0.25 M Na2HPO4, pH 7.2, 7% sodium dodecyl sulfate [SDS]) at 65°C. After thorough washing, hybridized RNA species were visualized by exposure to x-ray film (Hyperfilm MP; Amersham Pharmacia Biotech) at −70°C.
Western blot analysis
Cells (107) were washed in phosphate-buffered saline (PBS) and lysed in 200 μL RIPA buffer (1% NP40, 0.5% sodium deoxycholate, 0.1% SDS in PBS). Cellular debris was removed by centrifugation, and the cell extract was kept frozen at −80°C. Total protein concentration was measured with the Bio-Rad (Hercules, CA) protein assay. A loading solution (65 mM Tris, pH 6.8, 10% glycerol, 3.8% SDS, 5% 2-mercaptoethanol, 0.003% bromophenol blue) containing proteins from TS-1HTLV-1, PHA-PBMC, and HEKC293 cells transfected with wild-type or mutated IL-12Rβ1–chain gene was boiled for 5 minutes. Each sample (25 μL) was subjected to SDS-7.5% polyacrylamide gel electrophoresis (90 minutes), and proteins were transferred to the polyvinylidene difluoride membrane (Millipore, Bedford, MA). The membrane was blocked overnight in Tris-buffered saline containing 5% skim milk (Wako, Osaka, Japan), then incubated at room temperature for 1 hour with a goat antibody (polyclonal) against the extracellular domain of human IL-12Rβ1 protein (Genzyme/Techne, Cambridge, MA) or a rabbit antibody (polyclonal) against the carboxy terminus of human IL-12Rβ1 protein (Santa Cruz Biotechnology, Santa Cruz, CA) (dilution 1:500). Then the membrane was washed with Tris-buffered saline containing 0.05% Triton X-100 and incubated with the peroxidase-conjugated antigoat or antirabbit immunoglobulin (dilution 1:1000; Santa Cruz Biotechnology) for 1 hour. Again the membrane was washed, and the bands were detected using ECL-Plus (Amersham Pharmacia Biotech, Arlington Heights, IL) Western blotting detection reagents.
Statistical analysis
To examine whether the observed polymorphisms of IL-12Rβ1 were related to susceptibility to mycobacterial infection, the Fisher exact probability test was performed. Statistical analysis was carried out with Statview (Abacus Concepts, Berkeley, CA).
Results
Absence of IL-12Rβ1 chains on PBMCs from the patient
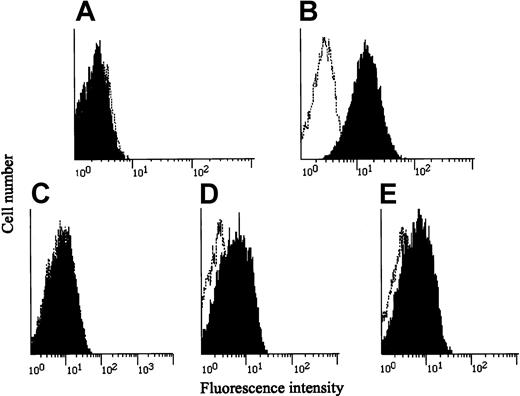
Unusual disseminated MAC infection without HIV infection or any apparent cause implied that a genetic alteration(s) was responsible for the patient's immunodeficient state. Taking into consideration that IL-12 and IFN-γ play major roles in the defense against mycobacterial infections, we first examined whether IL-12Rβ1 chains were present on PHA-PBMCs from the patient by using an IL-12Rβ1–specific monoclonal antibody and flow cytometry. No IL-12Rβ1 chains were seen on the surfaces of the PHA-PBMCs (Figure 1A), though they were readily detected on those from healthy subjects (Figure 1B). We immortalized the patient's T cells with HTLV-1 by co-culturing with lethally irradiated HTLV-1–producing MT-2 cells in the presence of IL-2 as previously described.14 The resultant HTLV-1–transformed T cells (TS-1HTLV-1) were also found to lack cell surface IL-12Rβ1 chains (Figure 1C). Three unrelated T-cell populations immortalized by HTLV-1 proved to fully express cell surface IL-12Rβ1 chains, verifying that immortalization by HTLV-1 per se does not affect the expression of IL-12Rβ1 chains (data not shown). We next examined whether the patient's cells bore IFN-γR1 on their surfaces by using an IFN-γR1–specific monoclonal antibody and flow cytometry. As shown in Figure 1D-E, the patient's and a healthy subject's PBMCs displayed comparable levels of IFN-γR1.
Expression of IL-12Rβ1 chains and IFN-γ receptors in various cells.
(A-C) PBMCs from the patient and a healthy subject were stimulated with PHA, cultured for 3 days, washed, and incubated with a monoclonal antibody against IL-12Rβ1 chain (black) or a control IgG-1 monoclonal antibody (0.5β), stained with an FITC-labeled, goat-antimouse IgG, and subjected to flow cytometry. The patient's HTLV-1–immortalized T (TS-1HTLV-1) cells were treated and analyzed similarly. (D-E) PBMCs from the patient and a healthy subject were stained with an IFN-γR1–specific monoclonal antibody and subjected to flow cytometry.
Expression of IL-12Rβ1 chains and IFN-γ receptors in various cells.
(A-C) PBMCs from the patient and a healthy subject were stimulated with PHA, cultured for 3 days, washed, and incubated with a monoclonal antibody against IL-12Rβ1 chain (black) or a control IgG-1 monoclonal antibody (0.5β), stained with an FITC-labeled, goat-antimouse IgG, and subjected to flow cytometry. The patient's HTLV-1–immortalized T (TS-1HTLV-1) cells were treated and analyzed similarly. (D-E) PBMCs from the patient and a healthy subject were stained with an IFN-γR1–specific monoclonal antibody and subjected to flow cytometry.
We also asked whether PBMCs from the patient were capable of producing IFN-γ because they were stimulated with PHA in vitro. PHA-PBMCs from all 3 healthy subjects produced substantial amounts of IFN-γ in vitro (833 ± 289 pg/mL); however, those from the patient failed to produce a significant level of IFN-γ (40.7 pg/mL). These data indicated that the patient's PBMCs lacked the expression of IL-12Rβ1 chains and had an impaired ability to produce IFN-γ.
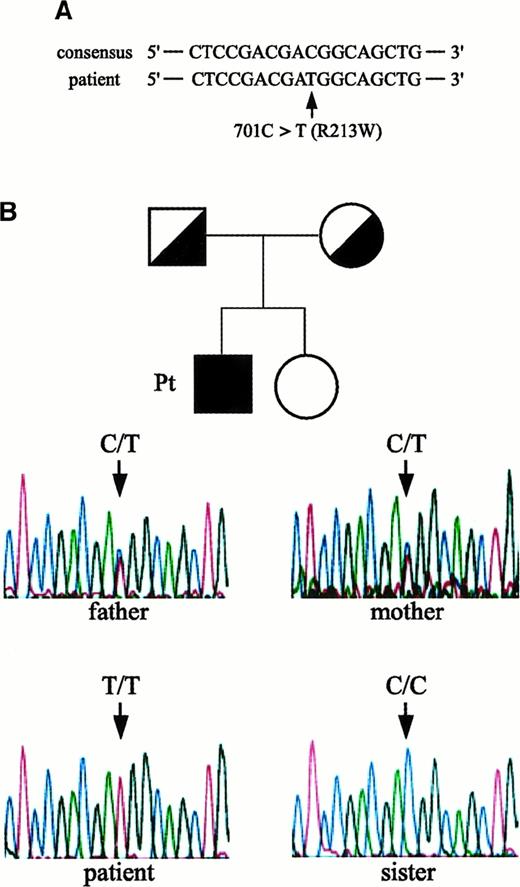
Genetic changes in the patient's IL-12Rβ1 chain–encoding gene
We next asked whether the IL-12Rβ1–encoding gene in cells from the patient had a genetic change(s) and determined its entire nucleotide and amino acid sequences. As shown in Figure2A, a missense mutation (C to T) at nucleotide position 701, which results in the substitution of arginine (CGG) with tryptophan (TGG) at amino acid position 213 (designated R213W), was identified. The human IL-12R consists of 2 distinct chains, β1 and β2, and forms high-affinity receptors to IL-12. It was possible that the lack of cell surface expression of IL-12Rβ1 chains in cells from the patient was due to a mutation(s) in the IL-12Rβ2 chain–encoding gene. However, we found no substitutions in the nucleotide or amino acid sequence of his IL-12Rβ2 chain–encoding gene compared to a consensus sequence.6Study of the family members of the index patient with respect to the IL-12Rβ1 chain–encoding gene revealed that both his parents were heterozygous for R213W, whereas the expression level of IL-12Rβ1 chain on their PHA-PBMCs was within a normal range (Figure 2B). The patient's sister did not carry this mutation, and all family members had no episodes of mycobacterial infections. It was possible that the observed R213W substitution represented a polymorphism in the IL-12Rβ1 chain–encoding gene in the Japanese population; however, the R213W substitution was not seen in cells from 32 healthy Japanese subjects examined.
Missense mutation in the IL-12Rβ1 chain–encoding gene identified in the patient and his pedigree.
(A) A missense mutation identified in the patient: an amino acid substitution from arginine (CGG) to tryptophan (TGG) at position 213 (designated R213W). (B) Study of family members of the patient. Family pedigree and electropherograms for nucleotide 701 position (amino acid 213 position) in the IL-12Rβ1 chain–encoding gene of the index patient, his parents, and sister are shown. Note that the patient was homozygous for 701T (T/T), whereas both his parents were heterozygous (C/T) and his sister did not carry R213W (C/C).
Missense mutation in the IL-12Rβ1 chain–encoding gene identified in the patient and his pedigree.
(A) A missense mutation identified in the patient: an amino acid substitution from arginine (CGG) to tryptophan (TGG) at position 213 (designated R213W). (B) Study of family members of the patient. Family pedigree and electropherograms for nucleotide 701 position (amino acid 213 position) in the IL-12Rβ1 chain–encoding gene of the index patient, his parents, and sister are shown. Note that the patient was homozygous for 701T (T/T), whereas both his parents were heterozygous (C/T) and his sister did not carry R213W (C/C).
Altare et al9 have recently reported that certain patients deficient in IL-12Rβ1 chains and with impaired antimycobacterial immunity had a missense mutation at 214 (Q214R) that was associated with an impaired expression of IL-12Rβ1 chains. However, we observed that, of 32 healthy Japanese subjects examined in this study, 6 and 13 were homozygous and heterozygous for the Q214R substitution, respectively (Table 1). Of 33 healthy white subjects, 6 and 4 were heterozygous and homozygous for the Q214R substitution, respectively. These data strongly suggest that the Q214R substitution represents a polymorphism and is not directly linked to the impaired cell surface expression of IL-12Rβ1 chains.
Nucleotide and amino acid substitutions identified in the IL-12Rβ1 chain–encoding gene in Japanese persons
| . | 705A > G (Q214R) . | 1158T > C (M365T) . | 1196G > C (G378R) . | 1376C > T (H438Y) . | 1637G > A (A525T) . | 1845G > A (G594E) . |
|---|---|---|---|---|---|---|
| Healthy subjects | 13/13/6 | 20/6/6 | 13/14/5 | 32/0/0 | 31/1/0 | 32/0/0 |
| Patients with tuberculosis | 12/3/4 | 11/4/4 | 11/5/3 | 18/1/0 | 18/1/0 | 18/1/0 |
| Patients with atypical mycobacterium infection | 3/3/0 | 4/2/0 | 2/4/0 | 6/0/0 | 5/1/0 | 6/0/0 |
| . | 705A > G (Q214R) . | 1158T > C (M365T) . | 1196G > C (G378R) . | 1376C > T (H438Y) . | 1637G > A (A525T) . | 1845G > A (G594E) . |
|---|---|---|---|---|---|---|
| Healthy subjects | 13/13/6 | 20/6/6 | 13/14/5 | 32/0/0 | 31/1/0 | 32/0/0 |
| Patients with tuberculosis | 12/3/4 | 11/4/4 | 11/5/3 | 18/1/0 | 18/1/0 | 18/1/0 |
| Patients with atypical mycobacterium infection | 3/3/0 | 4/2/0 | 2/4/0 | 6/0/0 | 5/1/0 | 6/0/0 |
The entire nucleotide sequence of the IL-12Rβ1 chain–encoding gene (1989 bp) was determined, and the reduced amino acid sequence was defined. Only missense substitutions are shown: nucleotide substitutions are on the top lines, and amino acid substitutions are in parentheses. The 3 numbers illustrate wild type (wt) wt-homozygotes/wt-mutant (mt) heterozygotes/mt-mt homozygotes are shown at each amino acid position.
R213W substitution failed IL-12Rβ1 chain expression
To determine whether the R213W substitution was directly responsible for the absence of cell surface IL-12Rβ1 chains, an expression vector carrying a mutated IL-12Rβ1 chain gene with the R213W substitution (IL-12Rβ1R213W) was transfected into a human embryonal kidney cell line 293 (HEKC293) that completely lacked the cell surface expression of IL-12Rβ1 chains (Figure3A, broken line). As shown in Figure 3A, HEKC293 cells, as transfected with an expression vector carrying a wild-type IL-12Rβ1 chain–encoding gene, successfully expressed IL-12Rβ1 chains on their surfaces. However, when transfected with IL-12Rβ1R213W, no HEKC293 cells expressed IL-12Rβ1 chains on their surfaces (Figure 3B).
Cell surface IL-12Rβ1 chain expression in HEKC293 cells transfected with wild-type and mutated IL-12Rβ1 genes.
An expression vector carrying the wild-type (A) or mutated (B) IL-12Rβ1 chain–encoding gene was transfected into HEKC293 cells. Transfected HEKC293 cells were selected with hygromycin, stained with a monoclonal antibody against IL-12Rβ1 chain (black) or a control IgG-1 monoclonal antibody 0.5β (white), stained with an FITC-labeled, goat-antimouse IgG, and subjected to flow cytometry.
Cell surface IL-12Rβ1 chain expression in HEKC293 cells transfected with wild-type and mutated IL-12Rβ1 genes.
An expression vector carrying the wild-type (A) or mutated (B) IL-12Rβ1 chain–encoding gene was transfected into HEKC293 cells. Transfected HEKC293 cells were selected with hygromycin, stained with a monoclonal antibody against IL-12Rβ1 chain (black) or a control IgG-1 monoclonal antibody 0.5β (white), stained with an FITC-labeled, goat-antimouse IgG, and subjected to flow cytometry.
Northern and Western blot analyses of IL-12Rβ1 chain expression
Surface IL-12Rβ1 chain-negative HEKC293 cells (broken line, Figure 3A) lacked the expression of the IL-12Rβ1 chain mRNA and proteins (Figure 4, lane 1; Figure5A, lane 3). Unexpectedly, with Northern blot analysis using a whole IL-12Rβ1 cDNA probe, comparable levels of transcripts of the IL-12Rβ1 chain gene were detected in HEKC293 cells transfected with either wild-type or mutated IL-12Rβ1 chain gene (Figure 4, lanes 2 and 3). In the patient's PHA-PBMCs, IL-12Rβ1 transcripts were detected at distinct but lower levels than in those from a healthy subject (Figure 4, lanes 4 and 5). However, in TS-1HTLV-1 cells, which were completely negative for IL-12Rβ1 chains (Figure 1C), a substantial level of the transcripts was detected (Figure 4, lane 6).
Presence of transcripts of the IL-12Rβ1 gene in the patient's cells.
The presence of transcripts of the IL-12Rβ1 chain gene was examined with Northern blot analysis using a whole IL-12Rβ1 cDNA probe. Lane 1, HEKC293 cells transfected with a vector only; lane 2, HEKC293 cells transfected with the wild-type IL-12Rβ1 chain gene; lane 3, HEKC293 cells transfected with the mutated IL-12Rβ1 chain gene; lane 4, PHA-PBMCs from the patient; lane 5, PHA-PBMCs from a healthy subject; lane 6, patient's HTLV-1–immortalized TS1HTLV-1 cells. Lower panel shows 18S and 28S RNA species visualized by ethidium bromide staining, confirming that an approximately equal amount of RNA was applied to each lane. Note that the patient's PHA-PBMCs had IL-12Rβ1 transcripts at a distinct but lower level, but TS-1HTLV-1 cells had a substantial level of transcripts.
Presence of transcripts of the IL-12Rβ1 gene in the patient's cells.
The presence of transcripts of the IL-12Rβ1 chain gene was examined with Northern blot analysis using a whole IL-12Rβ1 cDNA probe. Lane 1, HEKC293 cells transfected with a vector only; lane 2, HEKC293 cells transfected with the wild-type IL-12Rβ1 chain gene; lane 3, HEKC293 cells transfected with the mutated IL-12Rβ1 chain gene; lane 4, PHA-PBMCs from the patient; lane 5, PHA-PBMCs from a healthy subject; lane 6, patient's HTLV-1–immortalized TS1HTLV-1 cells. Lower panel shows 18S and 28S RNA species visualized by ethidium bromide staining, confirming that an approximately equal amount of RNA was applied to each lane. Note that the patient's PHA-PBMCs had IL-12Rβ1 transcripts at a distinct but lower level, but TS-1HTLV-1 cells had a substantial level of transcripts.
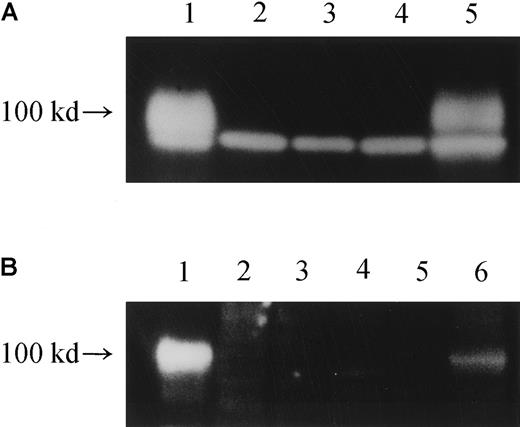
Western blot analysis of the production of IL-12Rβ1 chains.
(A) Cell lysates from various cells were subjected to Western blot analysis using an antibody against the carboxy terminus of human IL-12Rβ1 protein (polyclonal), and the production of an IL-12Rβ1 chain protein of approximately 100 kd was examined. Lane 1, HEKC293 transfected with wild-type IL-12Rβ1wt gene; lane 2: HEKC293 transfected with mutated IL-12Rβ1R213W gene; lane 3, HEKC293 mock transfected with an expression vector only; lane 4, TS-1HTLV-1 cells; lane 5, cells from a healthy subject. The lower band represents a nonspecific band. (B) Cell lysates were similarly treated, but a different anti–IL-12Rβ1 antibody (polyclonal) was used to confirm the authenticity of the 100-kd band visualized. Lane 1, HEKC293 cells transfected with an expression vector carrying the wild-typeIL-12Rβ1wt gene; lane 2, HEKC293 cells transfected with expression vector carrying a mutated IL-12Rβ1 gene; lane 3, HEKC293 cells transfected with expression vector only (mock); lane 4, TS-1HTLV-1 cells, an HTLV-I–immortalized T-cell line derived from the index patient; lane 5, PHA-PBMCs from the patient; lane 6, PHA-PBMCs from a healthy subject. Note that the same 100-kd protein profile was seen in panels A and B.
Western blot analysis of the production of IL-12Rβ1 chains.
(A) Cell lysates from various cells were subjected to Western blot analysis using an antibody against the carboxy terminus of human IL-12Rβ1 protein (polyclonal), and the production of an IL-12Rβ1 chain protein of approximately 100 kd was examined. Lane 1, HEKC293 transfected with wild-type IL-12Rβ1wt gene; lane 2: HEKC293 transfected with mutated IL-12Rβ1R213W gene; lane 3, HEKC293 mock transfected with an expression vector only; lane 4, TS-1HTLV-1 cells; lane 5, cells from a healthy subject. The lower band represents a nonspecific band. (B) Cell lysates were similarly treated, but a different anti–IL-12Rβ1 antibody (polyclonal) was used to confirm the authenticity of the 100-kd band visualized. Lane 1, HEKC293 cells transfected with an expression vector carrying the wild-typeIL-12Rβ1wt gene; lane 2, HEKC293 cells transfected with expression vector carrying a mutated IL-12Rβ1 gene; lane 3, HEKC293 cells transfected with expression vector only (mock); lane 4, TS-1HTLV-1 cells, an HTLV-I–immortalized T-cell line derived from the index patient; lane 5, PHA-PBMCs from the patient; lane 6, PHA-PBMCs from a healthy subject. Note that the same 100-kd protein profile was seen in panels A and B.
We therefore examined whether IL-12Rβ1 chain proteins were detected in various cells with Western blot analysis. In the lysates of PHA-PBMCs from a healthy subject and HEKC293 cells transfected with the wild-type IL-12Rβ1–chain gene, substantial levels of IL-12Rβ1–chain proteins of approximately 100 kd were detected as either of 2 different IL-12Rβ1 chain–specific monoclonal antibodies was used (Figure 5A-B). However, no such proteins were detected in the patient's PHA-PBMCs, TS 1HTLV-1 cells, or HEKC293 cells transfected with IL-12Rβ1R213W (Figure 5). These data strongly suggest that a post-translational mechanism is at work for the null IL-12Rβ1–chain phenotype in the index patient.
Polymorphism of IL-12Rβ1–chain gene
Polymorphism of the IL-4Rα subunit has been reported to be linked to a gain-of-function mutation and to be associated with atopy.18 It is possible that certain polymorphisms, which alter the function of IL-12Rβ1 chain, may be associated with increased susceptibility to mycobacterial infection. Although certain genetic alterations linked to increased susceptibility to mycobacterial infection have been reported,19 it remains to be determined whether a polymorphism within the IL-12Rβ1 chain-encoding gene affects sensitivity to mycobacterial infection. We therefore determined the nucleotide sequence of the IL-12Rβ1 chain encoding gene in 32 healthy Japanese subjects, 19 Japanese patients with pulmonary tuberculosis, and 6 Japanese patients with nontuberculous mycobacterial infection. Six amino acid substitutions (Q214R, M365T, G378R, H438Y, A525T, and G594E) were identified, as shown in Table 1. There were no apparent accumulations in any of the observed amino acid substitutions, and the incidences of the occurrence of such substitutions were not significantly different among the groups.
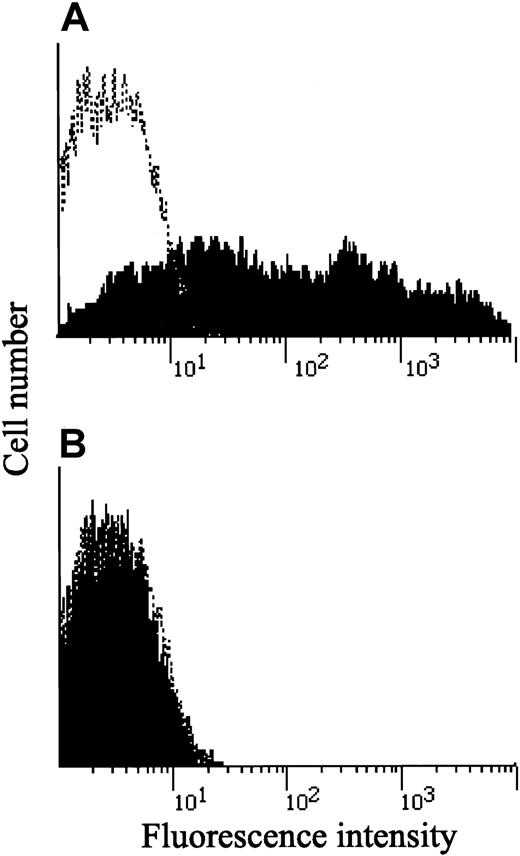
As described earlier, Q214R, which was thought to be a candidate substitution responsible for the IL-12Rβ1–chain deficiency,9 was frequently found in persons examined in this study. As shown in Table 1, 19 and 10 of 57 Japanese persons and 6 and 4 of 33 healthy white persons were heterozygous and homozygous for the Q214R substitution, respectively. Moreover, PHA-PBMCs from 2 healthy subjects, who were homozygous for the Q214R substitution, fully expressed IL-12Rβ1 chains on their surfaces (Figure6), and those 2 PHA-PBMC preparations produced IFN-γ (greater than 1000 and 3180 pg/mL) comparably, as did PHA-PBMCs from those with wild-type IL-12Rβ1 (833 ± 289 pg/mL; n = 3). It should also be noted that the R213W substitution was not found in any of the 90 healthy subjects we examined. Taken together, the data presented in this study strongly suggest that the R213W substitution is responsible for IL-12Rβ1 deficiency in the present patient, but the Q214R substitution represents a polymorphism and is unlikely to be related to IL-12Rβ1 deficiency.
Expression of IL-12Rβ1 chains in PHA-PBMC from 2 persons carrying the Q214R substitution.
PHA-PBMCs from 2 healthy Japanese persons who were homozygous for the Q214R substitution were examined for the presence of cell surface IL-12Rβ1 chains, as described in the legend to Figure 1. Note that both fully expressed IL-12Rβ1 chains on their surfaces.
Expression of IL-12Rβ1 chains in PHA-PBMC from 2 persons carrying the Q214R substitution.
PHA-PBMCs from 2 healthy Japanese persons who were homozygous for the Q214R substitution were examined for the presence of cell surface IL-12Rβ1 chains, as described in the legend to Figure 1. Note that both fully expressed IL-12Rβ1 chains on their surfaces.
Discussion
Patients with IFN-γR deficiency are highly susceptible to mycobacterial infection at young ages and most often die prematurely.3,4 It is noteworthy that the onset of infection in the patient examined in this study did not occur until he was 30 years of age, indicating that the production of IFN-γ minimally required for protection against mycobacterial infection was maintained until the age of 30 when mycobacterial infection became frank. It is possible that an apparently normal level of natural killer activity seen in this patient had been maintained by this minimally retained level of IFN-γ. Indeed, Altare et al9 have also described that patients with IL-12Rβ1–chain deficiency had a mild clinical course, as seen in our patient. Jouanguy et al20recently reported that partial deficiency of IFN-γR1, caused by a missense mutation in its extracellular domain, presumably results in a mild clinical course compared to that in patients with absolute IFN-γR1 deficiency. In fact, mycobacterial infection in patients with such mild clinical courses can be well controlled by antibiotics against such organisms, and IFN-γ supplementation often improves the clinical outcome. In those with absolute IFN-γR1 deficiency, however, the formation of granulomatous lesions is absent in lymph nodes, whereas granulomatous lesions are observed in patients with IL-12Rβ1 chain deficiency, IL-12 deficiency,21 and partial IFN-γR1 deficiency.15 These findings suggest that the production of even insignificant amounts of IFN-γ is sufficient for granuloma formation and for preventing or delaying the contraction of mycobacterial infection.
In the present patient, dinucleotide CG changed to TG, converting arginine (CGG) to tryptophan (TGG). The CG dinucleotide is known as a mutational hot spot that causes approximately one third of all transition mutations.22 It is thought that cytidine of the CG dinucleotide is often methylated, and the resultant 5-methylcytidine is susceptible to spontaneous deamination, which yields thymidine. Indeed, in a number of genes, including the p53 gene, the methylation of CG sites is thought to be responsible for mutations.23-25
The patient in this study did not show any disease susceptibility other than MAC infection. Considering that patients with IFN-γR1 deficiency, IL-12 deficiency, and IL-12Rβ1–chain deficiency are susceptible to Mycobacterium or Salmonellainfection though they do not contract viral, fungal, or bacterial (except Salmonella) infections, it appears that IL-12 may not be required for protection against the latter pathogens.2,9,10,26 27 IL-12 is a pleiotropic cytokine implicated in the development of type 1 helper T-cells (Th1), which are apparently critical for developing and maintaining cell-mediated immunity to certain pathogens and cancers. In fact, there are no reports that patients with IL-12Rβ1–chain deficiency experience distinct viral or malignant diseases. One can pose at least 3 possibilities to explain this selective susceptibility to mycobacterial infection. First, because the IL-12Rβ2 chain is critical for the development of Th1 cells, the expression of the IL-12Rβ2 chain without β1 chain is sufficient for the generation of Th1 cells. It is argued, however, that the expression of IL-12Rβ1 chain is essential for the production of IFN-γ and that a reduction of IFN-γ production results in an increased susceptibility to mycobacterial infection. Second, the production of IFNs other than IFN-γ or a minute amount of IFN-γ is sufficient for protecting a person against pathogens other than intracellular microorganisms. Third, the generation of Th1 cells sufficient for protecting a person against infection from pathogens other than mycobacteria is possible without IL-12 involvement.
Genetic polymorphism of cytokine receptors has been linked to certain diseases. For example, polymorphism of the IL-4Rα subunit has been shown to be associated with atopy.18 We hypothesized that genetic predisposition to mycobacterial infection was possible and examined the IL-12Rβ1 chain-encoding gene in 32 healthy Japanese subjects, 19 Japanese patients with pulmonary tuberculosis, and 6 Japanese patients with nontuberculous mycobacterial infections. Six amino acid substitutions (Q214R, M365T, G378R, H438Y, A525T, and G594E) were identified, though the incidences of such substitutions were not significantly different among the groups (Table 1). Of the 6 amino acid substitutions, Altare et al9 have reported that the Q214R substitution was singly responsible for IL-12Rβ1 chain deficiency, which was not in agreement with our present data. We found that 19 and 10 of 57 Japanese persons were heterozygous and homozygous for Arg-214, respectively. Because this particular substitution is possibly a polymorphism unique to the Japanese, we also examined the genotypic profile in 33 healthy white persons. The results that 6 and 4 of the 33 white persons were similarly heterozygous and homozygous for Arg-214, respectively, strongly suggested that the Q214R substitution represents a polymorphism. It is possible that a substitution(s) responsible for the illness other than Q214R was not identified in those patients examined by Altare et al9 or that they had other unidentified abnormality(ies) that caused IL-12Rβ1 deficiency. It is also possible that the abnormality caused by Q214R was compensated by other concomitant amino acid substitution(s) in the IL-12Rβ1 encoding gene. However, among 32 healthy Japanese persons examined, one carried the Q214R substitution (homozygous) with 2 substitutions (M365T and G378R; both heterozygous), suggesting that the Q214R substitution is unlikely to be responsible for IL-12Rβ1 deficiency.
By contrast, the R213W substitution identified in the index patient was not seen in any of the 90 persons examined. Hypothesizing that the R213W substitution observed in the patient was directly responsible for the IL-12Rβ1–chain deficiency, we transfected HEKC293 cells lacking IL-12Rβ1 mRNA or protein expression with either the wild-type or the mutated IL-12Rβ1 gene. The HEKC293 cells produced IL-12Rβ1 protein only when transfected with the wild-type gene but not with the mutated gene transfected (Figure 3), strongly suggesting that the R213W substitution was directly responsible for the IL-12Rβ1 deficiency. It was noted, however, that the patient's PHA-PBMC produced a limited but distinct level of IL-12Rβ1 mRNA and that the patient's T-cells immortalized with HTLV-1 (TS-1HTLV-1), which do not bear surface IL-12Rβ1 chains, produced a significant amount of IL-12Rβ1 mRNA. This feature was corroborated by the observation that the HEKC293 cells transfected with the mutated gene also produced IL-12Rβ1 mRNA comparable to that transfected with the wild-type gene (Figure 4, lanes 2 and 3).
These data suggest that the IL-12Rβ1–chain deficiency seen in the index patient was due mainly to a post-translational event(s), most likely a shortened turnover of the protein, though an increased IL-12Rβ1 mRNA turnover rate or a decreased IL-12Rβ1 mRNA translation rate may also be involved. Indeed, rapid intracellular proteolysis has been implicated in several diseases causing immunodeficiency, such as X-linked hyper IgM syndrome28and chronic granulomatous disease.29 For example, in patients with X-linked chronic granulomatous disease who are deficient in the β subunit of cytochrome b558, gp91phox amino acid substitutions in the cybb gene have been shown not to affect its mRNA expression but to cause rapid turnover of gp91phox proteins in cells, resulting in a lack of NADPH-oxidase activity.29Taken together, the observed R213W substitution is directly responsible for the IL-12Rβ1 deficiency seen in the index patient, and this particular substitution most likely causes an intracellular rapid protein turnover of the IL-12Rβ1 chain in cells, though the exact mechanism for such a proposed rapid proteolysis of mutant proteins remains to be elucidated.
We thank Drs Hiroyuki Nunoi and Sadahiro Tamiya for helpful discussions and comments.
Supported in part by a grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Masao Matsuoka, Laboratory of Virus Immunology, Research Center for AIDS, Institute for Virus Research, Kyoto University, Kyoto Japan 606-8506, Japan; e-mail:mmatsuok@virus1.virus.kyoto-u.ac.jp.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal