Abstract
Omenn syndrome (OS) is an inherited disorder characterized by an absence of circulating B cells and an infiltration of the skin and the intestine by activated oligoclonal T lymphocytes, indicating that a profound defect in the lymphoid developmental program could be accountable for this condition. Inherited mutations in either the recombination activating genes RAG1 orRAG2, resulting in partial V(D)J recombinase activity, were shown to be responsible for OS. This study reports on the characterization of new RAG1/2 gene mutations in a series of 9 patients with OS. Given the occurrence of the same mutations in patients with T-B–severe combined immune deficiency or OS on 3 separate occasions, the proposal is made that an additional factor may be required in certain circumstances for the development of the Omenn phenotype. The nature of this factor is discussed.
Introduction
The diversity of immunoglobulins and T-cell receptors (TCRs) is mediated by the somatic recombination of genes encoding variable (V), diversity (D), and joining (J) segments by a mechanism known as V(D)J recombination.1 The RAG1 and RAG2 proteins, the expression of which is restricted to immature lymphocytes, initiate the reaction. The RAG1/2 complex introduces a DNA double-strand break (dsb) in the recombination signal sequences (RSSs), composed of conserved heptamer and nonamer separated by either 12 or 23 bp, that flank all V, D, and J segments.2-6 During the subsequent steps, a nonlymphoid-specific machinery is responsible for the repair of this DNA damage.7,8 Faulty V(D)J recombination generally results in the arrest of both B- and T-cell development, leading to severe combined immune deficiency (SCID). This was documented in several settings including the targeted deletion of the RAG1 and RAG2 genes,9,10 the inactivation of other known components of the DNA repair machinery,11-15 as well as the murine and equine SCID conditions in which the DNA-dependent protein kinase (DNA-PKcs) encoding gene is mutated.16,17 A similar condition exists in humans, characterized by a complete absence of both B and T cells (T-B-SCID).18,19 We have previously shown that 2 subsets can be individualized within this group of patients depending on cell sensitivity to ionizing radiation.20,21 In the first one (OMIM no. 601457) the initial phase of the V(D)J recombination is impaired, owing to mutations in either the RAG1 orRAG2 gene,22,23 although cell sensitivity to radiation is normal. The second subset (OMIM no. 602450), the affected gene of which is not known yet,24 is characterized by a defect in DNA-break repair as judged by the increased γ-ray sensitivity and the inability to rejoin coding ends during V(D)J recombination.25
Omenn syndrome (OS) (OMIM no. 603554) is yet another SCID condition characterized by the early occurrence in life of diffuse erythrodermia, hepatosplenomegaly, protracted diarrhea, and failure to thrive.26,27 No circulating mature B cells are found in these patients despite the usually high level of serum IgE. This is in sharp contrast with the detection of a large number of poorly functional, activated (HLA-DR+) T lymphocytes in blood, together with eosinophils. These T cells, which are not of maternal origin, produce TH2-type cytokines28-32 and infiltrate the skin, the gut, the liver, and the spleen, causing a graft-versus-host (GVH)–like disease.27,33-37 The T-cell population in patients with OS exhibits an extremely restricted TCR heterogeneity34,36-38 in the periphery as well as in the thymus,32 which is strongly suggestive of a defect in the lymphoid developmental program. In addition, the finding in the same kindred of siblings with either OS or T-B-SCID strengthened this hypothesis and suggested that an impaired V(D)J recombination could be the underlying molecular defect in this condition.27Indeed, mutations in both RAG1 and RAG2 genes were described in patients with OS.32 38-40 We report here the analysis of RAG1/2 genes in a series of 9 OS patients. Three of these mutations, causing OS in some patients, were also associated with typical T-B-SCID condition in other patients. Altogether, this result suggests that a low level of V(D)J recombination caused by a leaky mutation in either RAG1 orRAG2 gene may not always be sufficient to account for the Omenn condition.
Patients, materials, and methods
Patients
Patients OM1 to OM9 are suffering from typical OS (Table1) as defined by early onset of diffuse erythrodermia, protracted diarrhea, eosinophilia, failure to thrive, and expansion of oligoclonal, activated, patient-derived T lymphocytes.26,27 P27, P42, and P52 presented with a typical phenotype of autosomal T-B-recessive SCID18 with no circulating B cells, no circulating T cells for P42, and few T cells of patient origin in P27 and P52 (150/μL in each case); natural killer (NK) cells were present in all of them. Patient 42 and OM9 are siblings and were described previously.27 Patients OM1, OM2, OM3, OM5, and P52 were from consanguineous families. Informed consent was obtained from the patients' parents prior to this study.
Clinical and biologic characteristics of patients with Omenn syndrome
| . | Patients . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| OM1 . | OM2 . | OM3 . | OM4 . | OM5 . | OM6 . | OM7 . | OM8 . | OM9 . | |
| Sex | F | F | M | M | F | F | F | F | M |
| Age (mo) | 4 | 4 | 2 | 2.5 | 0.2 | 3 | 5 | 4 | 2 |
| Consanguinity | + | + | + | − | + | − | − | − | − |
| Lymph (/μL) | 8160 | 9500 | 19 500 | 3000 | 12 700 | 6000 | 1500 | 22 400 | 10 000 |
| CD3 (/μL) | 6000 | 7700 | 17 000 | 2800 | 6800 | 4800 | 1000 | 18 400 | 7400 |
| DR+ (/μL) | 1800 | 3700 | 12 000 | nd | 5100 | nd | 850 | 14 700 | nd |
| CD19 (/μL) | 0 | 190 | 0 | 0 | 127 | 60 | 0 | 0 | 0 |
| CD56 (/μL)* | nd | nd | 1380 | (+) | 1900 | nd | 90 | 380 | nd |
| IgE (KU/L) | 215 | >3000 | 38 300 | 64 | nd | 20 000 | >1000 | nd | nd |
| Eosino (/μL) | 952 | 1390 | 10 700 | 3200 | 5600 | 6700 | 500 | nd | nd |
| Erythrodermia | + | + | + | + | + | + | + | + | + |
| RAG1/2† | C2306T | G3030A | ΔT631 | 1)C1322T | ΔT631 | 1)G1533A | 1)G1983A | 1)ΔAA368 | 1)A1316G |
| mutations | L732F | R973H | T173FS | R404W | T173FS | R474H | R624H | K86FS | R39G |
| 2)ΔT2735 | 2)G2301T | 2)A3086G | 2)T1111A | 2)G1887A | |||||
| S875FS | G730F | K992E | Y333X | R229Q | |||||
| Mutation in T cells | + | + | + | + | + | nd | nd | nd | nd |
| . | Patients . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| OM1 . | OM2 . | OM3 . | OM4 . | OM5 . | OM6 . | OM7 . | OM8 . | OM9 . | |
| Sex | F | F | M | M | F | F | F | F | M |
| Age (mo) | 4 | 4 | 2 | 2.5 | 0.2 | 3 | 5 | 4 | 2 |
| Consanguinity | + | + | + | − | + | − | − | − | − |
| Lymph (/μL) | 8160 | 9500 | 19 500 | 3000 | 12 700 | 6000 | 1500 | 22 400 | 10 000 |
| CD3 (/μL) | 6000 | 7700 | 17 000 | 2800 | 6800 | 4800 | 1000 | 18 400 | 7400 |
| DR+ (/μL) | 1800 | 3700 | 12 000 | nd | 5100 | nd | 850 | 14 700 | nd |
| CD19 (/μL) | 0 | 190 | 0 | 0 | 127 | 60 | 0 | 0 | 0 |
| CD56 (/μL)* | nd | nd | 1380 | (+) | 1900 | nd | 90 | 380 | nd |
| IgE (KU/L) | 215 | >3000 | 38 300 | 64 | nd | 20 000 | >1000 | nd | nd |
| Eosino (/μL) | 952 | 1390 | 10 700 | 3200 | 5600 | 6700 | 500 | nd | nd |
| Erythrodermia | + | + | + | + | + | + | + | + | + |
| RAG1/2† | C2306T | G3030A | ΔT631 | 1)C1322T | ΔT631 | 1)G1533A | 1)G1983A | 1)ΔAA368 | 1)A1316G |
| mutations | L732F | R973H | T173FS | R404W | T173FS | R474H | R624H | K86FS | R39G |
| 2)ΔT2735 | 2)G2301T | 2)A3086G | 2)T1111A | 2)G1887A | |||||
| S875FS | G730F | K992E | Y333X | R229Q | |||||
| Mutation in T cells | + | + | + | + | + | nd | nd | nd | nd |
nd indicates not determined; IgE, immunoglobulin E; X, nonsense; FS, frameshift.
(+) functional NK activity in OM5.
RAG1 mutations in OM1-8, RAG2 mutations in OM9.
RAG1 and RAG2 gene sequencing and cloning
The RAG1 gene coding sequence was amplified by polymerase chain reaction (PCR) on genomic DNA obtained from whole blood using R1F7B 5′-CGGGATCCTTATAAGATACATCAGTGGG-3′ and R1R6X 5′-GCTCTAGAGCCCCATACACAGCAGTAA-3′ primers and the expand high fidelity PCR reaction system (Roche Molecular Biochemicals, Meylan, France) according to the manufacturer's recommendations. Nucleotide primers were chosen on the HuRAG1 sequence from Schatz and colleagues.2 The RAG2 coding sequence was amplified as previously described.23 PCR products were directly sequenced using internal primers and the dRhodamine terminator cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom). For V(D)J recombination assays, RAG1 and RAG2 PCR products were subcloned together with a c-myc epitope asBamHI-XbaI fragments in pcDNA1.1 vector (Invitrogen, Groningen, The Netherlands).
Western blot
Wild-type (wt) and mutated RAG1 expression constructs were transfected into 293T cells by conventional CaPO4precipitation. Forty-eight hours after transfection, cells were lyzed in 500 μL lysis buffer, 10 mM Tris (pH 8, 1% NP-40, 10 mM NaCl, 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) and spun 2 minutes at 15 000 rpm as described.41 The nuclear fractions were solubilized in nuclear lysis buffer (10 mM Tris [pH 8], 1% NP-40, 0.4 mM NaCl, 20 mM HEPES, 0.2 mM PMSF, 10 μg/mL pepstatin and aprotinin) and clarified by centrifugation 15 minutes at 15 000g. The pellet representing the insoluble fraction was resuspended in 100 μL 2 × Laemmli loading buffer and sonicated. Then, 10 μL was loaded on 8% sodium dodecyl sulfate (SDS)–polyacrylamide gel. After transfer, RAG1 proteins were detected by Western blot with an anti-myc antibody (clone 9E10, Santa Cruz Biotechnology, Santa Cruz, CA).
V(D)J recombination assay
The V(D)J recombination assay was carried out in fibroblasts as described previously.25 Briefly, 5 × 106exponentially simian virus 40 (SV40)–transformed fibroblasts were electroporated in 400 μL complete culture medium (RPMI 1640, 10% fetal calf serum [FCS]) with 6 μg RAG1 and 4.8 μg RAG2 encoding plasmids, carrying either the wt or the mutated sequences, together with 2.5 μg pHRecCJ, pHRecSJ, or pH2V14CJ (see below) V(D)J extrachromosomal substrates. Following transfection and recovery of the extrachromosomal constructs, the V(D)J recombination frequency was assessed by plating bacteria on X-Gal containing plates.
Vβ14-RSS sequence analysis and cloning
Vβ14 RSS sequences were PCR-amplified from genomic DNA using V14-F1 5′-CAGCCCCAACCAGACCTCT-3′ and V14-R1 5′-CTGCCCAACTTTGAAACCTCA-3′ primers and directly sequenced using dRhodamine terminator cycle sequencing kit. pH2V14CJ construct was derived from pHRecCJ by replacing the RSS-23 (SalI-EcoRI) with that of Vβ14 obtained by annealing the following oligos: 5′-AATTCCAAGCTTATCGATACCGTCGAGTGTTTTTGTGCAGAGAGCAGCTGGCTGTGCAACACTGTGGATAAACTGCTGGCACAGG-3′ and 5′-TCGACCTGTGCCAGCAGTTTATCCACAGTGTTGCACAGCCAGCTGCTCTCTGCACAAAAACACTCGACGGTATCGATAAGCTTGG-3′. The Vβ14-RSS oligos include 19 Vβ14-encoding nucleotides (in bold) upstream of the RSS heptamer motif, because coding bases flanking the RSSs were found to modify the V(D)J recombination efficiency in the context of particular RAG1 mutations.42 43
Results
RAG1 mutations in OS
The RAG1 and RAG2 genes were analyzed by genomic sequencing in a series of 9 patients with typical characteristics of OS including a high number of T lymphocytes (1500-22 400/μL) bearing HLA-DR activation marker, an almost complete absence of circulating B cells, a hypereosinophilia in 7 of 9 cases and an erythrodermia in all cases (Table 1). Eleven mutations inRAG1 and 2 mutations in RAG2 were found either as homozygous or compound heterozygous. The mutations were always found inherited from both parents. In OM3, OM5, and OM8, mutations were either nonsense (Y333X in OM8) or involved deletion of one (ΔT631 in OM3 and OM5) or 2 (ΔAA368 in OM8) nucleotides resulting in a frameshift and the appearance of premature stop codons early in the coding sequence, before the active core of the protein (amino acids 384-1009).44-46 These mutations are, in theory, not compatible with any activity of the RAG1 protein and should have resulted in a complete T-B-SCID phenotype when present on both chromosomes, which is the case for OM3 and OM5. Altogether, the homozygous ΔT631 mutation was found in 3 siblings of OM3 family, in OM5, in another unrelated patient (data not shown) as well as in a recently described OS patient.40 All these OS patients originate from the Mediterranean border. To explain the “leaky” V(D)J recombination phenotype in these patients, one has to assume that the premature stop codon generated by the ΔT631 mutation is bypassed by an alternative translation initiation through a downstream AUG codon, giving rise to an N-terminal truncated protein still containing the entire active core domain. Translation reinitiation is not unprecedented and has previously been described in mammalian cells to abrogate nonsense-mediated messenger RNA (mRNA) decay.47Noordzij and colleagues recently demonstrated that a natural alternative translation initiation site exists in human RAG1, at position 202, that leads to a N-terminal truncated protein of 100 kd.40 They demonstrated that this alternative AUG codon allows for the production of a truncated RAG1 in the context of the ΔT631 mutation protein, albeit at a very low level when compared to full-length wt RAG1. In case of OM4, the premature stop codon resulting from the ΔT2735 mutation almost certainly corresponds to a null RAG1 allele because it is located within the active core. The Omenn phenotype likely results from the missense mutation (R404W) on the other allele in this patient.
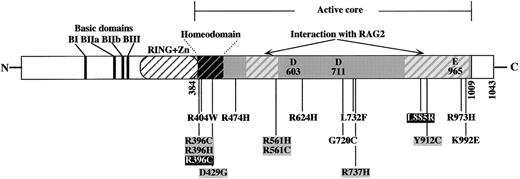
Seven of the RAG1 gene mutations (Figure1) were single nucleotide changes leading to amino acid substitutions (L732F, R973H, R404W, R474H, G720C, R624H, and K992E) (single-letter amino acid codes). All these mutations are located in the core region of RAG1. To formally exclude the possibility that these mutations could represent polymorphisms, we performed V(D)J recombination assays in fibroblasts with mutated RAG1 constructs (Table 2). The recombination frequencies obtained were dramatically reduced compared to those obtained with the wt RAG1 plasmid for all the mutations tested except for the ΔT631 (see above). This was true when using reporter construct specific for either the coding (pHRecCJ) or the signal (pHRecSJ) joint formation. Missense mutations did not result in unstable proteins as judged by Western blot analysis (Figure2). One mutation (R404W) is located within the homeodomain of RAG1,48 a region involved in DNA binding and previously associated with other mutations in patients with OS.38 R973H and K992E missense mutations are located in one of the 2 domains of RAG1 previously shown to be required for a proper interaction with RAG2.49 Finally, it is interesting to note that none of the mutations described to date in OS patients directly affect the 3 recently defined acidic residues (D603, D711, and E965) composing the catalytic active site of the protein.50-52 However, 10 of them are located close to these residues, whereas the others are clustered in the homeodomain, therefore sharpening the critical regions of RAG1. The emergence of few T cell clones in OS patients could have theoretically been the result of the reversion of the RAG1 gene mutations in precursor cells as previously described in another SCID condition.53DNA sequence analysis of purified T cells from patients OM1 through OM5 revealed the presence of the RAG1 mutations on both alleles, ruling out the possibility of a reversion and therefore attesting that residual V(D)J activity in the context of “leaky” RAG1mutations was responsible for the emergence of few T-cell clones in these patients. This is also likely to be true for B lymphocytes because OS patients secrete immunoglobulins (hyper IgE), although the reason for an absence of detectable B cells in the periphery is not yet fully understood.
Structure of the human RAG1 protein and localization of missense mutations causing OS.
Mutations are from Villa and colleagues38 (black on gray), Wada and coworkers39 (white on black), and this report (black on white). All the mutations are located within the active core (amino acids 384-1009) of the protein and some of them involve the homeodomain or one of the RAG2 interaction domains.49 Many of the mutations map close to the 3 acidic residues (DDE) defining the catalytic site of RAG1.
Structure of the human RAG1 protein and localization of missense mutations causing OS.
Mutations are from Villa and colleagues38 (black on gray), Wada and coworkers39 (white on black), and this report (black on white). All the mutations are located within the active core (amino acids 384-1009) of the protein and some of them involve the homeodomain or one of the RAG2 interaction domains.49 Many of the mutations map close to the 3 acidic residues (DDE) defining the catalytic site of RAG1.
In vitro V(D)J recombination assay with RAG1mutants causing Omenn syndrome
| RAG1 . | pHRecCJ (coding joints) . | pHRecSJ (signal joints) . | ||||
|---|---|---|---|---|---|---|
| Blue col . | Total . | R(× 10−3)* . | Blue col . | Total . | R(× 10−3)† . | |
| Wild type | 39 | 11 800 | 9.9 | 560 | 53 360 | 10.5 |
| L732F | 0 | 66 500 | < 0.01 | 0 | 930 000 | < 0.001 |
| R973H | 0 | 66 000 | < 0.01 | 1 | 17 220 | 0.06 |
| ΔT631 | 24 | 44 320 | 1.6 | 42 | 36 000 | 1.16 |
| R404W | 0 | 64 000 | < 0.01 | 0 | 876 000 | < 0.001 |
| S875FS3 | 0 | 117 000 | < 0.008 | 1 | 36 960 | 0.03 |
| R474H | 8 | 78 300 | 0.30 | 0 | 24 400 | < 0.04 |
| G720C | 0 | 27 000 | < 0.03 | 0 | 22 000 | < 0.04 |
| R624H | 0 | 117 000 | < 0.008 | 0 | 7000 | < 0.14 |
| K992E | 0 | 31 500 | < 0.03 | 0 | 15 000 | < 0.06 |
| K86FS | 0 | 19 840 | < 0.05 | 0 | 15 400 | < 0.06 |
| Y333X | 1 | 67 800 | 0.04 | 0 | 27 600 | < 0.03 |
| RAG1 . | pHRecCJ (coding joints) . | pHRecSJ (signal joints) . | ||||
|---|---|---|---|---|---|---|
| Blue col . | Total . | R(× 10−3)* . | Blue col . | Total . | R(× 10−3)† . | |
| Wild type | 39 | 11 800 | 9.9 | 560 | 53 360 | 10.5 |
| L732F | 0 | 66 500 | < 0.01 | 0 | 930 000 | < 0.001 |
| R973H | 0 | 66 000 | < 0.01 | 1 | 17 220 | 0.06 |
| ΔT631 | 24 | 44 320 | 1.6 | 42 | 36 000 | 1.16 |
| R404W | 0 | 64 000 | < 0.01 | 0 | 876 000 | < 0.001 |
| S875FS3 | 0 | 117 000 | < 0.008 | 1 | 36 960 | 0.03 |
| R474H | 8 | 78 300 | 0.30 | 0 | 24 400 | < 0.04 |
| G720C | 0 | 27 000 | < 0.03 | 0 | 22 000 | < 0.04 |
| R624H | 0 | 117 000 | < 0.008 | 0 | 7000 | < 0.14 |
| K992E | 0 | 31 500 | < 0.03 | 0 | 15 000 | < 0.06 |
| K86FS | 0 | 19 840 | < 0.05 | 0 | 15 400 | < 0.06 |
| Y333X | 1 | 67 800 | 0.04 | 0 | 27 600 | < 0.03 |
FS indicates frameshift mutation; X, nonsense mutation.
R (coding joints) = 3 × (Blue col)/(Total) × 1000.
R (signal joints) = (Blue col)/(Total) × 1000.
Western blot analysis of RAG1 mutants.
The wt and RAG1 mutants were expressed in 293T cells and revealed by Western blot using an anti-myc antibody.
Western blot analysis of RAG1 mutants.
The wt and RAG1 mutants were expressed in 293T cells and revealed by Western blot using an anti-myc antibody.
Some RAG1/2 mutations can cause both OS and T-B-SCID
We have previously described a family with both T-B-SCID (P42) and OS (OM9) affected siblings.27 Two identical compound heterozygous mutations in the RAG2 gene (R39G and R229Q) were found in these 2 patients (Tables 1 and3). R229Q substitution was previously described in another OS patient32 as well as in a T-B-SCID patient bearing a RAG1 deletion on the other allele.22 The V(D)J recombination assay using RAG2 expression plasmid harboring these mutations clearly established their deleterious effect on recombination in vitro (Corneo et al23 and data not shown). Altogether, RAG2-R229Q mutation is equally associated with OS (2 of 4 cases) and T-B-SCID (2 of 4 cases). In addition, the RAG1 ΔT631 mutation, present in several OS patients, was also responsible for the SCID phenotype in P52 (Table 3). Interestingly, the presence of few autologous T cells (150/μL) in this patient, which attests for the leakiness of the mutation, was not accompanied by the clinical and biologic manifestations of OS. Lastly, we found the same homozygous RAG1-R561H missense mutation, first identified in the OS patient OS2 described by Villa and coworkers,38 in a T-B-SCID patient (P27 in Table 3) who had also very few circulating autologous T lymphocytes (150/μL) without OS manifestation. These 3 observations clearly demonstrate that residual V(D)J recombinase activity in the context of mutated RAG1 or RAG2 proteins may not always be sufficient to cause the OS phenotype, leaving the possibility for the existence of additional factors required for the development of OS. Because OS is caused by “leaky” mutations, which are by essence variables, one can imagine that depending on the degree of leakiness, an additional factor may or may not be required for full OS phenotype expression. A precise survey ofRAG1 and RAG2 mutations in new T-B-SCID patients and OS should clarify this issue in the future.
The same mutation in RAG1 or RAG2can cause severe combined immune deficiency or Omenn syndrome
| Patients . | OS23-150 . | P27 . | OM3/5 . | P52 . | OM93-151 . | P423-151 . |
|---|---|---|---|---|---|---|
| Phenotype | Omenn | SCID | Omenn | SCID | Omenn | SCID |
| T lymphocyte (/μL) | 150 | 19 500/12 700 | 150 | 6300 | 0 | |
| B lymphocyte (/μL) | 0 | 0/0 | 0 | 0 | 0 | |
| Mutation | RAG1 | RAG1 | RAG2 | |||
| (R561H) | (ΔT631) | (R39G, R229Q) | ||||
| Patients . | OS23-150 . | P27 . | OM3/5 . | P52 . | OM93-151 . | P423-151 . |
|---|---|---|---|---|---|---|
| Phenotype | Omenn | SCID | Omenn | SCID | Omenn | SCID |
| T lymphocyte (/μL) | 150 | 19 500/12 700 | 150 | 6300 | 0 | |
| B lymphocyte (/μL) | 0 | 0/0 | 0 | 0 | 0 | |
| Mutation | RAG1 | RAG1 | RAG2 | |||
| (R561H) | (ΔT631) | (R39G, R229Q) | ||||
Discussion
Hypothesis for an additional factor involved in the development of OS
If “leaky” mutations in RAG1 or RAG2 are not solely responsible for the development of OS, what could be the additional factor(s) required to switch from T-B-SCID to OS? Two important characteristics of T cells from OS patients have to be considered in trying to address this issue. First, despite the important restriction in the T-cell repertoire in OS patients, some TCR-Vβ such as Vβ14 are often represented at high frequency in these patients.36 This finding could suggest that Vβ14 is preferentially rearranged in the context of suboptimal recombinase activity in OS patients. To test this hypothesis we first cloned the Vβ14 RSS (together with 19 nucleotides of the flanking Vβ14 coding sequence) in the pHRecCJ V(D)J reporter plasmid and performed the V(D)J recombination assay with either wt RAG1 or mutated forms of RAG1 (Table4). The pH2V14CJ reporter construct did not show a higher in vitro recombination frequency over pHRecCJ when using wt RAG1/2. Moreover, this construct was not rearranged in the context of the Omenn-specific mutated RAG1 or RAG2 proteins. We also ruled out the possibility, by DNA sequencing, of a polymorphism in the Vβ14 RSS that would be specifically present in OS patients and would render this Vβ prone to V(D)J recombination in the context of “leaky” RAG1/2 mutations (data not shown). The second striking peculiarity of the few T-cell clones present in OS patients is their continuous in vivo activation and their presence in tissues such as skin and gut, thus causing GVH-like disease.32,33,36 We and others proposed that in the context of a defective T-cell developmental program, few emerging T cells, already present in the thymus,32 could expand in the absence of retrocontrol by other populations of T lymphocytes.27 One challenging hypothesis would be that a particular antigen, present in some OS patients but not in T-B-SCID patients, triggers the few clones emerging in these patients leading to their expansion and constant activation in vivo. This antigenic hypothesis is further strengthened by the fact that several TCR characteristics, such as the nature of the Vβ segment as well as the length and amino acid composition of the CDR3 loop, observed in OS patients are recurrent.32 36 These clones would otherwise die in the absence of the specific antigen. Although the exact nature of this putative “Omenn” antigen is at present totally unknown, one can assume that it should be present in the epithelia of the skin and the gut, the 2 major sites of infiltration by activated T cells in OS patients. This antigen could either be genetically encoded (autoantigen) or provided by the environment (exoantigen).
In vitro V(D)J recombination assay with Vβ14–recombination signal sequences
| . | pHRecCJ . | . | pH2V14CJ . | ||||
|---|---|---|---|---|---|---|---|
| RAG1 | wt | wt | R973H | L732F | R404Q | wt | wt |
| RAG2 | wt | wt | wt | wt | wt | R39G | R229Q |
| Blue col | 270 | 9 | 0 | 0 | 0 | 0 | 0 |
| Total | 37 800 | 7800 | 34 500 | 20 700 | 88 800 | 156 000 | 117 000 |
| R(× 10−2)4-150 | 2.1 | 0.1 | < 0.002 | < 0.004 | < 0.001 | < 0.0006 | < 0.0008 |
| . | pHRecCJ . | . | pH2V14CJ . | ||||
|---|---|---|---|---|---|---|---|
| RAG1 | wt | wt | R973H | L732F | R404Q | wt | wt |
| RAG2 | wt | wt | wt | wt | wt | R39G | R229Q |
| Blue col | 270 | 9 | 0 | 0 | 0 | 0 | 0 |
| Total | 37 800 | 7800 | 34 500 | 20 700 | 88 800 | 156 000 | 117 000 |
| R(× 10−2)4-150 | 2.1 | 0.1 | < 0.002 | < 0.004 | < 0.001 | < 0.0006 | < 0.0008 |
wt indicates wild type.
R (coding joints) = 3 × (Blue col)/(Total) × 10.
Supported by institutional grants from Institut National de la Santè et de la Recherche Mèdicale and Ministëre de l'Education Nationale de la Recherche et de la Technologie, and grants from Association de Recherche sur le Cancer (ARC), Association contre les myopathies (AFM) Commissariat l'Energie Atomique (CEA-LRC 7V). D.M. is supported by scholarships from ARC and the Deutsche Forschungsgemeinschaft. B.C. is supported by scholarships from ARC and Ligue Contre le Cancer.
B.C. and D.M. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jean-Pierre de Villartay, INSERM U429, Hôpital Necker Enfants Malades, 149 rue de Sèvres, 75015 Paris, France; e-mail: devillar@infobiogen.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal