Abstract
The (11;19)(q23;p13.1) translocation in acute leukemia leads to the generation of a chimeric protein that fuses MLL to the transcriptional elongation factor ELL. A novel protein was isolated from a yeast 2-hybrid screen with ELL that was named EAF1 for ELL-associated factor 1. Using specific antibodies, the endogenous EAF1 and ELL proteins were coimmunoprecipitated from multiple cell lines. In addition, endogenous EAF1 also exhibited the capacity to interact with ELL2. Database comparisons with EAF1 identified a region with a high content of serine, aspartic acid, and glutamic acid residues that exhibited homology with the transcriptional activation domains of several translocation partner proteins of MLL, including AF4, LAF4, and AF5q31. A similar transcriptional activation domain has been identified in this region of EAF1. By confocal microscopy, endogenous EAF1 and ELL colocalized in a distinct nuclear speckled pattern. Transfection of theMLL-ELL fusion gene delocalized EAF1 from its nuclear speckled distribution to a diffuse nucleoplasmic pattern. In leukemic cell lines derived from mice transplanted withMLL-ELL–transduced bone marrow, EAF1 speckles were not detected. Taken together, these data suggest that expression of the MLL-ELL fusion protein may have a dominant effect on the normal protein-protein interactions of ELL.
Introduction
The ELL gene was first identified as a fusion partner gene of MLL in the (11;19)(q23;p13.1) translocation, a recurring chromosomal aberration in acute myeloid leukemia.1 Subsequent studies revealed that ELL functions as an RNA polymerase II (Pol II) transcriptional elongation factor.2 As a result of the (11;19)(q23;p13.1) translocation, an MLL-ELL chimeric protein is formed that contains the amino-terminal region of MLL, including its AT hooks, methyltransferase domain, and repression domain, fused to amino acids 46 to 621 of ELL, including its elongation domain and a lysine-rich region within ELL. ELL2 was identified by sequence homology to ELL and exhibits similar activities to ELL in transcriptional elongation assays.3However, ELL2 has not been observed in association with chromosome translocations in leukemia or in other malignancies. In addition to ELL and ELL2, several different factors with elongation activity have been identified including TFIIS, P-TEFb, TFIIF, Elongin, and FACT. TFIIS and P-TEFb each prevent specific types of transcriptional arrest. TFIIS is involved in the maintenance of transcriptional fidelity and P-TEFb protects against the inhibition of elongation by DSIF.4,5FACT facilitates elongation through its interactions with chromatin.6 In contrast, ELL, ELL2, TFIIF, and Elongin function as general elongation factors and serve to prevent transient pausing by Pol II.7
More than 30 different recurring cytogenetic aberrations that affect the MLL gene at 11q23 have been described.8,9The critical feature of these chromosomal rearrangements is the generation of a chimeric transcript consisting of 5′ MLL and 3′ sequences of the gene on the partner chromosome. The functions of most MLL partner genes are not yet known. Although no consistent homologies or motifs among the partner gene sequences have been identified that might explain how their fusion to MLLresults in leukemia, certain groups of partner genes have similar features. These include ENL and AF9, which are serine and proline rich and share extensive amino acid homology.10,11AF4, LAF4, and AF5q31are also rich in serines and prolines and exhibit homology withENL and AF9.12,13AF4,LAF4, ENL, and AF9 contain transcriptional activation domains with similar properties in reporter gene assays.14,15 Other families of MLLpartner genes include CBP and P300, which function as transcriptional coactivators and have intrinsic histone acetyltransferase activity.16,17 Expression of theMLL-AF9, MLL-ENL, MLL-CBP, andMLL-ELL fusion genes in mouse models results in the development of acute myeloid leukemia.18-21
The pathways perturbed by the expression of MLL fusion genes remain unclear. In view of the large number and diverse nature ofMLL partner genes, one hypothesis is that MLLfusion genes disrupt normal functions of MLL. Alternatively, these MLL fusions may dominantly affect functions of the partner genes. In this study, we characterize a novel protein named EAF1 for ELL-associated factor 1 that coimmunoprecipitated as a complex with ELL and colocalized with ELL in a distinct nuclear speckled pattern in multiple cell types. The EAF1 speckles were delocalized by transient expression of MLL-ELL and were not detectable in cell lines derived from MLL-ELL leukemic mice. These data suggest that MLL fusion genes have the capacity to exhibit dominant effects on the normal protein-protein interactions of MLL partner genes.
Materials and methods
Yeast 2-hybrid interaction assay
The full-length open reading frame of ELL was cloned in the pAS2-1 vector and used as the bait to screen for interacting proteins in a yeast 2-hybrid screen. Using the lithium acetate method, yeast strain Y190 was sequentially transformed with pAS2-1-ELL and then subsequently with a human bone marrow complementary DNA (cDNA) library (Clontech, Palo Alto, CA) fused to the GAL4 transactivation domain in the pGAD10 vector.22 Approximately 5 × 106 independent clones were screened. To exclude false positives, cDNA clones isolated from the library screening were retransformed in yeast strains Y190 and CG1945 along with positive and negative controls and assessed for β-gal activity and for growth on media lacking histidine in the presence of 30 mM 3-AT. The bait plasmid, pAS2-1-ELL, was used as a positive control. As negative controls, we used the pAS2-1 vector alone, and pAS2 fused to lamin C, p53, CD30, and SNF1.
Nucleotide sequencing
The positive cDNA clones were sequenced on both strands using cycle sequencing with ABI BigDye Terminators (PE Applied Biosystems, Foster City, CA). Database searches were conducted with the BLAST search algorithm and with the DeCypher II Similarity Search System.23 Three expressed sequence tag (EST) clones, 384278, 729867, and 298276, that contained matches to the sequence isolated from the library screen were obtained from American Type Culture Collection (Bethesda, MD) and sequenced. The nucleotide sequence of the EAF1 cDNA clone has been deposited in GenBank under accession number AF272973.
Northern blot analysis
Multiple tissue Northern blots (Clontech) containing approximately 2 μg per lane of purified poly(A)+ RNA from different human tissues were hybridized for 1 hour with an [α-32P] dCTP-labeled EAF1 cDNA probe prepared using random primers (Stratagene, La Jolla, CA). Filters were washed with 2 × saline sodium citrate (SSC) and 0.1% sodium dodecyl sulfate (SDS) for 40 minutes with agitation at room temperature, and with 0.1 × SSC and 0.1% SDS for 40 minutes at 50°C, and then autoradiographed. To normalize for the relative amount of RNA in each lane, the blots were stripped and then reprobed with a human β-actin cDNA probe.
Production of a monoclonal antibody to EAF1
To produce a histidine-tagged EAF1 protein in bacteria, the full-length open reading frame of EAF1 was cloned into the pET-19b expression vector (Novagen, San Diego, CA) and transformed in the Escherichia coli strain BL21(DE3). The histidine-tagged EAF1 fusion protein was purified on a nickel column and eluted in 1 M imidazole, 500 mM NaCl, 20 mM Tris-HCl, pH 7.9. A monoclonal antibody to EAF1 was generated using standard methods.
Affinity purification of rabbit polyclonal ELL antisera
Approximately 1 mg GST-ELL protein was electrophoresed on a preparative gel, transferred to nitrocellulose, blocked in tris buffered saline with 0.05% Tween 20 (TBST) containing 10% normal goat serum, and incubated with 2 mL ELL antiserum overnight at 4°C. The strips were then washed in 1% Tween and then phosphate-buffered saline (PBS). Bound antibody was eluted in 0.2 M glycine, pH 2.8, neutralized with 1 M Tris, pH 10.5, and dialyzed against PBS.
Cell culture, transient transfection, and immunoprecipitation
Human 293 and HeLa cells were transiently transfected by the calcium phosphate method using 20 μg plasmid DNA. Cell pellets were resuspended in 1 mL TEN (40 mM Tris, 1 mM EDTA, 150 mM NaCl) buffer, centrifuged for 5 minutes at 1200g at 4°C, lysed with 500 μL NETN (100 mM NaCl, 20 mM Tris, pH 8.0, 1 mM EDTA, and 0.2% NP-40) containing a cocktail of protease inhibitors (Sigma Chemical, St Louis, MO), incubated on ice for 10 minutes, and centrifuged at 2500g for 30 minutes at 4°C. To precipitate the complexes, supernatants were precleared with 30 μL protein A/G agarose beads (Santa Cruz, Santa Cruz, CA) for 30 minutes and then incubated for 1 hour with the indicated antibody. We then added 30 μL of a 50% slurry of protein A/G agarose beads, incubated overnight at 4°C, washed 5 times at 4°C with lysis buffer, boiled in Laemmli sample buffer, fractionated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes (Biorad). Blots were blocked in Tris-buffered saline (TBS) with 0.05% Tween 20 and 5% nonfat milk followed by incubations with the indicated primary and secondary antibodies in this buffer. To obtain normal murine hematopoietic progenitor cells, bone marrow cells were harvested from C57Bl/6 mice, incubated with magnetically labeled antibodies to CD5, B220, Cd11b, Gr-1, and TER119, followed by depletion of labeled differentiated cells by passage through a magnetic column (Stem Cell Technologies, Vancouver, BC, Canada).
Immunofluorescence
HeLa cells were grown for 24 hours on glass coverslips coated with 0.2% gelatin, washed with PBS, and then fixed in one of 4 conditions: 3.7% formaldehyde in PBS at room temperature, 1.75% paraformaldehyde in PBS at room temperature, −20°C acetone, or a 1:1 mixture of methanol and acetone at room temperature. Incubation with the primary and secondary antibodies, quenching, and staining with 4′6-diamidino-2-phenylindole (DAPI) was performed as previously described.24 Fluorescence images were obtained with a Zeiss Axiophot microscope and confocal images were obtained with a Zeiss/NORAN system.
Antibodies
We used each of the 4 different fixation methods outlined above with the affinity purified polyclonal anti-ELL antisera at a 1:500 dilution and the anti-EAF1 monoclonal antibody at a 1:100 dilution. For dual labeling of HeLa cells, the following antibody dilutions and fixative conditions were used: MLL (kindly provided by Dr D. Mason) at 1:4 with paraformaldehyde, PML (Santa Cruz) at 1:100 with paraformaldehyde, CBP (Santa Cruz) at 1:200 with formaldehyde, AF4 (kindly provided by Dr J. Kersey) at 1:50 with formaldehyde, nucleoli (Chemicon, Temecula, CA) at 1:1000 with paraformaldehyde, SC35 (Pharmingen, San Diego, CA) at 1:2000 with −20°C acetone, 8WG16 (BAbCO, Richmond, CA) against the large subunit of Pol II at 1:2000 with paraformaldehyde, and H5 (BAbCO) against the hyperphosphorylated CTD of Pol II at 1:1000 with −20°C acetone. For the immunoprecipitations, the cell extracts were incubated with the EAF1 monoclonal or the isotype control monoclonal at 1:10, and with the anti-FLAG monoclonal (Sigma) at 1:500. For the Western blots, the membranes were incubated with the preimmune and polyclonal anti-ELL antisera at 1:000, anti-FLAG-M2 at 1:1000, anti-EAF1 at 1:10, isotype control at 1:10, anti-GAL4 at 1:500.
Chloramphenicol acetyltransferase assays
The 293 cells were transfected with Lipofectamine (Gibco BRL, Grand Island, NY) using 1.5 μg of a GAL4-E1bCAT or E1bCAT reporter plasmid, 0.5 μg of a β-gal plasmid, and 2 μg of a plasmid containing the GAL4 DNA binding domain fused to EAF1, truncations of EAF1, or the activation domains of AF4 and LAF-4. The GAL4 DNA binding domain consists of GAL4 amino acids 1 to 147, the GAL4-E1bCAT reporter plasmid contains 5 GAL4 binding sites 5′ to an E1bTATA element, and the E1bCAT reporter plasmid contains the E1bTATA element alone.25 Total cell lysates were prepared from cells harvested 48 hours after transfection, and expression of each construct was confirmed by Western blot. The β-galactosidase values were used to normalize the amount of cell lysate for enzyme-linked immunosorbent assay (ELISA) analysis. CAT activity was assayed using an ELISA kit following the manufacturer's recommendations (Boerhringer Mannheim, Sunnyvale, CA). CAT activity was measured using a Molecular Dynamics ELISA reader. Transfections and CAT assays were repeated at least 3 times with each set of constructs. Fold activation was expressed as the ratio of the absolute values obtained from samples cotransfected with a reporter plasmid containing GAL4-E1bCAT versus those cotransfected with a reporter plasmid containing E1bCAT (lacking GAL4-DNA binding sites).
Results
Isolation of EAF1
We used a yeast 2-hybrid screen to identify proteins that interact with ELL. We isolated 6 identical clones that contained the open reading frame of a novel gene that we named EAF1 for ELL-associated factor 1. To verify the interaction of EAF1with ELL and to exclude the possibility that EAF1 might either interact with other proteins in a nonspecific manner or activate the HIS3 orβ-gal reporters by itself, we retransformed pGAD10-EAF1 with pAS2-1–ELL and a series of 4 unrelated proteins cloned in the pAS2-1 vector. These assays confirmed the specificity of the interaction of EAF1 with ELL in the yeast 2-hybrid system.
Searches of GenBank with the EAF1 sequence revealed that this gene had not been previously identified. The clones that were isolated from the screen contained 989 nucleotides with a predicted open reading frame of 804 nucleotides with a 5′ untranslated region of 157 nucleotides and a 3′ untranslated region of 28 nucleotides. We compared EAF1 sequence to the EST database and identified several matching clones. Using 3 EST clones, we were able to obtain approximately 1784 nucleotides of additional untranslated sequence 5′ to the predicted open reading frame. The predicted protein of EAF1 contains 268 amino acids and has an estimated pI of 5.06 (Figure 1A). Database searches using the BLASTP and DeCypher II Smith Waterman algorithms revealed a region of limited homology to the LAF4, AF4, and AF5q31 proteins. This homology is highest from amino acids 188 through 216 of EAF1, a region rich in serine, aspartic acid, and glutamic acid residues (Figure 1B).
Amino acid sequence, homology, and Northern blot analysis.
(A) Amino acid sequence of EAF1. The underlined sequence is displayed in panel B. (B) Homology of EAF1 to LAF4 and AF4. The region of homology identified from the DeCypher II search is rich in serine, aspartic acid, and glutamic acid residues. Identical residues are indicated in black boxes and conservative residues by gray boxes. (C) Northern blot analysis. Human multiple tissues were hybridized with anEAF1 cDNA probe. EAF1 is broadly expressed in multiple tissues except in thymus. Splice variants are seen in skeletal muscle and testis. The same blot was probed with human β-actin as a control for RNA loading.
Amino acid sequence, homology, and Northern blot analysis.
(A) Amino acid sequence of EAF1. The underlined sequence is displayed in panel B. (B) Homology of EAF1 to LAF4 and AF4. The region of homology identified from the DeCypher II search is rich in serine, aspartic acid, and glutamic acid residues. Identical residues are indicated in black boxes and conservative residues by gray boxes. (C) Northern blot analysis. Human multiple tissues were hybridized with anEAF1 cDNA probe. EAF1 is broadly expressed in multiple tissues except in thymus. Splice variants are seen in skeletal muscle and testis. The same blot was probed with human β-actin as a control for RNA loading.
Northern blot analysis
To determine the pattern of expression of EAF1messenger RNA (mRNA) in different tissues, a human multiple tissue Northern blot was hybridized with the 989-bp EAF1 cDNA fragment isolated from the bone marrow library. A single 4.5-kb transcript could be visualized in all tissues examined except for the thymus, which demonstrated only minimal expression (Figure 1C). An additional smaller transcript of approximately 2.0 kb could be detected in testis and skeletal muscle. The size of the predominant message detected on the Northern blots exceeds the size of the EAF1 contig that we assembled from the bone marrow library screen and the EST clones, indicating that an additional 5′ or 3′ untranslated sequence has not yet been isolated.
EAF1 interacts with ELL and ELL2 in vivo
To examine the potential of the endogenous EAF1 protein to interact with ELL and ELL2, we transfected 293 cells with FLAG-tagged ELL and ELL2 constructs. As a control, we also transfected 293 cells with FLAG-tagged versions of ENL and AF4. The cell lysates were immunoprecipitated with either the EAF1 antibody or an isotype control antibody and then immunoblotted with the FLAG antibody. We observed that the EAF1 antibody could coprecipitate ELL and ELL2, but not the ENL and AF4 controls (Figure2A). In a reciprocal experiment, we immunoprecipitated the cell lysates with the FLAG antibody and immunoblotted with either the EAF1 antibody or an isotype control. The transfected ELL and ELL2 proteins coimmunoprecipitated with endogenous EAF1, which migrated at approximately 43 kd (Figure2B). However, no band was detectable in the ENL and AF4 controls. In the t(11;19), amino acids 46 to 621 of ELL fuse to the N-terminus of MLL. To determine whether the capacity to bind EAF1 was retained by these ELL sequences, we transfected a FLAG-tagged construct containing amino acids 46 to 621 of ELL. Similar to full-length ELL, amino acids 46 to 621 retained the capacity to precipitate endogenous EAF1 (Figure2C). To determine the region of ELL that interacts with EAF1, we transfected a series of FLAG-tagged constructs that contained different regions of ELL. The 293 cells were immunoprecipitated with the FLAG antibody and probed with the EAF1 monoclonal antibody. Endogenous EAF1 coprecipitated with the region of ELL (amino acids 46-621) that fuses to MLL in (11;19)(q23;p13.1) translocations. The interaction domain localized to amino acids 401 to 621 of ELL (Figure 2C).
EAF1 interacts with ELL and ELL2.
(A) The 293 cells were transfected with FLAG-ELL (lanes 1, 5, and 9), FLAG-ELL2 (lanes 2, 6, and 10), FLAG-ENL (lanes 3, 7, and 11), or the FLAG-tagged AF4 activation domain (lanes 4, 8, and 12). Expression of these constructs was demonstrated by Western blot analysis of cell lysates with the FLAG antibody (lanes 1-4). Cell extracts were immunoprecipitated with an isotype control antibody (lanes 5-8) or with the EAF1 monoclonal antibody (lanes 9-12). Endogenous EAF1 coprecipitated FLAG-ELL (lane 9) and FLAG-ELL2 (lane 10), as detected by Western blot analysis using the anti-FLAG antibody. However, EAF1 did not associate with FLAG-ENL or FLAG-AF4 (lanes 11 and 12). The lower band in lanes 5 to 12 is immunoglobulin heavy chain. (B) Transfected 293 cell extracts were immunoprecipitated with the anti-FLAG antibody, divided into 2 lanes each, and probed with either an isotype control antibody or with the EAF1 monoclonal antibody. Endogenous EAF1 coprecipitated with FLAG-ELL and FLAG-ELL2 (lanes 5 and 6), but not with FLAG-ENL or FLAG-AF4 (lanes 7 and 8). As an additional control, the immunoprecipitates were also probed with an isotype control antibody. (C) The carboxy-terminus of ELL binds to EAF1. The 293 cells were transfected with FLAG-tagged constructs containing multiple regions of ELL, immunoprecipitated with the FLAG antibody, and probed with the EAF1 antibody. Endogenous EAF1 coprecipitated with ELL amino acids 46 to 621, the region of ELL contributed to the MLL-ELL fusion protein. Coprecipitation of endogenous EAF1 could also be detected with amino acids 376 to 621 and 401 to 621 of ELL, but not with amino acids 46 to 208 or 207 to 411. An arrowhead indicates the EAF1 band.
EAF1 interacts with ELL and ELL2.
(A) The 293 cells were transfected with FLAG-ELL (lanes 1, 5, and 9), FLAG-ELL2 (lanes 2, 6, and 10), FLAG-ENL (lanes 3, 7, and 11), or the FLAG-tagged AF4 activation domain (lanes 4, 8, and 12). Expression of these constructs was demonstrated by Western blot analysis of cell lysates with the FLAG antibody (lanes 1-4). Cell extracts were immunoprecipitated with an isotype control antibody (lanes 5-8) or with the EAF1 monoclonal antibody (lanes 9-12). Endogenous EAF1 coprecipitated FLAG-ELL (lane 9) and FLAG-ELL2 (lane 10), as detected by Western blot analysis using the anti-FLAG antibody. However, EAF1 did not associate with FLAG-ENL or FLAG-AF4 (lanes 11 and 12). The lower band in lanes 5 to 12 is immunoglobulin heavy chain. (B) Transfected 293 cell extracts were immunoprecipitated with the anti-FLAG antibody, divided into 2 lanes each, and probed with either an isotype control antibody or with the EAF1 monoclonal antibody. Endogenous EAF1 coprecipitated with FLAG-ELL and FLAG-ELL2 (lanes 5 and 6), but not with FLAG-ENL or FLAG-AF4 (lanes 7 and 8). As an additional control, the immunoprecipitates were also probed with an isotype control antibody. (C) The carboxy-terminus of ELL binds to EAF1. The 293 cells were transfected with FLAG-tagged constructs containing multiple regions of ELL, immunoprecipitated with the FLAG antibody, and probed with the EAF1 antibody. Endogenous EAF1 coprecipitated with ELL amino acids 46 to 621, the region of ELL contributed to the MLL-ELL fusion protein. Coprecipitation of endogenous EAF1 could also be detected with amino acids 376 to 621 and 401 to 621 of ELL, but not with amino acids 46 to 208 or 207 to 411. An arrowhead indicates the EAF1 band.
Coimmunoprecipitation of endogenous ELL and EAF1
To exclude an artifact related to transfection and to establish whether endogenous ELL and EAF1 interact in vivo, we examined untransfected 293 and HeLa cells. Cell lysates were immunoprecipitated with the EAF1 antibody or with an isotype control. These immunoprecipitates were then immunoblotted with the ELL antiserum or with its preimmune serum. We detected a single band corresponding to endogenous ELL in the cell lysates immunoprecipitated with the EAF1 monoclonal antibodies and probed with the ELL antiserum, whereas the control lanes were negative (Figure 3A). Thus, ELL and EAF1 have a direct physical interaction in vivo.
Endogenous ELL is in a complex with endogenous EAF1 in untransfected cells.
(A) The 293 cell extracts were immunoprecipitated with an isotype control antibody or with the EAF1 monoclonal antibody. Using an affinity-purified polyclonal ELL antibody, endogenous ELL was detected in the lysates precipitated by the EAF1 antibody but not in the lysates precipitated by the isotype control. As an additional control, the immunoprecipitates were also probed with the rabbit preimmune serum. (B) Endogenous ELL and EAF1 form a stable complex. The 293 cell extracts were immunoprecipitated with the EAF1 monoclonal antibody and washes were performed with increasing concentrations of SDS, NP-40, or NaCl. Washes with standard NETN are shown in the far right lane. The precipitated ELL protein was detected using the affinity-purified polyclonal ELL antibody. The ELL/EAF1 complex was stable in washes containing up to 2% NP-40 and 1.2 M NaCl. The complex began to dissociate in washes containing 0.5% SDS and was almost completely dissociated at 1% SDS.
Endogenous ELL is in a complex with endogenous EAF1 in untransfected cells.
(A) The 293 cell extracts were immunoprecipitated with an isotype control antibody or with the EAF1 monoclonal antibody. Using an affinity-purified polyclonal ELL antibody, endogenous ELL was detected in the lysates precipitated by the EAF1 antibody but not in the lysates precipitated by the isotype control. As an additional control, the immunoprecipitates were also probed with the rabbit preimmune serum. (B) Endogenous ELL and EAF1 form a stable complex. The 293 cell extracts were immunoprecipitated with the EAF1 monoclonal antibody and washes were performed with increasing concentrations of SDS, NP-40, or NaCl. Washes with standard NETN are shown in the far right lane. The precipitated ELL protein was detected using the affinity-purified polyclonal ELL antibody. The ELL/EAF1 complex was stable in washes containing up to 2% NP-40 and 1.2 M NaCl. The complex began to dissociate in washes containing 0.5% SDS and was almost completely dissociated at 1% SDS.
ELL forms a stable complex with EAF1
To determine the stability of the ELL/EAF1 complex, we examined the stringency of this interaction under conditions of high salt, increased levels of nonionic detergent, or the addition of ionic detergent. We varied the concentration of NaCl or NP-40 in the NETN buffer or added SDS to NETN for the washes of the immunoprecipitated complexes. The ELL/EAF1 complex remained intact when the cell lysates were washed with NETN buffer containing up to 2% of the nonionic detergent NP-40 or in conditions of high salt up to 1.2 M NaCl (Figure3B). The ELL/EAF1 complex was also stable when washed with NETN with the addition of the ionic detergent SDS at 0.1%. At concentrations of 0.5% SDS added to the NETN, the complex was only partially dissociated, and disruption of the ELL/EAF1 complex required the addition of 1% SDS to the NETN buffer.
EAF1 contains a transactivation domain in its region of homology to AF4 and LAF4
To investigate the potential of EAF1 to function as a transcriptional activator, we generated fusions to the GAL4 DNA binding domain with full-length EAF1, EAF1 deletion mutants, and the previously described activation domains of AF4 and LAF4 (Figure4A). These constructs were then cotransfected into 293 cells with either a GAL4-E1bCAT or a control E1bCAT reporter construct and a β-galactosidase plasmid. Cell lysates were analyzed in a CAT ELISA assay, normalizing the amount of lysate for each assay by measuring β-galactosidase expression. Expression of each effector construct was confirmed by Western blot analysis (Figure4B). We observed that full-length EAF1 could function as a transactivator. Using the EAF1 deletion mutants, we mapped this activity to a region spanning 86 amino acids that includes the region rich in serine, glutamic acid, and aspartic acid residues that is homologous to AF4 and LAF4 (Figure 4C). This region showed the strongest activity, exhibiting a 29.4 ± 8.9-fold increase in activation compared to the control. The homologous regions of AF4 and LAF4 exhibited comparable activity, namely 34 ± 12.7- and 25.5 ± 1.01- fold activation, respectively.
EAF1 contains a transactivation domain that localizes to its C-terminus that is homologous to the LAF4 and AF4 transactivation domains.
(A) Schematic diagram of the GAL4-EAF1, AF4, and LAF4 constructs used for the CAT-ELISA assay. (B) Western blot of protein extracts from 293 cells transfected with GAL4 fusions. (C) Transcriptional activation assay. The indicated fold activation levels are relative to those of the controls. Each bar represents the mean ± SD of at least 3 independent experiments.
EAF1 contains a transactivation domain that localizes to its C-terminus that is homologous to the LAF4 and AF4 transactivation domains.
(A) Schematic diagram of the GAL4-EAF1, AF4, and LAF4 constructs used for the CAT-ELISA assay. (B) Western blot of protein extracts from 293 cells transfected with GAL4 fusions. (C) Transcriptional activation assay. The indicated fold activation levels are relative to those of the controls. Each bar represents the mean ± SD of at least 3 independent experiments.
Colocalization of ELL and EAF1
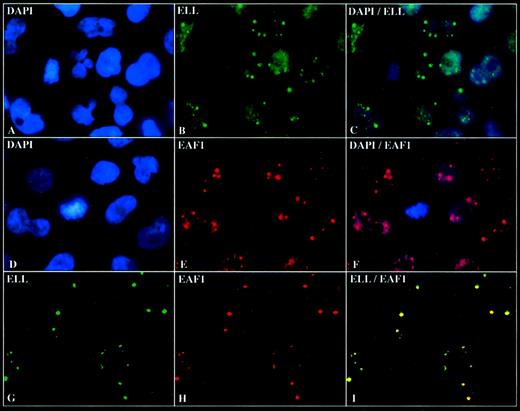
To determine the subcellular localization of EAF1, we incubated the adherent cell lines HeLa and 293 with the EAF1 antibody and observed that EAF1 exhibited a speckled nuclear pattern in nondividing cells and a diffuse pattern in dividing cells (Figure5E,F). Costaining with DAPI revealed that EAF1 localized exclusively to the nucleus. Using a polyclonal antiserum to ELL, we had previously observed that ELL also localized to the nucleus. To assess the subnuclear localization of ELL more precisely, we used the affinity-purified ELL antiserum and observed a speckled pattern in the nucleus similar to that of EAF1 (Figure 5B,C). To determine whether ELL and EAF1 might colocalize, we undertook confocal microscopy. To exclude artifact, we used the fluorochromes Cy2 and Cy5, which have no overlap in spectral emission. We observed that the nuclear speckles of ELL and EAF1 colocalized; the pattern of the speckles was indistinguishable for both ELL and EAF1 (Figure 5I). Moreover, we could detect colocalization of ELL and EAF1 speckles using several different fixation techniques, including formaldehyde, paraformaldehyde, methanol, and acetone.
ELL colocalizes with EAF1 in nuclear speckles.
(A) DAPI staining of HeLa cells. (B) Immunofluorescence with the affinity-purified polyclonal ELL antibody detected with fluorescein isothiocyanate (FITC)-labeled goat antirabbit antibodies. (C) Dual fluorescence imaging of ELL and DAPI showing that the speckles are nuclear. (D) DAPI staining of HeLa cells. (E) Immunofluorescence with the monoclonal EAF1 antibody detected with RRX-labeled goat antimouse antibodies. (F) Dual fluorescence imaging of EAF1 and DAPI. (G) Confocal microscopy with the affinity-purified polyclonal ELL antibody detected with FITC-labeled goat antirabbit antibodies. (H) Confocal microscopy with the monoclonal EAF1 antibody detected with Cy5-labeled goat antimouse antibodies. (I) Merged confocal image of ELL and EAF1 revealed colocalization.
ELL colocalizes with EAF1 in nuclear speckles.
(A) DAPI staining of HeLa cells. (B) Immunofluorescence with the affinity-purified polyclonal ELL antibody detected with fluorescein isothiocyanate (FITC)-labeled goat antirabbit antibodies. (C) Dual fluorescence imaging of ELL and DAPI showing that the speckles are nuclear. (D) DAPI staining of HeLa cells. (E) Immunofluorescence with the monoclonal EAF1 antibody detected with RRX-labeled goat antimouse antibodies. (F) Dual fluorescence imaging of EAF1 and DAPI. (G) Confocal microscopy with the affinity-purified polyclonal ELL antibody detected with FITC-labeled goat antirabbit antibodies. (H) Confocal microscopy with the monoclonal EAF1 antibody detected with Cy5-labeled goat antimouse antibodies. (I) Merged confocal image of ELL and EAF1 revealed colocalization.
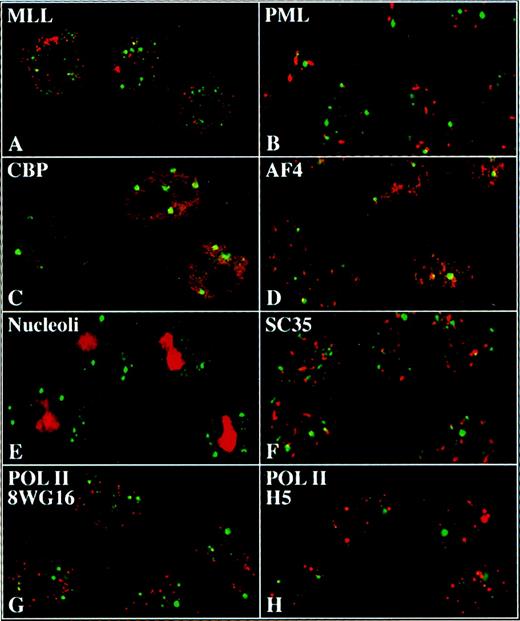
To investigate whether other proteins might also be components of the ELL/EAF1 speckles, we incubated HeLa cells with antibodies to several potential candidate proteins and assessed for colocalization by confocal microscopy (Figure 6). We examined antibodies to MLL, PML, CBP, SC-35, nucleoli, and to the hypophosphorylated and hyperphosphorylated forms of Pol II. However, we did not detect colocalization of ELL/EAF1 with any of these proteins.
ELL and EAF1 do not colocalize with candidate nuclear factors.
Merged images of immunofluorescence and confocal imaging of HeLa cells using ELL (A,B,E-H) or EAF1 (C,D) antibodies detected with an FITC-labeled second antibody. Antibodies against MLL (A), PML (B), CBP (C), AF4 (D), nucleoli (E), SC35 (F), 8WG16 (G), and H5 (H) were detected with Cy5 or RRX-labeled second antibodies.
ELL and EAF1 do not colocalize with candidate nuclear factors.
Merged images of immunofluorescence and confocal imaging of HeLa cells using ELL (A,B,E-H) or EAF1 (C,D) antibodies detected with an FITC-labeled second antibody. Antibodies against MLL (A), PML (B), CBP (C), AF4 (D), nucleoli (E), SC35 (F), 8WG16 (G), and H5 (H) were detected with Cy5 or RRX-labeled second antibodies.
Expression of the MLL-ELL fusion protein delocalizes EAF1 from nuclear speckles
To determine the effects of expression of the MLL-ELL fusion protein on the subcellular localization of endogenous EAF1, we transiently transfected HeLa cells with FLAG-tagged constructs containing full-length ELL, the amino-terminus of MLL, or the MLL-ELL fusion protein. We observed an increase in the number of both ELL and EAF1 speckles in cells transiently transfected with FLAG-ELL (Figure7). In the transiently transfected cells that overexpress ELL, numerous speckles appear to coalesce, resulting in a brighter, more diffuse nuclear pattern compared to that seen with endogenous ELL. We observed a similar nuclear staining pattern in cells transiently transfected with FLAG-ELL2 (data not shown). In cells transfected with the amino-terminus of MLL, we observed no change in the pattern of EAF1 speckles. In contrast, in cells transfected with the MLL-ELL fusion protein, we observed a fainter, more diffuse nucleoplasmic distribution of EAF1, with an absence of bright speckles (Figure 7). Recently, we have undertaken retroviral bone marrow infection of MLL-ELL to immortalize hematopoietic progenitor cells in vitro and to generate a mouse model of MLL-ELLleukemia. To determine the subcellular distribution of EAF1 in cells that stably express MLL-ELL, we examined cell lines derived from leukemic mice and from immortalized hematopoietic progenitor cells. As in the transiently transfected cells, we observed only a diffuse nucleoplasmic pattern of expression for EAF1, but no detectable speckles (Figure 8C,D). However, a punctate pattern of expression could be detected with antibodies to both MLL and ELL in these cells. A subset of the speckles colocalized, suggesting that these antibodies were detecting the MLL-ELL fusion protein in addition to the wild-type MLL and ELL proteins (Figure8E,F). To confirm the normal distribution of EAF1 and ELL in murine hematopoietic cells, we incubated normal murine bone marrow progenitor cells as well as the murine hematopoietic progenitor cell line BaF/3 with antibodies to ELL and EAF1 and observed that ELL and EAF1 colocalized in nuclear speckles (Figure 8A,B).
Delocalization of EAF1 by transient expression of MLL-ELL.
HeLa cells were transiently transfected with FLAG-tagged ELL, MLL-ELL, and the amino-terminus of MLL. Expression of these constructs was visualized by confocal microscopy using a biotinylated anti-FLAG antibody and streptavidin-FITC, and expression of endogenous EAF1 was visualized using the EAF1 monoclonal and Cy-5 labeled goat antimouse antibodies. In the upper row, an overall increase in the number of EAF1 speckles was observed in the cells transfected with FLAG-ELL. In the middle row, EAF1 was delocalized from its typical speckled distribution by expression of FLAG-MLL-ELL. In the lower row, the EAF1 pattern was not affected by expression of the amino-terminus of MLL.
Delocalization of EAF1 by transient expression of MLL-ELL.
HeLa cells were transiently transfected with FLAG-tagged ELL, MLL-ELL, and the amino-terminus of MLL. Expression of these constructs was visualized by confocal microscopy using a biotinylated anti-FLAG antibody and streptavidin-FITC, and expression of endogenous EAF1 was visualized using the EAF1 monoclonal and Cy-5 labeled goat antimouse antibodies. In the upper row, an overall increase in the number of EAF1 speckles was observed in the cells transfected with FLAG-ELL. In the middle row, EAF1 was delocalized from its typical speckled distribution by expression of FLAG-MLL-ELL. In the lower row, the EAF1 pattern was not affected by expression of the amino-terminus of MLL.
Delocalization of EAF1 in murine MLL-ELL leukemic cell lines.
Murine hematopoietic cells were examined with antibodies to ELL, EAF1, or MLL as indicated in the upper left of each image. To confirm the normal expression patterns of ELL and EAF1 in murine hematopoietic cells, confocal microscopy was used to examine normal bone marrow progenitor cells (A) and the BaF/3 cell line (B). ELL and EAF1 colocalized in a nuclear speckled pattern similar to that observed in human cells. To determine the distribution of EAF1 in cells that stably expressed the MLL-ELL fusion protein, we examined a cell line derived from hematopoietic progenitors that were immortalized in vitro by MLL-ELL (C) and a cell line derived from MLL-ELL leukemic mice (D). Confocal microscopy in both of these cell lines showed expression of ELL in a punctate pattern. In contrast, EAF1 exhibited a fainter, diffuse nucleoplasmic pattern, with no speckles detected. These cell lines were also incubated with antibodies to MLL and ELL, which revealed a punctate pattern for both proteins (E,F). A subset of the MLL and ELL speckles colocalized, suggesting that both antibodies were detecting the MLL-ELL fusion protein.
Delocalization of EAF1 in murine MLL-ELL leukemic cell lines.
Murine hematopoietic cells were examined with antibodies to ELL, EAF1, or MLL as indicated in the upper left of each image. To confirm the normal expression patterns of ELL and EAF1 in murine hematopoietic cells, confocal microscopy was used to examine normal bone marrow progenitor cells (A) and the BaF/3 cell line (B). ELL and EAF1 colocalized in a nuclear speckled pattern similar to that observed in human cells. To determine the distribution of EAF1 in cells that stably expressed the MLL-ELL fusion protein, we examined a cell line derived from hematopoietic progenitors that were immortalized in vitro by MLL-ELL (C) and a cell line derived from MLL-ELL leukemic mice (D). Confocal microscopy in both of these cell lines showed expression of ELL in a punctate pattern. In contrast, EAF1 exhibited a fainter, diffuse nucleoplasmic pattern, with no speckles detected. These cell lines were also incubated with antibodies to MLL and ELL, which revealed a punctate pattern for both proteins (E,F). A subset of the MLL and ELL speckles colocalized, suggesting that both antibodies were detecting the MLL-ELL fusion protein.
Discussion
We have isolated and characterized EAF1, a novel protein that interacts with ELL. Northern blot analysis revealed that theEAF1 gene transcript is expressed broadly with the exception of the thymus. Using a monoclonal antibody generated to EAF1 and an affinity-purified polyclonal antiserum to ELL, we have coimmunoprecipitated the endogenous ELL and EAF1 proteins, demonstrating that these proteins have a direct physical interaction in vivo. The ELL/EAF1 complex remained stable in conditions of high salt and in high levels of nonionic detergents. In transient transfections of epitope-tagged ELL2, we also found that endogenous EAF1 has the potential to bind to ELL2, suggesting that EAF1 binding may be important to the functions of both ELL and ELL2.
Using the BLASTP and the DeCypher II algorithms to compare the predicted amino acid sequence of EAF1 to that of known proteins, we identified several translocation partner proteins of MLL including LAF4, AF4, and AF5q31. The homology within these proteins is limited to a region of EAF1 that is rich in serine, aspartic acid, and glutamic acid residues. In the database searches, the LAF4 protein exhibited the greatest homology to this domain of EAF1. LAF4 was identified as a lymphoid-specific homologue of AF4, and was recently found to be involved in the t(2;11) chromosome translocation.26,27FMR2, another member of the AF4 family, is associated with the X-linked mental retardation fragile site syndrome (FRAXE), but is not known to be involved in chromosome translocations.28 In contrast to EAF1, members of the AF4 family share extensive homology with each other outside of their respective transactivation domains. In addition to the homologous amino acid sequence, EAF1, LAF4, AF5q31, and AF4 exhibit a similar overall amino acid composition within the region of homology. Moreover, a transcriptional activation domain has been mapped to this region in each of these proteins.
In view of the homology of EAF1 to the transcriptional activation domains of several MLL partner proteins, we examined EAF1 for the potential to function as a transactivator. We found that the full-length EAF1 protein could function as a relatively weak activator of transcription. This activity mapped to the C-terminal one third of EAF1, which contains the region of homology to AF4 family members. Compared to the full-length EAF1 protein, the potency of transcriptional activation was much greater in the C-terminal third of EAF1. A direct comparison of the homologous regions of EAF1, LAF4, and AF4 revealed that the transcriptional activation potential of these proteins was quite similar.
Recently, ELL was found to interact with p53 in a yeast 2-hybrid screen.29,30 Confirmation of this interaction was obtained by pull-down assays using transiently transfected epitope-tagged versions of ELL and p53. In addition, 3 proteins with approximate molecular weights of 20, 30, and 45 kd were found to coprecipitate with ELL from rat liver nuclear extracts.31 The 30-kd protein, named EAP30, was purified and peptide microsequences obtained. EAP30 exhibits amino acid homology with the yeast SNF8 protein. Comparison of the amino acid sequence reveals no homology between EAF1 and EAP30. It is unclear whether EAF1 represents the 45-kd protein identified as part of the complex with ELL in the rat liver nuclear extract. However, this protein appears to migrate slightly above a 45-kd marker, whereas EAF1 migrates at approximately 43 kd on SDS-PAGE.
By confocal microscopy, ELL and EAF1 speckles colocalized within the nucleus, indicating that ELL and EAF1 exist in a distinct nuclear complex. However, we cannot exclude that small amounts of these 2 proteins may exist outside of the nuclear speckles. At this time, it is not clear whether the ELL/EAF1 nuclear speckles represent storage forms of these proteins or a distinct substructure within the nucleus. In dividing cells, ELL and EAF1 were distributed diffusely, consistent with the dissolution of the nuclear membrane during cell division. The disappearance of the speckles in dividing cells may be associated with the lack of transcription by Pol II during mitosis.
The ELL/EAF1speckles were distinct from the other nuclear proteins that we have examined. We found no colocalization with the MLL protein, which exhibits a punctate nuclear distribution, suggesting that ELL and MLL are in distinct compartments within the nucleus.32 We examined several other proteins involved in chromosome translocations that did not colocalize with ELL and EAF1, including PML, AF4, and CBP. The AF4 protein exhibits a punctate nuclear pattern by immunofluorescence and is a partner protein of MLL in (4;11) translocations.33 CBP is a transcriptional coactivator and a partner protein of MLL in (11;16) translocations. Recently, several lines of evidence have linked RNA processing and splicing to transcription by Pol II. In addition, splicing factors have been shown to localize in speckled domains within the nucleus.34However, we found that ELL/EAF1 speckles did not colocalize with the SC-35 splicing factor. We also found that neither phosphorylated form of Pol II colocalized with ELL/EAF1. Hyperphosphorylation of Pol II is associated with transcriptional elongation. The hypophosphorylated form of Pol II localizes relatively diffusely within the nucleus, whereas the hyperphosphorylated form exhibits a speckled nuclear distribution.35 Because other proteins involved in transcription have been identified in a speckled distribution in the nucleus, the ELL/EAF1 speckles may have functional importance. The nuclear speckles that contain splicing factors such as SF2/ASF or SC-35 appear to supply these factors to genes actively transcribed by Pol II.34 Similarly, a potential role for the ELL/EAF1 speckles may be to supply elongation factors to the transcription complex.
We observed an increase in the number of EAF1 speckles in cells transfected with full-length ELL, suggesting that overexpression of ELL may affect the expression of endogenous EAF1. Strikingly, EAF1 speckles were not observed in HeLa cells transiently transfected with the MLL-ELLfusion gene. In contrast, transfection of the amino-terminus ofMLL did not affect the distribution of EAF1 nuclear speckles. At this time, human cell lines derived fromMLL-ELL leukemia cells have not been generated. Thus, we examined cell lines derived from leukemic mice transplanted with bone marrow infected with an MLL-ELL retrovirus or from primary hematopoietic progenitor cells that were immortalized in vitro with theMLL-ELL retrovirus. In both of these cell lines, expression of MLL and ELL could be detected with specific antibodies. We observed partial colocalization of MLL and ELL, suggesting that these antibodies were both recognizing the MLL-ELL fusion protein. In addition, we could detect nonoverlapping nuclear staining for both MLL and ELL, indicating the presence of wild-type MLL and ELL expression in these cells. However, we observed a diffuse nucleoplasmic staining pattern for EAF1, with no detectable speckles. A similar phenomenon is observed in acute promyelocytic leukemia as a result of the (15;17) translocation. In acute promyelocytic leukemia cell lines, nuclear body structures are disrupted with delocalization of PML and other nuclear body proteins.36,37 This occurs despite the expression of wild-type PML in addition to PML-RARα. The delocalization of EAF1 by MLL-ELL suggests that expression of MLL fusion proteins may dominantly affect the normal protein-protein interactions of MLL partner proteins. Expression of the amino-terminus of MLL by itself is not sufficient to generate leukemia in mouse models.18 19 Thus, it is not clear whether MLL fusion proteins act solely through pathways normally regulated by MLL. In these experiments, we observed that expression of MLL-ELL disrupted the normal subnuclear distribution of EAF1. Thus, expression of MLL fusion proteins may affect the normal functions of MLL, its partner proteins, or their associated factors.
Using a hematopoietic progenitor immortalization assay, we have recently undertaken a structure-function analysis of the ELLgene.38 We sequentially fused individual regions ofELL to 5′ MLL and examined their capacity to immortalize progenitor cells in vitro. Whereas the amino-terminal elongation domain within ELL was dispensable for immortalization, the carboxy-terminus of ELL was necessary and sufficient to immortalize hematopoietic progenitor cells. The carboxy-terminus of ELL contains its EAF1 interaction domain, suggesting that retention of this protein-protein interaction domain may be relevant to leukemic transformation by MLL-ELL. However, this region of ELL may also mediate interactions with other proteins or contain additional functional motifs.
The functional significance of the interaction of ELL with EAF1 remains to be defined. ELL has been shown to function as a Pol II elongation factor in vitro, and we now show that EAF1 has the capacity to function as a transcriptional activator. Previously, the functions of transcriptional activation and elongation factors have been found to overlap. In assays of processivity through transcriptional termination sites within c-myc and human immunodeficiency virus 2, transactivators including VP-16 and E1a have been shown to stimulate elongation by Pol II.39 In addition, the transcriptional activation domains of VP16 and heat shock factor 1 have been found to function not only in initiation but also in elongation by Pol II on anhsp70 gene template.40 However, mutational analysis within these activation domains revealed that distinct residues were essential for either initiation or elongation. Similarly, the ELL/EAF1 complex may function in both transcriptional activation and elongation. Future studies of ELL, EAF1, and other MLL partner proteins will be necessary to define the specific pathways that are regulated by these factors.
We are grateful to Dr D. Mason, Dr J. Kersey, and Dr P. Domer for generously providing reagents. We would like to thank Carol McShan and the University of Chicago Frank Fitch Monoclonal Antibody Facility (supported by grant CA14599 from the National Cancer Institute) and the Al Robin Laser Scanning Confocal Microscopy Core of the University of Chicago Digestive Disease Center.
Supported by grants from the National Institutes of Health (CA78431), Burroughs Wellcome Fund, Cancer Research Foundation, American Cancer Society Illinois Division, and the family of Robert A. Chapski.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michael J. Thirman, Section of Hematology/Oncology, University of Chicago, 5841 S Maryland Ave, MC2115, Chicago, IL 60637; e-mail: mthirman@medicine.bsd.uchicago.edu.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal