Abstract
Human immunodeficiency virus type 1 (HIV-1) uses the chemokine receptors CCR5 and CXCR4 as coreceptors for entry. It was recently demonstrated that HIV-1 glycoprotein 120 (gp120) elevated calcium and activated several ionic signaling responses in primary human macrophages, which are important targets for HIV-1 in vivo. This study shows that chemokine receptor engagement by both CCR5-dependent (R5) and CXCR4-dependent (X4) gp120 led to rapid phosphorylation of the focal adhesion-related tyrosine kinase Pyk2 in macrophages. Pyk2 phosphorylation was also induced by macrophage inflammatory protein-1β (MIP-1β) and stromal cell–derived factor-1α, chemokine ligands for CCR5 and CXCR4. Activation was blocked by EGTA and by a potent blocker of calcium release–activated Ca++(CRAC) channels, but was insensitive to pertussis toxin (PTX), implicating CRAC-mediated extracellular Ca++ influx but not Gαi protein-dependent mechanisms. Coreceptor engagement by gp120 and chemokines also activated 2 members of the mitogen-activated protein kinase (MAPK) superfamily, c-Jun amino-terminal kinase/stress-activated protein kinase and p38 MAPK. Furthermore, gp120-stimulated macrophages secreted the chemokines monocyte chemotactic protein-1 and MIP-1β in a manner that was dependent on MAPK activation. Thus, the gp120 signaling cascade in macrophages includes coreceptor binding, PTX-insensitive signal transduction, ionic signaling including Ca++ influx, and activation of Pyk2 and MAPK pathways, and leads to secretion of inflammatory mediators. HIV-1 Env signaling through these pathways may contribute to dysregulation of uninfected macrophage functions, new target cell recruitment, or modulation of macrophage infection.
Introduction
Human immunodeficiency virus type 1 (HIV-1) infection is initiated by the formation of a trimolecular complex on the cell surface, consisting of the HIV-1 envelope (Env) glycoprotein 120 (gp120), the principal cellular receptor CD4, and a chemokine receptor that functions as a coreceptor (reviewed by Berger1). Conformational changes induced in gp120 on binding to CD4 increase its affinity for the coreceptor, leading to fusion between the viral and cellular membranes. The chemokine receptors that serve as HIV coreceptors are members of the 7 transmembrane G protein–coupled receptor (GPCR) family. CCR5 and CXCR4 play a dominant role in HIV-1 entry into primary cells, and are the principal coreceptors used by macrophage (M)-tropic (R5) and T-cell line (T)–tropic (X4) isolates, respectively.
Given their normal cell signaling function, the interaction of gp120 with the chemokine receptors may, in addition to facilitating viral entry, also result in activation of cellular responses that could modulate the activation status of the cells and affect postentry stages of HIV replication (reviewed by Popik and Pitha2). Although chemokine receptors and their ligands play central roles in both HIV infection and in immune regulation, how signaling pathways mediated by CCR5 and CXCR4 contribute to the immunopathogenesis of HIV infection is not well defined. In T cells, gp120 activation of the chemokine receptors has been shown to elevate calcium3 and activate the focal adhesion tyrosine kinases Pyk2 and FAK4-6 as well as the mitogen-activated protein kinase (MAPK) pathway.7 8
Macrophages are important targets for HIV-1 in vivo. Infected macrophages may serve as a reservoir for viral persistence and participate in person-to-person transmission and are central to the pathogenesis of brain, lung, and other end-organ disease in acquired immunodeficiency syndrome (AIDS).9-12 In addition, aberrant functions of both infected and uninfected macrophages have been implicated in pathogenesis.13-15 Macrophages express CCR5, which is used for entry by primary and prototype M-tropic R5 HIV-1 variants. Macrophages also express CXCR4, which mediates entry by some X4 and R5X4 primary isolates, although laboratory-adapted X4 strains are restricted in their ability to use macrophage CXCR4 for reasons that are not yet clear.16-18 However, relatively little is known about how the chemokine receptors are coupled to cellular signaling pathways in primary macrophages. Because the cellular signaling pathways activated through 7-transmembrane GPCRs may differ depending on the cell type,19 we have examined whether gp120 engagement of the chemokine receptors leads to activation signals in primary human macrophages. Recently we reported that Env activates several ion channels and elevates calcium in macrophages through CCR5 and CXCR4.20 In this study we examined the ability of chemokine receptor stimulation by gp120 to activate protein kinase pathways in primary macrophages, because protein phosphorylation is an important molecular mechanism by which extracellular signals produce biologic responses in cells. In addition, to address potential functional consequences of Env signaling in macrophages, we examined β-chemokine secretion after gp120 stimulation. Our studies show that HIV-1 gp120 activates the focal adhesion–related kinase Pyk2 in primary human macrophages via CCR5 and CXCR4, stimulates phosphorylation of p38 and c-Jun amino terminal kinase/stress-activated protein kinase (JNK/SAPK) MAPKs, and leads to β-chemokine secretion. These results help elucidate the pathways by which the chemokine receptors are linked in primary macrophages and provide insight into mechanisms by which HIV-1 may dysregulate macrophage functions and contribute to pathogenesis.
Materials and methods
Primary human macrophages, Env glycoproteins, and chemokines
Monocytes were isolated from peripheral blood mononuclear cells of healthy donors by a 2-stage selective adherence procedure as previously described.21 Monocytes were plated at 5 × 105 cells/well in 24-well tissue culture plates and maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10% horse serum,l-glutamine (1 mM), penicillin (100 U/mL), and streptomycin (100 μg/mL), without exogenous cytokines or growth factors. Cells were cultured at 37°C under 5% CO2 for 1 week prior to study to allow differentiation into monocyte-derived macrophages (MDMs). Donors were screened for the CCR5 Δ32 mutation by polymerase chain reaction,22 and only donors homozygous for the wild-type allele were used for these studies, except for specified experiments in which CCR5-deficient macrophages were obtained from donors homozygous for the mutant allele.
Recombinant gp120 was produced as previously described23in 293T cells infected with gp120-expressing recombinant vaccinia viruses vCB28 (JRFL) or vSC60 (IIIB, BH8 clone) and was a generous gift of R. Doms (University of Pennsylvania). Supernatants were clarified by centrifugation and filtered (0.45 μm pore size). Virus was inactivated (0.1% Triton X-100) and gp120 was purified usingGalanthus nivalis lectin-coupled agarose beads (Vector Labs, Burlingame, CA) followed by protein concentration and buffer exchange. Env integrity was confirmed by Western blot with a rabbit polyclonal antibody as described.23 Chemokines signaling was investigated with macrophage inflammatory protein-1β (MIP-1β) and stromal cell–derived factor-1α (SDF-1α) (Peprotech, Rocky Hill, NJ).
Cell treatment
Twenty-four hours before exposure to gp120 or chemokines, MDM culture medium was removed and replaced with fresh media lacking serum to maintain the cells in a quiescent state. Macrophages were then exposed for the indicated periods of time at 37°C to either gp120 or chemokines (2.5 μg/mL and 200 nM, respectively, unless otherwise specified). Cells were then lysed and subjected to immunoprecipitation and immunoblot analysis. Cells were lysed in ice-cold RIPA buffer (50 mM Tris-HCl, pH 7.4; 1% NP-40; 0.5% sodium deoxycholate; 150 mM NaCl; 0.1% sodium dodecyl sulfate [SDS]) supplemented with protease and phosphatase inhibitors (5 mM phenylmethylsulfonyl fluoride; 5 μg/mL pepstatin; 5 μg/mL leupeptin; 5 μg/mL aprotinin; 1 mM sodium vanadate; 1 mM sodium fluoride; 1 mM sodium pyrophosphate) and incubated for 10 minutes at 4°C. Cell lysates were then clarified by centrifugation at 13 000 rpm for 30 minutes at 4°C. Protein concentrations were determined by protein assay (Bio-Rad Laboratories, Hercules, CA).
To test the effect of specific inhibitors, cells were treated prior to and during gp120 exposure with the CXCR4 antagonist AMD3100 (1 μg/mL); Bordetella pertussis toxin (PTX; 100 ng/mL; obtained from both Sigma, St Louis, MO, and Calbiochem, San Diego, CA); EGTA (2 mM; Sigma); and the calcium release–activated Ca++(CRAC) channel blocker lanthanum chloride (100 nM; Sigma). Inhibitors were added to MDMs 20 to 60 minutes before gp120 exposure, except for PTX, which was added up to 18 hours before exposure. To ensure that inhibitory effects were not due to nonspecific toxicity, signaling antagonists were tested for cytotoxicity on human macrophages by vital staining using a methyl tetrazolium (MTT)–based assay (CellTiter AQ; Promega, Madison, WI). MDMs were plated at 5 × 104cells/well in 96-well tissue culture. After 1 week in culture, medium was replaced with low serum medium (10% FBS) that was lacking phenol red. Test agents were added and cells were incubated for 18 hours. Tetrazolium reagent was then added and incubated for an additional 1 hour, and the optical density (OD; 492 nm) was determined using a plate spectrophotometer. Within each experiment a standard curve was established on the same plate by seeding MDMs at varying concentrations from 1 × 104 through 8 × 104 cells/well. The standard curve demonstrated a linear relationship between cell number and OD, and cytotoxicity was determined based on comparison with the standard curve. Five replicate wells were used to derive each data point.
Immunoprecipitation and immunoblot analysis
For Pyk2 immunoprecipitation, cell lysates containing identical amounts of protein (400 μg) from cells subjected to each treatment were clarified by incubation for 1 hour at 4°C with protein A+G-Sepharose (Santa Cruz Biotechnology; Santa Cruz, CA). Protein A+G-Sepharose was removed by centrifugation, and the supernatants were then incubated with 1 μg of the anti-Pyk2 monoclonal antibody (mAb) Pyk2/CAKβ (Transduction Laboratories; Lexington, KY) for 2 hours at 4°C. Antibody-antigen complexes were then immunoprecipitated by incubation for 2 hours at 4°C with 50 μL protein A+G-Sepharose (50% suspension) and then washed 3 times with lysis buffer and once with phosphate-buffered saline (PBS). Immunocomplexes were resuspended in Laemmli buffer, boiled for 5 minutes, and subjected to electrophoresis and immunoblotting.
Immunoblot analysis was carried out on immunoprecipitates generated as described above or directly on cell lysates containing 40 μg protein that were mixed with an equal volume of 2 × Laemmli buffer and boiled for 5 minutes. Samples were subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. The membranes were blocked overnight with 5% nonfat milk protein in PBS. They were then incubated sequentially with the primary specific antibody for 3 hours at room temperature or overnight at 4°C, washed, and then incubated with peroxidase-conjugated secondary antibody. The immunoreactive bands were visualized using enhanced chemiluminescence Western blotting system according to the manufacturer's instructions (Amersham, Piscataway, NJ). Blots were then stripped (2% SDS, 62.5 mM Tris, 100 mM β-mercaptoethanol) for 30 minutes at 50°C and washed in PBS containing 0.05% Tween 20, before blocking and reprobing with primary antibody.
Immunoblot detection was carried out using a 1:1000 dilution of antiphosphotyrosine mAb 4G10 (Upstate Biotechnology, Lake Placid, NY) in the case of Pyk2 immunoprecipitates. Whole cell extracts were analyzed using 0.5 μg/mL of a polyclonal antibody Anti-Pyk2 [pY402], which is specific for the Tyr402 phosporylated form of Pyk2 (Biosource International, Camarillo, CA), or 0.25 μg/mL mAb Pyk2/CAKβ, which detects total level of Pyk2 (Transduction Laboratories). MAPKs were detected using a 1:1000 dilution of polyclonal antibodies specific for Phospho-p38 [Thr180/Tyr182], Phospho-JNK/SAPK [Thr183/Tyr185], and Phosphop44/42 [Thr202/Tyr204], or a 1:1000 dilution of mAbs that detect total p38, JNK/SAPK, and ERK1/2 (all from Cell Signaling Technology, Beverly, MA). Peroxidase-conjugated secondary antibodies were used at a 1:3000 dilution (Amersham). Electrophoresis reagents and nitrocellulose membranes were obtained from Bio-Rad Laboratories.
Measurement of chemokine secretion
One-week-old macrophages were treated with gp120 (1 μg/mL) for 24 hours, in the presence or absence the MAPK-specific inhibitor SB-202190 (1 μg/mL; Calbiochem). Supernatant was clarified by centrifugation (5000 rpm for 5 minutes at 4°C) and stored at −80°C until analysis. Monocyte chemotactic protein-1 (MCP-1) and MIP-1β production in supernatant was measured by enzyme-linked immunoadsorbent assay (ELISA) as described by the manufacturer (R & D Systems; Minneapolis, MN). The detection limit for MCP-1 was 5 pg/mL and for MIP-1β 4 pg/mL.
Results
JRFL gp120 activates Pyk2 kinase in primary human macrophages
Pyk2 is a nonreceptor tyrosine kinase related to focal adhesion kinase (FAK) that is expressed mainly in cells of hematopoietic and neuronal lineage.24-26 Pyk2 can be activated by several growth factors, chemokines, and GPCR ligands, and on activation is phosphorylated at 4 tyrosine phosphorylation sites (Tyr402, 579, 580, 881), with Tyr402 as the major autophosphorylation site.27-31
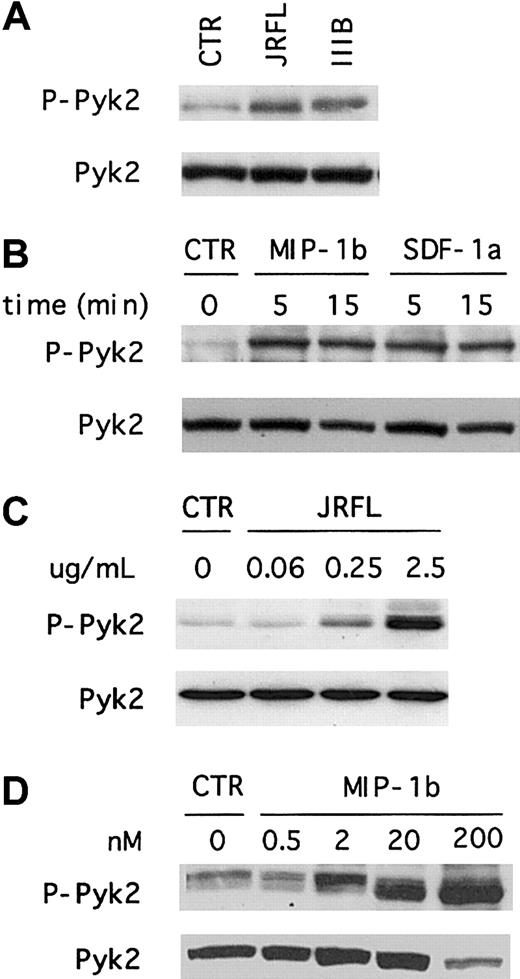
To determine whether HIV-1 Env activates Pyk2 in primary human macrophages, we treated MDMs with recombinant gp120 from the R5 isolate JRFL and analyzed the levels of total and Tyr402-phosphorylated forms by combined immunoprecipitation and immunoblot using specific antibodies (Figure 1A). This analysis showed that MDMs express an abundance of total Pyk2 protein that had a molecular mobility of approximately 110 kd, in accord with prior studies of Pyk2 expression in monocytes.32 Exposure of macrophages to gp120 resulted in rapid Pyk2 tyrosine phosphorylation. We next determined the time course of Pyk2 activation using immunoblot analysis of whole cell lysates with antibodies specific for the total or activated forms of Pyk2. As shown in Figure 1B, MDMs responded to JRFL Env in a time-dependent manner. Pyk2 tyrosine phosphorylation generally peaked within 5 minutes and decreased to nearly background levels by about 30 minutes. Analysis of earlier time points demonstrated activation as early as 1 minute after stimulation and showed that maximal activation occurred approximately 5 minutes after treatment (data not shown).
Tyrosine phosphorylation of Pyk2 in primary human macrophages occurs in response to HIV-1 R5 Env.
(A) Week-old cultures of MDMs were unstimulated (CTR) or exposed for 5 minutes to JRFL gp120 (2.5 μg/mL), lysed, and subjected to immunoprecipitation with an anti-Pyk2 antibody. Immune complexes were resolved on 8% SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to serial immunoblotting with an antiphosphotyrosine 4G10 antibody (upper panel) and an anti-Pyk2 antibody (lower panel). (B) MDMs were unstimulated (CTR) or exposed to JRFL gp120 (2.5 μg/mL) for the indicated times (minutes). Cell lysates were resolved by 8% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-Pyk2 phosphospecific [pY402] antibody (upper panel, P-Pyk2). The same blot was then reprobed with an antibody specific for total Pyk2 (lower panel). Blots shown are representative of 5 independent experiments using different donors.
Tyrosine phosphorylation of Pyk2 in primary human macrophages occurs in response to HIV-1 R5 Env.
(A) Week-old cultures of MDMs were unstimulated (CTR) or exposed for 5 minutes to JRFL gp120 (2.5 μg/mL), lysed, and subjected to immunoprecipitation with an anti-Pyk2 antibody. Immune complexes were resolved on 8% SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to serial immunoblotting with an antiphosphotyrosine 4G10 antibody (upper panel) and an anti-Pyk2 antibody (lower panel). (B) MDMs were unstimulated (CTR) or exposed to JRFL gp120 (2.5 μg/mL) for the indicated times (minutes). Cell lysates were resolved by 8% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-Pyk2 phosphospecific [pY402] antibody (upper panel, P-Pyk2). The same blot was then reprobed with an antibody specific for total Pyk2 (lower panel). Blots shown are representative of 5 independent experiments using different donors.
Although MDMs from most donors had little or no basal Pyk2 phosphorylation and demonstrated a robust response to gp120 stimulation, there was some donor-to-donor variability in the level of constitutive Pyk2 phosphorylation under unstimulated conditions and the magnitude of phosphorylation response to gp120. In general, MDMs from donors that showed high levels of basal Pyk2 phosphorylation had relatively little up-regulation elicited by gp120 (data not shown). Interestingly, the higher levels of constitutive Pyk2 activation were seen primarily among MDMs from cigarette smokers, the significance of which is presently unknown, but for that reason MDMs from nonsmoker donors were used for further analysis.
X4 gp120 as well as chemokines MIP-1β and SDF-1α activate Pyk2 in MDMs
Even though primary macrophages are not permissive for prototype CXCR4-dependent HIV-1 strains, we and others recently showed that MDMs express CXCR4 and that macrophage CXCR4 is able to support entry by some primary X4 isolates.16-18 Furthermore, CXCR4 in macrophages is functionally coupled to ionic signaling pathways including those that regulate intracellular calcium.20 33Therefore, we investigated whether X4 gp120 would also induce Pyk2 tyrosine phosphorylation in primary human macrophages. As shown in Figure 2A, Pyk2 phosphorylation was readily detected within 5 minutes after exposure to IIIB gp120. The overall levels of phosphorylated protein and the kinetics of activation were similar to those observed after JRFL gp120 stimulation. Thus, both R5 and X4 Env activate Pyk2 in primary macrophages. We observed the same Pyk2 activation pattern using several independent preparations of JRFL and IIIB gp120, and Pyk2 activation was completely abrogated when MDMs were treated with gp120 that had been subjected to heat inactivation (data not shown).
X4 gp120 and chemokines induce macrophage Pyk2 tyrosine phosphorylation.
Macrophages were stimulated for 5 minutes with 2.5 μg/mL X4 (IIIB) and R5 (JRFL) gp120 (A), or stimulated for the indicated time periods with 200 nM of the chemokines MIP-1β or SDF-1α (B). Cell lysates were resolved by 8% SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis with an antibody specific for the phosphorylated form of Pyk2 (upper panels, P-Pyk2). The same blot was then reprobed with an antibody specific for total Pyk2 (lower panels). Each experiment was repeated at least 4 times and the presented blots are representative of these experiments. To test the effect of different agonist concentrations, macrophages were stimulated for 5 minutes with the indicated concentrations of gp120 (C) and MIP-1β (D), and analyzed by immunoblot as described above.
X4 gp120 and chemokines induce macrophage Pyk2 tyrosine phosphorylation.
Macrophages were stimulated for 5 minutes with 2.5 μg/mL X4 (IIIB) and R5 (JRFL) gp120 (A), or stimulated for the indicated time periods with 200 nM of the chemokines MIP-1β or SDF-1α (B). Cell lysates were resolved by 8% SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis with an antibody specific for the phosphorylated form of Pyk2 (upper panels, P-Pyk2). The same blot was then reprobed with an antibody specific for total Pyk2 (lower panels). Each experiment was repeated at least 4 times and the presented blots are representative of these experiments. To test the effect of different agonist concentrations, macrophages were stimulated for 5 minutes with the indicated concentrations of gp120 (C) and MIP-1β (D), and analyzed by immunoblot as described above.
We then tested whether macrophage chemokine receptor stimulation by the natural ligands would elicit Pyk2 activation responses similar to that seen with Env, using MIP-1β, the most specific of several chemokines that signal through CCR5, and SDF-1α, the only known CXCR4 ligand. Both MIP-1β and SDF-1α treatment led to Pyk2 tyrosine phosphorylation (Figure 2B). Thus, Pyk2 is activated in primary macrophages both by gp120, which binds CD4 and the chemokine receptors on the cell surface, and by MIP-1β and SDF-1α, which activate CCR5 and CXCR4 independently of CD4.
To address the concentration dependence of Pyk2 activation, we tested the effects of varying gp120 and chemokine concentrations (Figure 2C,D). For gp120, Pyk2 activation was seen with concentrations ranging from 0.25 to 2.5 μg/mL (2-20 nM), but not at 0.06 μg/mL (0.5 nM) or below (Figure 2C and data not shown). Pyk2 was activated by MIP-1β at concentrations of 200, 20, and 2 nM, but not by 0.5-nM concentration (Figure 2D). Thus, gp120 and MIP-1β at equimolar concentrations both activated Pyk2.
Macrophage Pyk2 activation by R5 and X4 gp120 is mediated by CCR5 and CXCR4
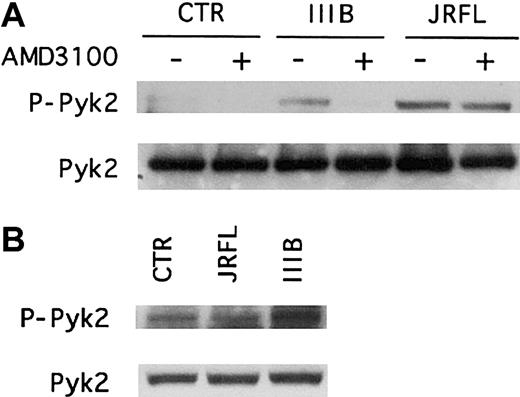
We next investigated whether the chemokine receptors were responsible for gp120-induced Pyk2 activation in macrophages (Figure3). To probe CXCR4 we used the bicyclam antagonist AMD3100, which blocks both the HIV-1 coreceptor function and SDF-1α signaling capability of CXCR4.34 The role of CCR5 was assessed using CCR5-deficient macrophages, obtained from donors homozygous for the CCR5 Δ32 allele. This frameshift mutation in the Δ32 allele abrogates CCR5 surface expression and eliminates both its chemokine signaling and coreceptor function.22 35
CCR5 and CXCR4 mediate Pyk2 phosphorylation in primary human macrophages treated with HIV-1 gp120.
(A) MDMs from a CCR5 wild-type donor were pretreated for 1 hour with (+) or without (−) the CXCR4 antagonist AMD3100 (1 μg/mL) before incubation for 5 minutes with R5 (JRFL) or X4 (IIIB) gp120 (2.5 μg/mL). (B) CCR5− MDMs derived from donors homozygous for the CCR5 Δ32 deletion allele were treated for 5 minutes with JRFL or IIIB gp120 (2.5 μg/mL). Whole cell lysates were analyzed by immunoblotting with an antibody specific for the phosphorylated form of Pyk2 (upper panel, P-Pyk2) or with antibody specific for total Pyk2 (lower panel). Each experiment was repeated at least 3 times and the blots shown are representative of these experiments.
CCR5 and CXCR4 mediate Pyk2 phosphorylation in primary human macrophages treated with HIV-1 gp120.
(A) MDMs from a CCR5 wild-type donor were pretreated for 1 hour with (+) or without (−) the CXCR4 antagonist AMD3100 (1 μg/mL) before incubation for 5 minutes with R5 (JRFL) or X4 (IIIB) gp120 (2.5 μg/mL). (B) CCR5− MDMs derived from donors homozygous for the CCR5 Δ32 deletion allele were treated for 5 minutes with JRFL or IIIB gp120 (2.5 μg/mL). Whole cell lysates were analyzed by immunoblotting with an antibody specific for the phosphorylated form of Pyk2 (upper panel, P-Pyk2) or with antibody specific for total Pyk2 (lower panel). Each experiment was repeated at least 3 times and the blots shown are representative of these experiments.
Immunoblot analysis of macrophage lysates using antibodies specific for the activated or total forms of Pyk2 showed that in wild-type MDMs, the CXCR4 antagonist AMD3100 completely inhibited IIIB gp120-induced Pyk2 activation but had no effect on the responses to JRFL gp120 (Figure3A). In contrast, macrophages lacking CCR5 exhibited reduced activation following JRFL gp120 exposure but responded to IIIB Env (Figure 3B). AMD3100 also blocked Pyk2 activation by SDF-1α but not by MIP-1β, whereas CCR5− MDMs responded to SDF-1α but not MIP-1β (data not shown). Thus, R5 and X4 gp120 activation of Pyk2 in macrophages is specifically mediated by the Env glycoproteins' cognate coreceptors. Furthermore, because neither AMD3100 nor the absence of functional CCR5 would interfere with gp120 binding to CD4, these results also show that CD4 binding in the absence of chemokine receptor stimulation is not sufficient to activate Pyk2.
Pyk2 activation is calcium-dependent but insensitive to PTX
We next wished to address the mechanisms involved in gp120-elicited chemokine receptor–mediated Pyk2 activation in macrophages. In some cell systems Pyk2 phosphorylation is regulated by calcium,25,26,30,31,36 and we and others recently found that gp120 induces intracellular calcium elevations in MDMs.20,37 We therefore determined whether Pyk2 activation by JRFL gp120 is regulated by [Ca++]ielevations. As shown in Figure 4A, immunoblot analysis of macrophage cell lysates showed that chelation of extracellular calcium with EGTA blocked gp120-elicited Pyk2 tyrosine phosphorylation (Figure 4A). We then tested the effect of lanthanum, a potent inhibitor of the CRAC channels38 that are known to be expressed in macrophages.39 Lanthanum also suppressed the Env-induced activation response (Figure 4B). Taken together, these results suggest that intracellular calcium elevations due to calcium influx, elicited by JRFL through CCR5, is responsible for Pyk2 phosphorylation.
Calcium signaling inhibitors but not PTX block gp120-induced tyrosine phosphorylation of Pyk2.
MDMs were exposed for 5 minutes to JRFL gp120 (2.5 μg/mL) following (A) pretreatment for 20 minutes with or without 2 mM EGTA or (B) following pretreatment for 20 minutes with 100 nM lanthanum or for 18 hours with 100 ng/mL PTX. Whole cell lysates were then analyzed by immunoblotting with an antibody specific for the phosphorylated form of Pyk2 (upper panels, P-Pyk2) or with an antibody specific for total Pyk2 (lower panels). Blots shown are representative of 4 independent experiments using different donors.
Calcium signaling inhibitors but not PTX block gp120-induced tyrosine phosphorylation of Pyk2.
MDMs were exposed for 5 minutes to JRFL gp120 (2.5 μg/mL) following (A) pretreatment for 20 minutes with or without 2 mM EGTA or (B) following pretreatment for 20 minutes with 100 nM lanthanum or for 18 hours with 100 ng/mL PTX. Whole cell lysates were then analyzed by immunoblotting with an antibody specific for the phosphorylated form of Pyk2 (upper panels, P-Pyk2) or with an antibody specific for total Pyk2 (lower panels). Blots shown are representative of 4 independent experiments using different donors.
We then tested the effect on gp120-induced Pyk2 phosphorylation of PTX, which uncouples Gαi proteins from G protein–linked 7-transmembrane receptors.40 JRFL Env-induced Pyk2 activation was not inhibited by PTX (Figure 4B). Furthermore, Pyk2 activation by MIP-1β was not blocked by PTX either (data not shown). Thus, CCR5 signaling in primary macrophages in response to both Env and chemokines is coupled through PTX-insensitive pathways and is therefore not mediated by Gαi.
JRFL gp120 and MIP-1β stimulate the MAP kinase pathway
To address potential downstream consequences of gp120-induced Pyk2 activation in macrophages, we next focused on the MAPK family of signaling molecules (Figure 5). The MAPKs are a group of protein kinases that are activated in response to extracellular stimuli through dual phosphorylation at conserved threonine and tyrosine residues (reviewed by Cobb and Goldsmith41). In several non-macrophage cell types, it has been shown that Pyk2 acts upstream of the MAPK cascade.28,31,36,42,43 In lymphocytes, binding of gp120 to CCR5 has been reported to induce MAPK activation.7 8
JRFL gp120 induces MAPK activation in macrophages.
MDMs were stimulated with JRFL gp120 (2.5 μg/mL) or MIP-1β (200 nM) for the indicated time periods. Cell lysates were resolved by 12% SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis with antibodies specific for the total or phosphorylated forms of (A) p38 MAPK (phospho-p38 MAPK [T183/Y185] antibody); (B) JNK/SAPK (phospho-JNK/SAPK [T180/Y182] antibody); and (C) ERK1/2 (p44/42) (phospho-p44/42 MAPK [T202/Y204] antibody). Blots shown are from one donor and are representative of 5 experiments using different donors that yielded similar results.
JRFL gp120 induces MAPK activation in macrophages.
MDMs were stimulated with JRFL gp120 (2.5 μg/mL) or MIP-1β (200 nM) for the indicated time periods. Cell lysates were resolved by 12% SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis with antibodies specific for the total or phosphorylated forms of (A) p38 MAPK (phospho-p38 MAPK [T183/Y185] antibody); (B) JNK/SAPK (phospho-JNK/SAPK [T180/Y182] antibody); and (C) ERK1/2 (p44/42) (phospho-p44/42 MAPK [T202/Y204] antibody). Blots shown are from one donor and are representative of 5 experiments using different donors that yielded similar results.
MDMs were stimulated with JRFL gp120 or MIP-1β and cell lysates were analyzed by Western blot using polyclonal antibodies specific for the dually phosphorylated forms of the 3 members of the MAPK superfamily, extracellular regulated kinase (ERK1/2), JNK/SAPK, and p38 MAPK. Stimulation of macrophages with both JRFL gp120 and MIP-1β resulted in the rapid phosphorylation of p38 MAPK (Figure 5A) and JNK/SAPK (Figure 5B). In contrast, we observed a high basal level of phosphorylation in the case of p44/42 MAPK, and no further activation was elicited by either Env or chemokine (Figure 5C). The kinetics of MAPK activation were similar to those observed for Pyk2, peaking at about 5 minutes of exposure. Stimulation with SDF-1α also led to p38 and JNK/SAPK MAPK phosphorylation, although activation by IIIB gp120 was inconsistent (data not shown).
Involvement of MAPK signaling pathway in gp120-mediated chemokine induction
We next addressed the important question of whether gp120-induced chemokine receptor–mediated signaling in macrophages results in functional consequences with potential biologic significance. Macrophages are a major source for the production of β-chemokines, which play a central role in inflammatory cell recruitment and activation.44 Changes in the production of specific chemokines have been demonstrated during the course of HIV infection and, particularly, in compartments where macrophages are believed to be important in pathogenesis.45-47 In addition, several groups have observed increased production of β-chemokines by macrophages after in vitro HIV infection or gp120 exposure.48-50 Therefore, we investigated gp120-induced chemokine secretion by human macrophages and whether the MAPK pathway might be involved.
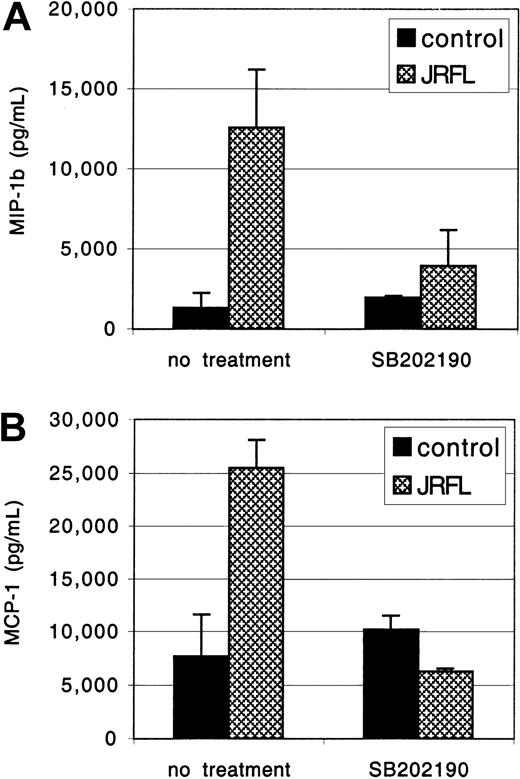
The MDMs were exposed to JRFL gp120 for 24 hours, and MIP-1β and MCP-1 concentrations in the clarified culture supernatants were measured by ELISA (Figure 6). Treatment with gp120 (1 μg/mL) up-regulated macrophage MCP-1 and MIP-1β production, up to 25 000 pg/mL and 12 000 pg/mL, respectively. No response was seen if gp120 was subjected to heat inactivation prior to MDM stimulation (data not shown). PTX did not inhibit gp120-induced MCP-1 and MIP-1β secretion, whereas induction was completely blocked by herbimycin, a broad-spectrum tyrosine kinase antagonist (data not shown). We then tested SB-202190, a specific inhibitor of MAPK activation.51 Pretreatment with SB-202190 completely abrogated gp120 induction of MIP-1β and MCP-1 in primary macrophages (Figure 6), without any evidence of cellular toxicity. Identical results were seen with gp120 at 2.5 μg/mL (data not shown). Thus, gp120 stimulates macrophage β-chemokine production through a MAPK-dependent pathway. Up-regulation of β-chemokine production by gp120 in macrophages may modulate subsequent rounds of infection by coreceptor blocking52 or, conversely, contribute to enhanced viral spread by new target cell recruitment and inflammatory consequences.
MAPK activation is involved in gp120-induced chemokine secretion.
MDMs were preincubated for 1 hour with or without the specific MAPK inhibitor SB-202190 (1 μg/mL) and then exposed to JRFL gp120 (1 μg/mL). After 24 hours of culture, supernatants were harvested. Levels of MIP-1β (A) and MCP-1 (B) and in the clarified supernatants were then detected by ELISA. Data are the mean of duplicate samples ± SD and one representative experiment of 4 is shown.
MAPK activation is involved in gp120-induced chemokine secretion.
MDMs were preincubated for 1 hour with or without the specific MAPK inhibitor SB-202190 (1 μg/mL) and then exposed to JRFL gp120 (1 μg/mL). After 24 hours of culture, supernatants were harvested. Levels of MIP-1β (A) and MCP-1 (B) and in the clarified supernatants were then detected by ELISA. Data are the mean of duplicate samples ± SD and one representative experiment of 4 is shown.
Discussion
Engagement of the chemokine receptor entry coreceptors by HIV-1 Env is essential for membrane fusion, but it may also initiate signaling events that alter cellular functions or even affect postentry stages of infection. Studies to date indicate that HIV-1 gp120 may participate in several signaling pathways through these receptors, including those involving G protein stimulation, ionic signaling, and protein phosphorylation.2 Macrophages are important targets for HIV-1 in vivo and express both CCR5 and CXCR4, the principal coreceptors for HIV-1 entry. However, relatively little is known about the pathways by which chemokine receptors signal in primary human macrophages in response to either gp120 or their natural chemokine ligands. We recently demonstrated that HIV-1 Env activates ionic signaling responses in macrophages through CCR5 and CXCR4.20 In the present study, we show that Env interaction with the coreceptors results in phosphorylation of the protein tyrosine kinase Pyk2, activation of the p38 and JNK/SAPK MAP kinases, and consequent induction of chemoattractant proinflammatory mediator secretion. These results begin to delineate the pathways by which the chemokine receptors are coupled in primary macrophages and, importantly, identify mechanisms by which HIV-1 may subvert the normal cellular signaling machinery.
Protein kinase activation by gp120 had not previously been examined in macrophages. We focused on the FAK-related protein Pyk2, also known as related adhesion focal tyrosine kinase (RAFTK),24calcium-dependent protein tyrosine kinase (CADTK),36 or cell adhesion kinase β (CAKβ),26 because this tyrosine kinase provides a potential link between chemokine receptor activation and multiple downstream intracellular pathways.53Moreover, Pyk2 activation may modulate ion channel function in certain cells,25,27 and we recently demonstrated ion channel activation by gp120 in macrophages.20 In this study we found that both R5 and X4 gp120 activate Pyk2 in primary macrophages. Activation is mediated specifically by CCR5 and CXCR4, and not by CD4, because no increase in phosphorylation was observed following Env binding to CD4 in CCR5− MDMs stimulated with JRFL and in AMD3100-treated macrophages stimulated with IIIB gp120. Supporting the idea that the chemokine receptors specifically mediate this signal, Pyk2 activation was also seen following stimulation by the natural chemokine ligands. Pyk2 activation has been reported in T cells following CCR5 and CXCR4 stimulation by chemokines and gp120,5,6,29,54 as has the related FAK following gp120 stimulation.4 Thus, our data extend these findings to primary macrophages, an important target for HIV-1 infection in vivo, to show that CCR5 and CXCR4 are linked to the protein kinase Pyk2 in macrophages, and that both R5 and X4 gp120 are functional agonists for this pathway.
We found that [Ca++]i elevation mediated by CRAC channels is a principal link between CCR5 stimulation and Pyk2 activation in macrophages, because both EGTA and the CRAC channel inhibitor lanthanum blocked gp120-induced Pyk2 phosphorylation. This result is in accord with our previous finding that gp120 elevates macrophage cytosolic calcium,20 and observations by others that Pyk2 activation is linked to [Ca++]i in various cell types.25,27,30,31,36 We also found that CCR5-mediated Pyk2 activation in macrophages was insensitive to PTX, indicating that it is not coupled through Gαi. Both PTX-sensitive and PTX-insensitive signaling through CCR5 and CXCR4 in lymphocytes has been reported for chemokines55-57 and gp120.4,5 Our finding that chemokine and Env signalings in macrophages were both insensitive to PTX suggests that these receptors are coupled in MDMs to G proteins other than Gαi, although it is also possible that alternative signaling pathways independent of G proteins may be responsible as has recently been described for several other GPCRs.58,59 Of note, it has been demonstrated that the G proteins with which a GPCR are coupled may differ depending on the cell type.19 Thus, our observation of PTX-insensitive CCR5 signaling in macrophages supports the notion that these signal transduction events are highly cell specific and emphasize the importance of defining these pathways in the relevant primary cell type of interest.
Because Pyk2 is implicated as an upstream regulator of the MAPK family of proteins in a variety of cell types,28,29,31,36,42,43we examined MAPK activity in macrophages and found that JRFL gp120 as well as chemokines triggered activation of p38 and JNK/SAPK MAPK. HIV-1– or gp120-induced MAP kinase activation has been reported in T cells, where it may play a role in regulating viral expression.7,8,60,61 Like Pyk2, activation of macrophage MAPK was not blocked by PTX, indicating that it is independent of Gαi. In other cell types, both Gαi- and Gαq-coupled pathways have been associated with GPCR activation of MAPKs.28 62 Because specific inhibitors of Pyk2 activation are currently not available, we were unable to definitively determine whether MAPK stimulation is Pyk2-dependent. We did find, however, that gp120-induced p38 phosphorylation was largely inhibited by blocking Ca++ influx (data not shown), which is consistent with the possibility that Pyk2 may be an upstream mediator of gp120-elicited MAPK activation.
A central question regarding these data involves the biologic significance of signaling cascades induced by gp120 in macrophages. MAPK activation plays an important role in regulating inflammatory responses, such as cytokine secretion in response to multiple stimuli.7,61,63-66 HIV-1 exposure or infection may induce a variety of functional and secretory responses in primary macrophages,13,48,50,67 including gp120-induced secretion of β-chemokines.49 Therefore, we investigated whether the MAPK signaling pathway might be involved in gp120-mediated β-chemokine induction. We found that gp120 stimulation of macrophages induced the production of both MIP-1β and MCP-1, potent chemoattractants for monocytes and macrophages, and that this response was blocked by MAPK inhibition, implicating this pathway in the macrophage secretory response to gp120. Thus, although the exact relationships between Pyk2, MAPK, ionic signaling, and [Ca++]i in coreceptor-mediated β-chemokine secretion in response to gp120 remains to be fully elucidated, the interaction of HIV-1 Env with the chemokine receptors on macrophages can result in the activation of signaling pathways and lead to proinflammatory responses even in the absence of infection.
Coreceptor signaling is dispensable for infection in cell lines,55,68 but indirect evidence has suggested that signaling might modulate infection in primary cells. Desensitization of CCR5 signaling has been associated with inhibition of lymphocyte infection,69 and chemokine receptor–mediated signaling was recently implicated in modulating productive versus nonproductive macrophage infection.37 Thus, whether signaling pathways elicited by Env in primary macrophages are involved in cell- and virus-specific mechanisms that modulate infection subsequent to entry is an important question that remains to be fully defined.
In addition to Env on infectious virions, macrophages may be exposed to gp120 on the surface of noninfectious virions or as free protein in tissues or into the circulation of infected subjects.70,71Concentrations of gp120 in circulation up to 92 ng/mL have been reported,71 which is close to the level seen to activate Pyk2 here. Furthermore, we found chemokine secretion induced by as little as 50 ng/mL gp120 (data not shown). Importantly, however, macrophages are likely to be exposed to Env glycoprotein in vivo within tissues, where higher concentrations of gp120 may be achieved locally at sites of viral replication and contribute to induction of kinase activation. Thus, the activation pathways identified in this report may also contribute to pathogenesis through interactions with uninfected cells. Various abnormalities of macrophage functions have been observed in HIV-1 infection in vivo and in vitro that in many cases can be induced by virions or soluble gp120 even in the absence of infection.13,15,72 Also, although β-chemokines have the potential to block R5 viral entry,52 dysregulated chemokine secretion may also promote the recruitment of uninfected cells to sites of active viral replication, promoting viral spread.73 In the central nervous system (CNS) of individuals with HIV encephalopathy (HIVE), infected macrophages and microglia are necessary for the development of HIVE, but clinical disease appears to be most closely related to the degree of macrophage and microglia activation.10,74 Elevated levels of β-chemokines are found in the CNS of individuals with HIVE45,46,50,75 and animals with simian immunodeficiency virus encephalopathy.47 Therefore, macrophage/microglia activation may contribute to pathogenesis through release of soluble mediators that themselves may be neurotoxic, including β-chemokines,76 or indirectly through leukocyte recruitment into the CNS.75 77 Thus, our studies elucidate specific gp120-mediated macrophage activation pathways that may lead to macrophage activation and dysfunction and contribute to pathogenesis in vivo.
We thank J. Cutilli, J. Riess and J. Moschella for technical assistance; R. Doms for Env proteins and valuable discussions; and blood donors who generously provided cells.
Supported in part by grants from the National Institutes of Health (MH 61139, AI 35502, and HL 58004) and from the Italian Ministry of Health (40D.7).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ronald Collman, 522 Johnson Pavilion, 36th and Hamilton Walk, Philadelphia, PA 19104; e-mail:collmanr@mail.med.upenn.edu.

![Fig. 1. Tyrosine phosphorylation of Pyk2 in primary human macrophages occurs in response to HIV-1 R5 Env. / (A) Week-old cultures of MDMs were unstimulated (CTR) or exposed for 5 minutes to JRFL gp120 (2.5 μg/mL), lysed, and subjected to immunoprecipitation with an anti-Pyk2 antibody. Immune complexes were resolved on 8% SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to serial immunoblotting with an antiphosphotyrosine 4G10 antibody (upper panel) and an anti-Pyk2 antibody (lower panel). (B) MDMs were unstimulated (CTR) or exposed to JRFL gp120 (2.5 μg/mL) for the indicated times (minutes). Cell lysates were resolved by 8% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-Pyk2 phosphospecific [pY402] antibody (upper panel, P-Pyk2). The same blot was then reprobed with an antibody specific for total Pyk2 (lower panel). Blots shown are representative of 5 independent experiments using different donors.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/10/10.1182_blood.v98.10.2909/5/m_h82211757001.jpeg?Expires=1768318826&Signature=k2QTmyhY8wbj47vOgQR8WOf2ptSy37qLRD-KGUfiGTeXgFrcvE3wovkn4qWoGBR~fgCHQxclbhZHXR6BpNrb5AEa0ffiX0eyhAw7aXou9kQ53pH9pPu9fl~vrCt6OxiL5RaskupadP1R7UN6GP7Ox7voOb4YDwtk0DXJSbW-Y14lYPObvxzLnz65gIg2RHi8TK3buNQxA4NLesZBKZsEoMKgvqv7uGneKICXXzMpFswZ4BIqs0DBFGrbtybbeylF~dx3YVmr3tE~7uUJYXLAm~BajUi6cH3XaIg7D8KcNtWbjtN4Ydjq~8acK~vcYQCym4qWxWk92MQncFNuaSxppA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 5. JRFL gp120 induces MAPK activation in macrophages. / MDMs were stimulated with JRFL gp120 (2.5 μg/mL) or MIP-1β (200 nM) for the indicated time periods. Cell lysates were resolved by 12% SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis with antibodies specific for the total or phosphorylated forms of (A) p38 MAPK (phospho-p38 MAPK [T183/Y185] antibody); (B) JNK/SAPK (phospho-JNK/SAPK [T180/Y182] antibody); and (C) ERK1/2 (p44/42) (phospho-p44/42 MAPK [T202/Y204] antibody). Blots shown are from one donor and are representative of 5 experiments using different donors that yielded similar results.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/10/10.1182_blood.v98.10.2909/5/m_h82211757005.jpeg?Expires=1768318826&Signature=HW1bPcpGfGmI-GtFzlEfRSMepjWae9wT~TvpiYZPoBvgCu8wd6bDCvuwbX4c632cZ58r2taQ6-roDZAdNAf4ITLZ3YzjZFN59frUf2AQdgldilQBaFUmqEMhrSbGXKtJ0Sn1NPhS5HHu1vPQzmWumfEYsgExwuuRF-QjGjmRfLJ~6C302Z0kgK20FlZv0NvAz2UsVwodRgfLF-rmp3yTClHf5KajVkkE5ZWbGuav5naHyjDu2EUCwBFamL2UmlhStYfE1JcigwUYI~-wy8aSicHDRWH56w~y95ejdGQMkDvuIwBhpzDZIiuUMtIlR1BYT-mDUuAQ2zoLe1~s6InHJQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal