Abstract
Endothelial cells are linked to each other through intercellular junctional complexes that regulate the barrier and fence function of the vascular wall. The nature of these intercellular contacts varies with the need for permeability: For example, in brain the impervious blood-brain barrier is maintained by “tight” contacts between endothelial cells. By contrast, in high endothelial venules (HEVs), where lymphocytes continuously exit the bloodstream, the contacts are generally leaky. The precise molecular components that define the type of junction remain to be characterized. An immunoglobulin superfamily molecule named JAM-2, specifically expressed in lymphatic endothelial cells and HEVs, was recently identified. JAM-3 was cloned and characterized in the current study, and JAM-1, -2, and -3 were shown to form a novel protein family belonging to the larger cortical thymocyte Xenopus (CTX) molecular family. Using antibodies specific for each of the 3 family members, their specific participation in different types of cell-cell contact in vivo and their specific and differential localization in lateral contacts or tight junctions were demonstrated. Furthermore, it was shown that JAM-1 and JAM-2 differentially regulate paracellular permeability, suggesting that the presence of JAM-1, -2, or -3 in vascular junctions may play a role in regulating vascular function in vivo.
Introduction
The immune surveillance of the body is carried out by lymphocytes constantly circulating from the blood to lymphoid or peripheral organs and back to the blood. Most lymphocytes extravasate through specialized vascular endothelium created by tissue microenvironment or inflammation. This suggests that a regional specialization of endothelial cells may control lymphocyte traffic. Postcapillary high endothelial venules (HEVs) are specialized sites along vessels where the migration of lymphocytes to lymphoid organs occurs.1,2 Migration occurs in a multistep process involving rolling of lymphocytes along the vessel wall, adhesion, and transmigration.3 The first and the second steps are well described and involve selectins, mucins, immunoglobulin (Ig) superfamily molecules, and integrins.4,5 The last transmigration step is less well understood and may occur through a transcellular pathway or by the paracellular route.6
Evidence to support the latter model comes from numerous studies suggesting that the transmigration of leukocytes involves the disruption of interendothelial junctions in a specific and localized manner.7-9 Nevertheless, the molecular events underlying the transmigration process are still controversial because other studies have demonstrated that transendothelial migration may occur without a widespread disruption of tight or adherens junctions.10,11 In this context, the molecules participating specifically in intercellular junctional complexes of endothelial cells may play a central role in regulating leukocyte transmigration and vascular functions.12,13 Although all endothelial cells are involved in exchanges of material from blood to tissue, there is a great degree of tissue-specific specialization of vascular junctions. This heterogeneity was described more than 20 years ago when the first electron microscopy studies pointed out structural differences in junctional complexes in different endothelial cells.14,15 Recently, the molecular characterization of proteins participating in intercellular junctional complexes has increased our understanding of vascular junction heterogeneity. The interendothelial adhesive structures include tight, adherens and gap junctions in which surface proteins such as occludin, claudins, cadherins, or connexins are specifically incorporated.16-19 Interestingly, some members of these protein families—among them Claudin-5 and VE-cadherin—have been found to be specifically expressed by endothelial cells.20,21Both molecules are involved in vascular integrity and normal vascular function.22,23 In addition, junctional proteins normally found outside the vascular system may participate to interendothelial junctions of specialized vessels in a tissue-specific manner. This is obvious for occludin, which is highly expressed by the brain vascular bed, but it is barely detectable in other interendothelial junctions.24 These results led to the concept that the tissue-specific specialization of blood vessels may be mediated by the molecular architecture of interendothelial junctions.12 25
We recently cloned and characterized a molecule called JAM-2, homologous to JAM and expressed by HEVs and lymphatic sinuses in murine mesenteric lymph nodes.26 JAM-2 is a transmembrane type 1 protein with 2 immunoglobulin domains that share its overall protein structure with JAM. The latter was shown to be present in tight junctions of epithelial and vascular endothelial cells.27,28 It has been found to interact with the junctional proteins ZO-1 and AF-6 through its cytoplasmic PDZ-binding domain.29,30 Furthermore, it was demonstrated that JAM plays a central role in the regulation of paracellular permeability and leukocyte transmigration across the endothelial barrier.28,31 More recently, Palmeri et al32characterized a novel human molecule, VE-JAM, which is structurally related to JAM and is expressed by vessels and HEVs. Independently, other authors have shown that VE-JAM supports leukocyte adhesion and interacts with a 43-kd molecule expressed by human leukocytes.33 For the sake of clarity, we will hereafter refer to JAM as JAM-1 and VE-JAM as JAM-3.
The contribution of JAM-1 to epithelial tight junctions and its expression by endothelial cells have been described,31,34but little is known about the participation of JAM-2 and JAM-3 in specialized interendothelial contacts. Therefore, the relative participation of JAM-1, JAM-2, and JAM-3 in different cellular junctions in vivo is central for understanding the specific function of each molecule. In the current study, cloning of murine JAM-3 allowed us to study and compare the tissue distribution of JAM-1, JAM-2, and JAM–3. We define the JAM family as a novel subset of immunoglobulin molecules that belong to the larger cortical thymocyteXenopus (CTX) family.35 36 Furthermore, using cells transfected with JAM-1, -2, or -3, we defined the respective properties of each family member such as cell-cell contact localization, participation in tight junctions, or receptor-ligand interactions. These results suggest that JAM family members share a similar structure and a common feature of homotypic interaction. Nevertheless, the demonstration of differential function of JAM-1 and JAM-2 in the control of paracellular permeability in vitro and the important differences in tissue distribution or subcellular localization observed in vivo suggest that they play specific roles in the control of vascular functions in vivo.
Materials and methods
Cell lines and antibodies
Murine thymic (tEnd.1) and embryonic (eEnd.2) endothelioma cell lines37 were provided by Dr W. Risau and Dr B. Engelhardt (Max Planck Institute, Bad-Nauheim, Germany). The murine SV40 transformed lymph node endothelial cell line TME was provided by Dr A.Hamann.38 The murine squamous cell carcinoma KLN 205 and MDCK cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). All cells were grown in Dulbecco modified essential medium (Gibco BRL, Paisley, Scotland), supplemented with 10% fetal calf serum (PAA Laboratories, Linz, Austria), 2 mM glutamine, 100 U/mL penicillin, and 100 U/mL streptomycin (all Gibco BRL). Rat monoclonal antibodies against MadCAM, PECAM, JAM-1 (H202.106.7.4), ZO-1 (R40/76), and JAM-2 (CRAM-18F26, referred as XVIIIF26) were previously described.26,39-42 Polyclonal antibody against JAM-3 has been described in detail elsewhere32 and was kindly provided by Dr Steven Rosen (Department of Anatomy, University of California, San Francisco). It was raised against a soluble form of huJAM-3 (VE-JAM.Ig), purified on the immunogen. Its specific reactivity on moJAM-3 is shown in the current study.
Sequence analysis
Cloning of JAM-2 has been previously described,26and one murine EST (accession number, AA445150; IMAGE, 848010) was identified as a related sequence. IMAGE Consortium cDNA clones were obtained from the United Kingdom HGMP Resource Center (Cambridge) and were resequenced. Sequencing the EST revealed an incomplete sequence and allowed us to run the 5′ RACE on cDNA of t-end endothelial cell line (Gibco BRL) using the following primers: AA44-R148, 5′-catctttaaaccagatgtactccgga-3′; AA44-R113, 5′-ccttctttatcatggcatcgtagc-3′; and AA44-R44, 5′-ggaacagcaggagccactagtactt-3′. Cloning and sequencing the obtained 5′ rapid amplification of cDNA ends (RACE)–polymerase chain reaction (PCR) product allowed us to assemble the full-length coding sequence of murine JAM-3 and to identify further the complete EST: AA690843 (IMAGE, 1195543) and W80145 (IMAGE, 402960). These 2 clones differed in the length of the 3′ untranslated region. All sequence analyses and sequence comparisons were performed using the applications available on the ExPASy Molecular Biology Server (Blast, Prosite, Swiss-Prot). The cDNA encoding murine JAM-1 was kindly provided by Pr Ph Naquet (CIML; Marseilles-Luminy).
Reverse transcription–polymerase chain reaction on cell lines
Specific PCR products were obtained using the following primer pairs: 5′-gtaactgtaatgggcaccgag-3′ and 5′-cacagcatccatgtgtgcagcctc-3′ for JAM-1, 5′-tcgacatggcgctgagc-3′ and 5′-cagtgttgccgtcttgcctacag-3′ for JAM-2, 5′-cctggactatcataaggcaaatgg-3′ and 5′-catctttaaaccagatgtactccgga-3′ for JAM-3. The PCR products were 704 base pairs (bp), 460 bp, and 457 bp long for JAM-1, JAM-2, and JAM-3, respectively. Control hypoxanthine phosphoribosyl transferase (HPRT) amplification was performed using 5′-gttggatacaggccagactttgttg-3′ and 5′-gagggtaggctggcctataggct-3′, a primer pair that resulted in a 350-bp-long PCR product.
Cloning and expression of chimeric proteins
Cloning of JAM-1 and JAM-2 in frame with enhanced green fluorescent protein (EGFP) was previously described.26 Briefly, it consisted of, respectively, the 3′HpaI or 3′ScaI of JAM-1 or JAM-2 sequences to fuse the coding sequence with EGFP in pcDNA3.43 Because no equivalent restriction sites were available in the sequence of JAM-3, we used a PCR approach to obtain the JAM-3–EGFP chimeric molecule using Pfu DNA polymerase and the following primers: 5′-tcagctaggcagccagct-3′ as forward primer and 5′-cgaccggtgtgactttagatgcaggactgcc-3′ as reverse primer. The reverse primer was modified by the AgeI site to clone the PCR product in frame with EGFP using the blunt EcoRI site on the 5′ extremity and the AgeI site on the 3′ extremity.
To insert a FLAG-tag at the N-terminus of murine JAM-3, a 2-step PCR-based strategy on the cloned full-length cDNA was used. The first step consisted of amplification and cloning of the sequence encoding the leader peptide with 5′-tcagctaggcagccagct-3′ and 5′-tcagaccggtcttatcgtcatcgtctttataatccccatttgccttatgatagtccagg-3′ as forward and reverse primer, respectively. The reverse primer encoded the FLAG-tag sequence in frame with the sequence of JAM-3 leader peptide and was modified to insert an AgeI site. Therefore, a second PCR product was obtained by amplification with 5′-tcagaccggtttttctgcatcaaaagaccaccgt-3′ and Sp6, and it encoded murine JAM-3 starting from the leader peptide. This PCR product was digested by AgeI and XhoI, and it was subcloned in the vector encoding the leader peptide fused to the FLAG-tag sequence. The integrity of the chimeric JAM-3 sequences was checked by sequencing the cloned product on both strands before transfection in MDCK cells using Fugene 6 (Roche Diagnostics, Rotkreuz, Switzerland). After 2 weeks of selection in medium containing 1 mg/mL G418, expressing cells were selected by fluorescence-activated cell sorter (Facstar; Becton Dickinson, Mountain View, CA) using EGFP fluorescence or immunostaining with appropriate antibody. Similar results were obtained with bulk sorted cells or clones.
Northern blot analysis
Total mRNA from cells or murine tissues was extracted using Trizol (Life Technologies AG, Basel, Switzerland) according to the manufacturer's instructions. Poly-A mRNA was extracted from 250 μg total RNA with the Oligotex mRNA Purification Kit (Qiagen, Zurich, Switzerland). Embryonic poly-A Northern blot was purchased from Clontech (PH Stehelin and Cie AG, Basel, Switzerland). Riboprobes were prepared from pcDNA3 vector (Invitrogen, Leek, Netherlands) and comprised the sequences encoding for the immunoglobulin domains of JAM-1, JAM-2, or JAM-3. Hybridization was performed at 62°C in buffer containing 50% formamide. Blots were washed twice (0.5 × standard sodium citrate [SSC], 0.1% sodium dodecyl sulfate [SDS], 67°C) and were autoradiographed on Kodak X-Omat at −80°C.
Immunostaining
For immunohistochemistry with anti–JAM-1 (H202-106) or double staining for JAM-1 and JAM-2, samples were fixed for 5 minutes with cooled methanol (−20°C), dried, and rehydrated in phosphate-buffered saline (PBS), 0.2% gelatin, and 0.05% Tween 20 (PGT). For single staining with polyclonal antibody against JAM-3 or anti–JAM-2 (XVIIIF26), acetone fixation for 5 minutes at −20°C was used. Stainings were visualized using secondary reagent coupled to Texas Red or to horseradish peroxidase (Jackson Immunoresearch Laboratories, West Grove, PA). Peroxidase activity was revealed using AEC as substrate, and sections were counterstained for 1 minute with Hemalum before they were mounted in Aquatex (Merck, Germany). For double staining, anti–JAM-2 was revealed using antirat Texas Red, followed by staining with biotinylated anti–JAM-1 in the presence of normal rat serum and streptavidin–fluorescein isothiocyanate (FITC). For triple staining, sections were fixed with 70% methanol–30% acetone at −20°C, dried, rehydrated, and incubated overnight with a mix of anti–JAM-1–Alexa584 (H202-106), anti–JAM-2–Alexa488 (CRAM-18F26), and rabbit polyclonal antibody against JAM-3 in PGT. After washing, JAM-3 staining was visualized with secondary reagent against rabbit coupled to DAMCA incubated for 1 hour in PGT in the presence of 0.2% normal rat serum. Pictures were acquired using an Axiocam or Axiovert fluorescence microscope (Zeiss, Oberkochen, Germany). For immunocytochemistry, 11 days before the experiment, cells were plated at 1 × 104 cells/cm2 on coated coverslips (1% fetal calf serum in PBS), and the medium was changed every day starting on day 4. Cells were fixed for 5 minutes with cooled methanol and were washed 3 times in PBS and 0.2% bovine serum albumin before incubation for 1 hour at room temperature with anti–JAM-1, JAM-2, or antiFLAG antibody (M2; Sigma). After 3 washes, primary staining was revealed using appropriate secondary reagent coupled to FITC (Jackson Immunoresearch Laboratories). After 3 washes, rabbit anti–ZO-1 (Zymed) was incubated for 1 hour at room temperature in the presence of 0.2% normal rat or mouse serum. After washing, ZO-1 staining was visualized using a probe antirabbit coupled to Texas Red (Jackson Immunoresearch Laboratories) incubated in the presence of 0.2% normal rat or mouse and 0.2% goat serum. Under these conditions, no signal in the red channel was observed when the anti–ZO-1 reagent was omitted, and the absence of leakage from red in green channel was checked by single staining for ZO-1. Pictures were acquired using LSM510 confocal microscope and LSM510 software (Zeiss).
Permeability assays
Permeability was measured using Transwell chambers (6.5-mm diameter polycarbonate filters, 0.4-μm pore size [Costar] or 10-mm diameter polycarbonate filters, 3-μm pore size [Nunc]). Briefly, 5 × 104 transfected or nontransfected MDCK cells were cultured to confluence on filters for 4 days. Medium was changed for prewarmed nutrient–F-12 medium (Life Technologies) without fetal calf serum (500 μL in the lower chamber and 350 μL containing 1 mg/mL FITC-dextran, Mr 42 000; Sigma-Aldrich Fluka Chemie AG, Buchs, Switzerland) in the upper chamber. After 4 hours, the chambers were removed and fluorescence was read directly in the lower chamber using Cytofluor II. Mean fluorescence intensity of 4 control wells (nontransfected MDCK cells) was calculated and referred to as 100% permeability. Values obtained with different transfected cells were then expressed as a percentage of control.
Results
JAM-1, JAM-2, and JAM-3 form a subset of the immunoglobulin family of CTX
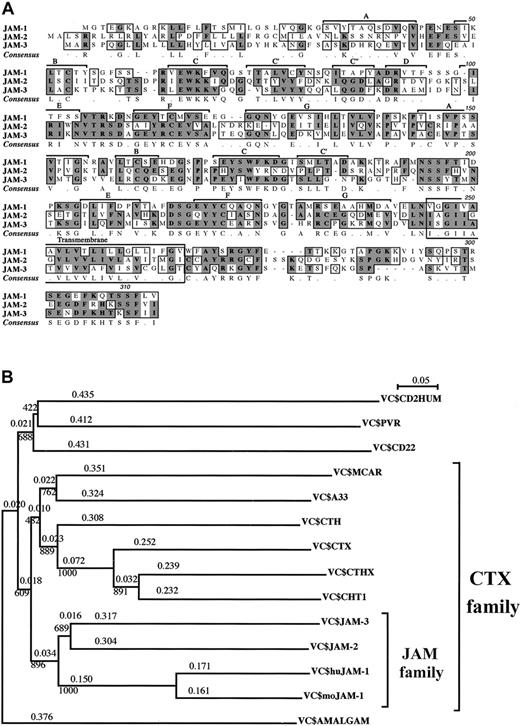
We recently identified JAM-2, a novel immunoglobulin Sf molecule expressed by HEVs and lymphatic endothelial cells in mice.26 By sequence comparison, we identified 2 related sequences—JAM-1 and a mouse EST (accession number, AA445150), encoding an incomplete protein sequence identified earlier as a member of theCTX gene family.36 The missing 5′ region was obtained with the rapid amplification of cDNA ends to give the full-length coding sequence that we called JAM-3. JAM-3 was subsequently identified as the murine equivalent of human VE-JAM, recently identified by Palmeri et al.32 The 3 JAM proteins were closely related: amino acid identities ranged from 31% for JAM-2 and JAM-1, up to 35% for JAM-2 and JAM-3, and the homologies were 51% and 54%, respectively (Figure 1). The 3 proteins were type 1 proteins with 2 extracellular immunoglobulin Sf domains, a transmembrane, and a short cytoplasmic segment. It is noteworthy that the cytoplasmic membrane proximal sequence (A-Y/Q-S/R-R/K-GYF) and the C-terminal sequence (T/K-S/K-SF-L/V/I-V/I) were almost identical for the 3 proteins. In JAM-2 and JAM-3, the extracellular domain of the protein distal to the membrane was of V type, and the proximal domain was of C2 type. Based on our analysis, we propose a similar structure for JAM-1. Nevertheless, this is still controversial because JAM-1 was originally described as a molecule comprising 2 V domains and was more recently proposed to present 2 C2 domains.27,44 This latter analysis was solely based on the number of amino acids between the cysteine residues contributing to the structure of the immunoglobulin domains. However, this criterion is invalid because the spacing between cysteine residues can vary in both V and C2domains.45,46 In addition, JAM-2 and JAM-3—but not JAM-1—contain an extra pair of cysteines in the A and G strands of their C2 domains, at the same position described in all CTX family members. This has been described as one of the characteristic features of C2 domains from CTX family members47; therefore, we postulate that the JAM family may be part of the larger CTX molecular family. Until now, this family of proteins comprised the following members: CTX (Xenopus); ChT1 (Gallus); CTH; A33; CAR, Coxsackie virus receptor; and CTHx (all in mammals). As illustrated by the subdivisions of the phylogenetic tree of the V+C domains (Figure 1B), JAM members form a subgroup within the CTX family. These subdivisions are valid for mouse and human molecules. The cytoplasmic tails of the CTX family members are heterogeneous. Some members have ITIM motifs (CTX, ChT1, CAR), others are not, and the length of the cytoplasmic tail varies. However, in the case of JAM-1, JAM-2, and JAM-3, this part of the molecule is relatively conserved in length and composition, and it contributes to the feature of the JAM subset. This suggests that members of the JAM subset may perform similar functions inside the cells once their external ligands are engaged. Other features of this subfamily seem to be conserved during evolution; 3 molecules of zebra fish andXenopus had been identified in the database, with the C strand (P/S-R-V/I/L-EWK-F/K) of their V domain remarkably similar to that of JAM-1, JAM-2, and JAM-3.47
JAM subfamily sequences and phylogenetic trees.
(A) Multiple alignment of murine JAM-1, JAM-2, and JAM-3 amino acid sequences. Identical and homologous amino acids are shaded in dark and light gray, respectively. The β-strand–loop immunoglobulin domain organization is indicated, and the letters refer to the strands of the V and C2 domains. Murine sequences for JAM-1 and JAM-2 are accessible from the database under the respective accession numbers, U89915 and AJ300304. JAM-3 has been submitted to EMBL Nucleotide Sequence Database and is accessible under the numberAJ291757. (B) Phylogenetic trees obtained with the Clustal X program using the neighbor-joining method. Positions with gaps were excluded, and bootstrap values are indicated at the knots and phylogenetic distances over the branches. Grouping of the JAM subfamily is obtained by comparison of the V+C extracellular domains, andDrosophila amalgam is used as an outgroup. The same grouping was obtained using full-length sequences.
JAM subfamily sequences and phylogenetic trees.
(A) Multiple alignment of murine JAM-1, JAM-2, and JAM-3 amino acid sequences. Identical and homologous amino acids are shaded in dark and light gray, respectively. The β-strand–loop immunoglobulin domain organization is indicated, and the letters refer to the strands of the V and C2 domains. Murine sequences for JAM-1 and JAM-2 are accessible from the database under the respective accession numbers, U89915 and AJ300304. JAM-3 has been submitted to EMBL Nucleotide Sequence Database and is accessible under the numberAJ291757. (B) Phylogenetic trees obtained with the Clustal X program using the neighbor-joining method. Positions with gaps were excluded, and bootstrap values are indicated at the knots and phylogenetic distances over the branches. Grouping of the JAM subfamily is obtained by comparison of the V+C extracellular domains, andDrosophila amalgam is used as an outgroup. The same grouping was obtained using full-length sequences.
Relative tissue distribution of JAM family members
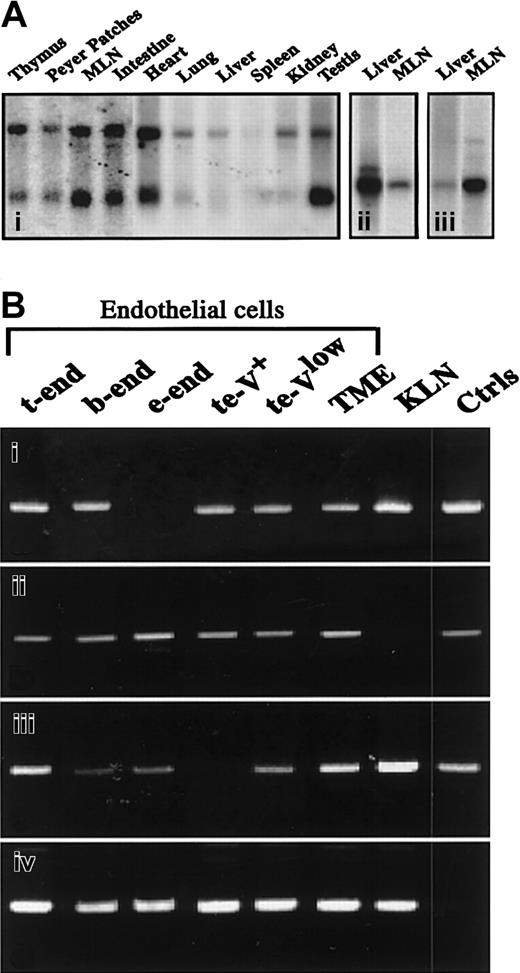
To further understand the relationship between JAM-1, JAM-2, and JAM-3, we explored the relative expression of the transcripts by Northern blot analysis. As shown in Figure2A, JAM-3 gives 2 hybridization signals at 1.8 kb and 4 kb, with mRNA obtained from different murine tissues (Figure 2Ai), whereas JAM-1 and JAM-2 give single signals at 2 kb (Figure 2Aii-iii). When the distribution of the 3 family members is compared, JAM-1 is abundantly expressed in liver, whereas JAM-2 and -3 are weakly expressed. In contrast, JAM-2 and JAM-3 are highly expressed in lymph nodes, which express only limited amounts of JAM-1 (Figure 2A, Table 1). Notably, differences in transcriptional expression of the 3 molecules were also observed during embryogenesis (Table 1). However, these relative differences must be carefully interpreted because the results were obtained with mRNA from whole embryos in which the ratio between developing organs and the entire embryo varied with age. Similarly, differences between the relative expression of JAM members in adult tissues might result from cell type–specific expression of the individual JAMs.
Relative expression of transcripts encoding JAM family members.
(A) Northern blot analysis of JAM-3 (i), JAM-1 (ii), or JΑΜ-2 (iii) transcripts on polyA mRNA extracted from indicated tissues. JAM-3 probe (i) gives 2 hybridization signals at ≈1.5 and 4.4 kb, whereas JAM-1 (ii) and JAM-2 (iii) probes give a single hybridization signal at ≈2 kb. (B) Results of reverse PCR analysis for JAM-1 (i), JAM-2 (ii), JAM-3 (iii), or HPRT (iv) on cDNA from indicated cell lines. Positive controls (Ctrls) were obtained by running the PCR with plasmids encoding full-length sequences of JAM-1, -2, and -3. Negative controls were obtained by omitting the reverse transcription step, resulting in an absence of detectable signals. MLN indicates mesenteric lymph nodes.
Relative expression of transcripts encoding JAM family members.
(A) Northern blot analysis of JAM-3 (i), JAM-1 (ii), or JΑΜ-2 (iii) transcripts on polyA mRNA extracted from indicated tissues. JAM-3 probe (i) gives 2 hybridization signals at ≈1.5 and 4.4 kb, whereas JAM-1 (ii) and JAM-2 (iii) probes give a single hybridization signal at ≈2 kb. (B) Results of reverse PCR analysis for JAM-1 (i), JAM-2 (ii), JAM-3 (iii), or HPRT (iv) on cDNA from indicated cell lines. Positive controls (Ctrls) were obtained by running the PCR with plasmids encoding full-length sequences of JAM-1, -2, and -3. Negative controls were obtained by omitting the reverse transcription step, resulting in an absence of detectable signals. MLN indicates mesenteric lymph nodes.
Northern blot analysis of JAM-1, JAM-2, and JAM-3 expression in murine tissues
| Tissue . | JAM-1 . | JAM-2 . | JAM-3 . |
|---|---|---|---|
| Embryo | |||
| E7 | − | + | ± |
| E11 | − | + | ± |
| E15 | − | ++ | ± |
| E17 | − | + | + |
| Adult | |||
| Heart | +++ | + | ++ |
| Lungs | +++ | ± | ± |
| Liver | +++ | ± | ± |
| Spleen | ± | − | − |
| Kidney | ++ | ++ | ± |
| Testis | + | +++ | +++ |
| Thymus | ++ | ± | + |
| Peyer patches | ++ | ++ | + |
| Lymph nodes | + | ++ | ++ |
| Tissue . | JAM-1 . | JAM-2 . | JAM-3 . |
|---|---|---|---|
| Embryo | |||
| E7 | − | + | ± |
| E11 | − | + | ± |
| E15 | − | ++ | ± |
| E17 | − | + | + |
| Adult | |||
| Heart | +++ | + | ++ |
| Lungs | +++ | ± | ± |
| Liver | +++ | ± | ± |
| Spleen | ± | − | − |
| Kidney | ++ | ++ | ± |
| Testis | + | +++ | +++ |
| Thymus | ++ | ± | + |
| Peyer patches | ++ | ++ | + |
| Lymph nodes | + | ++ | ++ |
− indicates absence of detectable signal; ±, +, ++, +++, gradual increases in hybridization signals. Results were normalized to the β-actin hybridization signal and represent relative intensities of the hybridization signal (see Figure 2A).
To test this, we analyzed the expression of the 3 molecules by reverse transcription–PCR in murine endothelial and epithelial cell lines. Transcripts of JAM-1, JAM-2, and JAM-3 are found in most endothelial lines (Figure 2B). Interestingly, JAM-3 is absent from the te-V+ cell line, which represents a variant of t-end cells with phenotypic and morphologic properties of angiogenic endothelial cells (Aurrand-Lions et al, manuscript in preparation). The transcript encoding JAM-2 is detected in all tested endothelial cells but is absent from the epithelial carcinoma cell line KLN 205, which expresses reasonable levels of JAM-1 and JAM-3 transcripts. This indicates that JAM-1 and JAM-3 expression are not exclusive to vascular endothelial cells.
Specificity of anti–JAM-1, -2, and -3 antibodies
To further address the question of the relative tissue distribution of the 3 JAM family members at the protein level, antibodies against each molecule were used. As shown in Figure3A, MDCK cells transfected with plasmids encoding murine JAM-1, JAM-2, or N-terminal FLAG-tagged JAM-3 (FgJAM-3) were specifically stained by antibodies to JAM-1 (H202-106), JAM-2 (XVIIIF26), JAM-3 (polyclonal antibody), or FLAG-tag (M2) (plain profiles), respectively. In addition, results showed that none of these reagents cross-reacted on another JAM family member.
Specificity of antibodies against JAM family members.
(A) Specificity of antibodies directed against JAM-1, JAM-2, and JAM-3. MDCK cells stably transfected with cDNA encoding murine JAM-1, JAM-2, or FLAG-tagged JAM-3 were stained, as indicated with, antibodies to JAM-1 (H202-106), JAM-2 (XVIIIF26), JAM-3 (polyclonal antibody), or FLAG peptide (M2). Dashed profiles represent negative controls obtained by omitting the primary antibody. (B) Expression of JAM members by KLN205 epithelial cells. Cells were stained with monoclonal antibodies against JAM-1 (i), JAM-2 (ii), or JAM-3 (iii) as indicated. Negative control (iv) was obtained by using an IgG fraction from nonimmune rabbit serum. Secondary antibodies were antirat and antirabbit coupled to FITC. Magnification × 630.
Specificity of antibodies against JAM family members.
(A) Specificity of antibodies directed against JAM-1, JAM-2, and JAM-3. MDCK cells stably transfected with cDNA encoding murine JAM-1, JAM-2, or FLAG-tagged JAM-3 were stained, as indicated with, antibodies to JAM-1 (H202-106), JAM-2 (XVIIIF26), JAM-3 (polyclonal antibody), or FLAG peptide (M2). Dashed profiles represent negative controls obtained by omitting the primary antibody. (B) Expression of JAM members by KLN205 epithelial cells. Cells were stained with monoclonal antibodies against JAM-1 (i), JAM-2 (ii), or JAM-3 (iii) as indicated. Negative control (iv) was obtained by using an IgG fraction from nonimmune rabbit serum. Secondary antibodies were antirat and antirabbit coupled to FITC. Magnification × 630.
A further specificity control was obtained with the KLN 205 squamous carcinoma cell line, which expressed high levels of JAM-1 and JAM-3 transcript and no detectable JAM-2 mRNA. Immunocytochemistry was performed on this cell line using monoclonal antibodies against JAM-1 and JAM-2 or affinity-purified polyclonal antibody against JAM-3.26,32 41 As expected, monoclonal antibody against JAM-1 stains the intercellular border with a sharp line resembling the pattern of tight junctions (Figure 3B, i). Anti–JAM-3 also stains cell borders but the pattern is hazy, indicating that this molecule is distributed over the entire lateral surface (Figure 3B, iii). In agreement with the absence of JAM-2 transcript in KLN 205, we did not detect a signal with anti–JAM-2 monoclonal antibody (Figure 3B, ii).
Endothelial expression of JAM family members in vivo
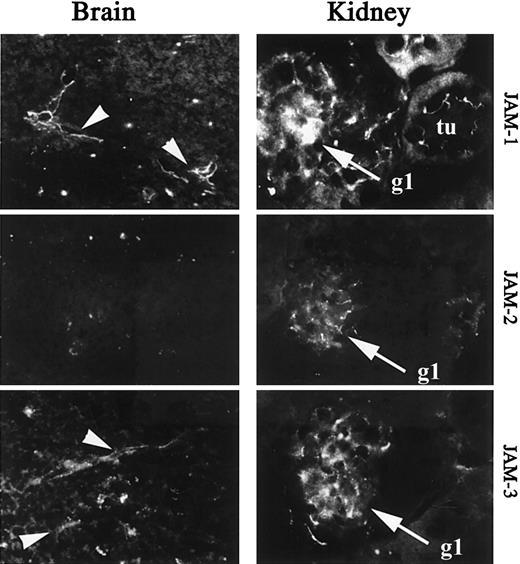
To further analyze the heterogeneity of JAM expression on endothelial cells in vivo, we performed immunohistochemical analysis of JAM-1, -2, and -3 expression on sections of brain or kidney. These organs are known to contain highly specialized vascular structures, such as glomerular endothelial cells in kidney or tightly sealed vascular beds of the blood barrier in the brain. Analysis of brain sections shows a restricted expression of JAM-1 and JAM-3 to vessels, whereas JAM-2 is not detected (Figure 4, left panel). In the kidney JAM-1, -2, and -3 are expressed in glomeruli (gl), whereas surrounding epithelial cells of tubules (tu) are solely stained for JAM-1 (Figure 4, right panel). This is in agreement with the previously reported expression of JAM-1 in tight junctions of epithelial cells.34 49
Immunohistologic analysis of JAM-1, -2, and -3 expression in murine brain and kidney.
Frozen section of brain and kidney were stained for JAM-1 (H202-106), JAM-2 (XVIIIF26), or JAM-3 (polyclonal antibody) as indicated. Arrowheads indicate the vessels on micrographs obtained from brain sections. tu, tubules on micrographs obtained from kidney sections; gl, glomeruli on micrographs obtained from kidney sections. Secondary antibodies were antirat or antirabbit coupled to FITC. Magnification × 400.
Immunohistologic analysis of JAM-1, -2, and -3 expression in murine brain and kidney.
Frozen section of brain and kidney were stained for JAM-1 (H202-106), JAM-2 (XVIIIF26), or JAM-3 (polyclonal antibody) as indicated. Arrowheads indicate the vessels on micrographs obtained from brain sections. tu, tubules on micrographs obtained from kidney sections; gl, glomeruli on micrographs obtained from kidney sections. Secondary antibodies were antirat or antirabbit coupled to FITC. Magnification × 400.
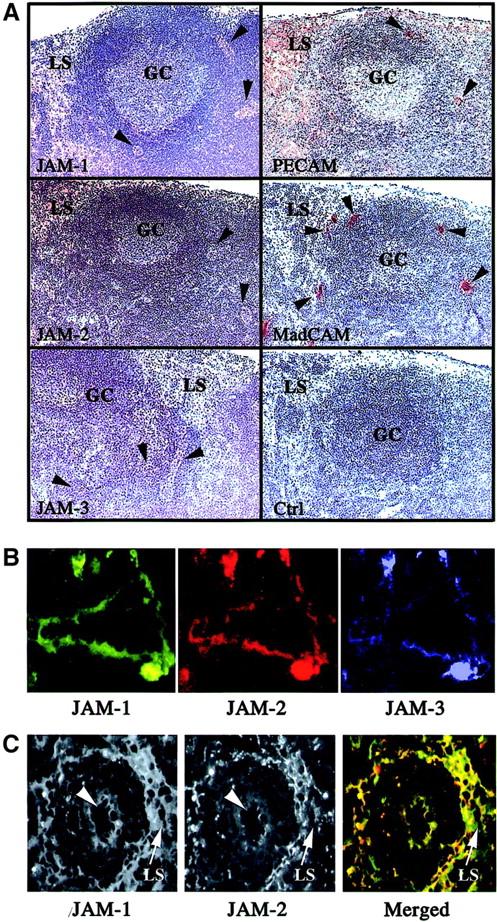
However, because the transcripts encoding JAM-1, -2, and -3 were also detected in RNA preparations of lymph nodes, we were interested in defining the cellular localization of the molecules in these organs. Peroxidase staining was performed on sections of murine mesenteric lymph nodes with antibodies directed against JAM-1, JAM-2, JAM-3, PECAM, and MAdCAM to identify the cellular compartment expressing the different family members. As expected, all these molecules are expressed in mesenteric lymph nodes with differences in their patterns of expression (Figure 5A). PECAM is used as a positive control to visualize the entire endothelial compartment, comprising the lymphatic sinuses, and MAdCAM is used to distinguish specifically the HEVs (arrowheads). When the staining obtained for JAM-1, -2, or -3 is compared to these 2 markers, it appears that all 3 molecules are expressed relatively weakly by HEVs (Figure 5A, arrowheads) and that JAM-1 and JAM-2 are expressed on lymphatic sinuses where JAM-3 is not detected. To confirm the coexpression of JAM-1, JAM-2, and JAM-3 by some endothelial cells of mesenteric lymph nodes, we performed triple immunofluorescence staining. Some vascular structures, presumably HEVs, expressed all 3 JAM family members (Figure 5B), whereas other endothelial structures such as lymphatic sinuses were solely stained with anti–JAM-1 and anti–JAM-2. The micrograph presented in Figure 5C shows a reticular network of lymphatic medullary sinus surrounding a blood vessel (arrowhead), diffusely stained for JAM-1, whereas JAM-2 is distributed in sharp lines at cell-cell borders. This characteristic staining pattern for the 2 molecules was also observed on lymphatic vessels located in the cortical area of Peyer patches (not shown).
Immunohistologic analysis of JAM-1, -2, and -3 expression in murine lymphoid organs.
(A) Frozen sections of murine mesenteric lymph nodes were stained for JAM-1, JAM-2, JAM-3, MAdCAm, or PECAM using peroxidase-coupled secondary antibodies. The negative control was obtained using an IgG fraction from nonimmune rabbit serum, followed by antirabbit coupled to peroxidase. Peroxidase-stained structures appear brown, and the hemalum counterstain of tissue appears blue. Germinal centers (GC), lymphatic sinuses (LS), and HEVs (arrowheads) are indicated. Magnification × 40. (B) Frozen sections of murine mesenteric lymph nodes were triple stained for JAM-1, JAM-2, and JAM-3 as indicated. Single-color images were acquired using specific excitation and emission filter sets, and pseudocolored images were acquired using Adobe Photoshop 5.5. The absence of fluorescence leakage in the different channels was checked by single staining using the same experimental setup. Magnification × 400. (C) Differential localization of JAM-1 and JAM-2 on lymphatic sinuses. Lymphatic sinuses indicated by arrows express JAM-1 diffusely, whereas JAM-2 is concentrated in cell-cell contacts. Arrowheads indicate a vascular structure expressing JAM-1 and JAM-2. Secondary antibodies were antirat Texas Red and streptavidin-FITC to visualize, respectively, anti–JAM-2 and biotinylated anti–JAM-1 reactivities. In the merged picture, JAM-1 appears in green and JAM-2 in red. Magnification × 160.
Immunohistologic analysis of JAM-1, -2, and -3 expression in murine lymphoid organs.
(A) Frozen sections of murine mesenteric lymph nodes were stained for JAM-1, JAM-2, JAM-3, MAdCAm, or PECAM using peroxidase-coupled secondary antibodies. The negative control was obtained using an IgG fraction from nonimmune rabbit serum, followed by antirabbit coupled to peroxidase. Peroxidase-stained structures appear brown, and the hemalum counterstain of tissue appears blue. Germinal centers (GC), lymphatic sinuses (LS), and HEVs (arrowheads) are indicated. Magnification × 40. (B) Frozen sections of murine mesenteric lymph nodes were triple stained for JAM-1, JAM-2, and JAM-3 as indicated. Single-color images were acquired using specific excitation and emission filter sets, and pseudocolored images were acquired using Adobe Photoshop 5.5. The absence of fluorescence leakage in the different channels was checked by single staining using the same experimental setup. Magnification × 400. (C) Differential localization of JAM-1 and JAM-2 on lymphatic sinuses. Lymphatic sinuses indicated by arrows express JAM-1 diffusely, whereas JAM-2 is concentrated in cell-cell contacts. Arrowheads indicate a vascular structure expressing JAM-1 and JAM-2. Secondary antibodies were antirat Texas Red and streptavidin-FITC to visualize, respectively, anti–JAM-2 and biotinylated anti–JAM-1 reactivities. In the merged picture, JAM-1 appears in green and JAM-2 in red. Magnification × 160.
Altogether, these results show that JAM-1 expression on brain endothelial cells and most epithelial cells is in agreement with its putative function in tight junctions. In contrast, JAM-2 is not detected on brain vasculature and is expressed by lymphatic sinuses known to be highly permeable. This suggests that JAM-1 and JAM-2 may play opposite roles in interendothelial junctional sealing.
JAMs differentially regulate paracellular permeability
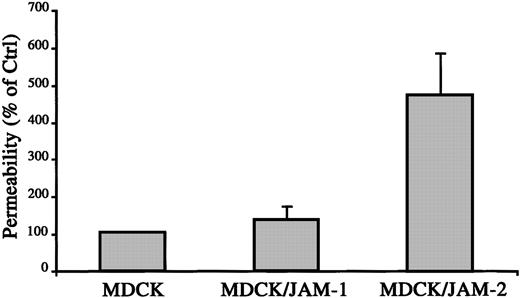
To directly test the hypothesis of opposite roles of JAM-1 and JAM-2 in interendothelial junctions and vascular functions, we performed paracellular permeability assays on MDCK cells stably transfected with JAM-1 or JAM-2. As shown in Figure6, the stable expression of JAM-1 did not affect the paracellular permeability assessed by FITC dextran (42 kd) diffusion across monolayer. Interestingly, the expression of JAM-2 protein on the surfaces of MDCK cells, with an expression level comparable to that of JAM-1, resulted in a 5-fold increase in FITC-dextran flux. These results indicate that JAM-1 and JAM-2, though highly related, may perform distinct functions in the regulation of intercellular sealing and vascular function.
JAM-2 increases paracellular permeability.
Diffusion of FITC-dextran (42 kd) across monolayers of MDCK, MDCK/JAM-1, or MDCK/JAM-2–transfected cells was measured after 3 hours. Quantification of FITC dextran in the lower compartment was measured using Cytofluor II, and the 100% diffusion was calculated according to the fluorescence observed in the lower compartment using a monolayer of nontransfected MDCK cells.
JAM-2 increases paracellular permeability.
Diffusion of FITC-dextran (42 kd) across monolayers of MDCK, MDCK/JAM-1, or MDCK/JAM-2–transfected cells was measured after 3 hours. Quantification of FITC dextran in the lower compartment was measured using Cytofluor II, and the 100% diffusion was calculated according to the fluorescence observed in the lower compartment using a monolayer of nontransfected MDCK cells.
Localization of JAM family members to cell-cell contacts
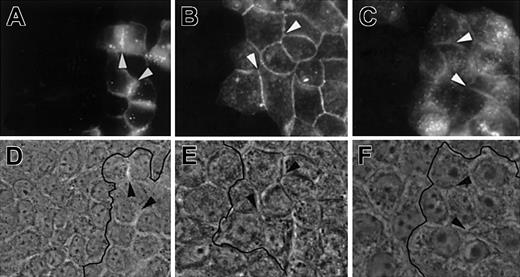
Because the barrier function and the control of permeability are known to depend on adherens and tight junctions, we addressed the issue of specific junctional and subcellular localization of JAM-1, JAM-2, and JAM-3. For this purpose, we used MDCK II cells that show the classical arrangement of apical tight and lateral adherens junctions. MDCK cells, transfected with full-length JAM-1, JAM-2, or FgJAM-3, were doubly stained with antibodies against each JAM in green and an antibody to ZO-1 in red to visualize the tight junctional regions (Figure 7A). Differences in the subcellular localization of the 3 family members were immediately apparent. JAM-1 colocalizes with ZO-1 at the most apical part of cell-cell contact, but it is also present in more basal locations (Figure 7A, z-axis). In contrast, JAM-2 is solely enriched in apical ZO-1–positive regions and is almost absent from more basal cell-cell contacts as depicted in yellow on the z-axis reconstitution. JAM-3 appears more diffuse and is not specifically enriched in ZO-1–positive tight junctional regions. The same results were obtained when experiments were performed on cells transfected with JAM molecules fused to EGFP on their carboxy terminal side (Figure 7B). Interestingly, this indicates that the C-terminal cytoplasmic PDZ binding motif is not required for the differential targeting of the 3 JAMs to different cell-cell contact regions of polarized cells because this motif was removed in the chimeric constructs. This prompted us to test the type of extracellular protein interactions required to target JAM family members to intercellular contacts. For this purpose, we mixed nontransfected MDCK cells with cells transfected with JAM-1–EGFP, JAM-2–EGFP, or JAM-3–EGFP chimeras. As illustrated in Figure 8, all JAM members are enriched at cell-cell borders between transfected cells but are absent from contacts between transfected and nontransfected cells (depicted by the black line on phase-contrast images). Although we cannot exclude that other ligands for JAM family members exist, these results indicate that each JAM member interacts in a homophilic way.
Subcellular localization of JAM members to epithelial cell-cell contacts.
(A) MDCK cells stably transfected with cDNA encoding murine JAM-1, JAM-2, or FLAG-tagged JAM-3 were stained for ZO-1 in red and with antibodies to JAM-1 (H202-106), JAM-2 (XVIIIF26), or FLAG peptide (M2) in green. Confocal images taken at the apical level of tight junctions and micrographs showing the Z-axis reconstitutions are shown. The latter were obtained from a stack of 40 pictures every 0.4 μm taken at the level of the arrowheads. Magnification × 630. (B) MDCK cells expressing EGFP fusion proteins for JAM-1 (i and iv), JAM-2 (ii and v), or JAM-3 (iii and vi). Staining with anti–ZO-1 followed by antirat Texas Red (iv-vi) are shown below the pictures obtained for green fluorescence (i-iii). Micrographs were acquired using specific filter sets and Openlab software. Magnification × 1000.
Subcellular localization of JAM members to epithelial cell-cell contacts.
(A) MDCK cells stably transfected with cDNA encoding murine JAM-1, JAM-2, or FLAG-tagged JAM-3 were stained for ZO-1 in red and with antibodies to JAM-1 (H202-106), JAM-2 (XVIIIF26), or FLAG peptide (M2) in green. Confocal images taken at the apical level of tight junctions and micrographs showing the Z-axis reconstitutions are shown. The latter were obtained from a stack of 40 pictures every 0.4 μm taken at the level of the arrowheads. Magnification × 630. (B) MDCK cells expressing EGFP fusion proteins for JAM-1 (i and iv), JAM-2 (ii and v), or JAM-3 (iii and vi). Staining with anti–ZO-1 followed by antirat Texas Red (iv-vi) are shown below the pictures obtained for green fluorescence (i-iii). Micrographs were acquired using specific filter sets and Openlab software. Magnification × 1000.
Homophilic interactions of JAM members.
MDCK cells transfected with JAM-1–EGFP, JAM-2–EGFP, or JAM-3–EGFP were mixed with nontransfected MDCK cells and analyzed for localization of EGFP chimeric molecules in A, B, and C, respectively. Corresponding phase-contrast images are shown in D, E, and F. Arrows highlight contacts between transfected cells enriched in fusion proteins. Black lines on phase-contrast pictures indicate the border between transfected and nontransfected cells. Magnification × 400.
Homophilic interactions of JAM members.
MDCK cells transfected with JAM-1–EGFP, JAM-2–EGFP, or JAM-3–EGFP were mixed with nontransfected MDCK cells and analyzed for localization of EGFP chimeric molecules in A, B, and C, respectively. Corresponding phase-contrast images are shown in D, E, and F. Arrows highlight contacts between transfected cells enriched in fusion proteins. Black lines on phase-contrast pictures indicate the border between transfected and nontransfected cells. Magnification × 400.
Discussion
The homology among JAM-1, JAM-2, and JAM-3 allowed their classification in a novel subfamily of adhesion molecules. The structural organization in the extracellular portions of JAM-2 and JAM-3 consists of one V-like and one C2 domain with 2 disulfide bridges. This structure has been shown to be specific for molecules of the CTX family, which present a typical exon-intron structure, an extracellular organization in V-C2 domains with J features, and an extra disulfide bridge within the C domain. Although JAM-1 was originally described as a protein with 2 V domains27 and later as a 2 C2-containing protein,44 our sequence alignments suggested better matching to a V-C2 structure with a single disulfide bridge. The structural data indicate that the 3 JAMs represent a subgroup of the CTX family. Furthermore, by database search, human JAM-1,48 JAM-2, and JAM-3 genomic sequences were localized, respectively, on chromosomes 1, 11, and 21 already outlined for their immunoglobulin Sf CTX subset contents.47 This may suggest that CTX and JAMs are present in gene clusters and that they originate from gene duplication. However, the high homology between the 3 JAMs argues for a recent functional adaptation and the existence of a unique, functionally specialized linkage group. This hypothesis is further supported by the fact that the homology between JAM-1, JAM-2, and JAM-3 concerns not only the overall protein structure but also the transmembrane and the cytoplasmic domains indicating functional adaptation.
The novelty of the JAM family consists in the fact that each member of a unique protein family participates specifically in different junctional compartments. When expressed in polarized MDCK cells, JAM-1, -2, or 3 is incorporated in different types of junctions. JAM-2 colocalizes with ZO-1, whereas JAM-1 is only partially incorporated in tight junctions as demonstrated by partial colocalization with ZO-1 in MDCK cells. Although we cannot exclude that JAM-1 localization was caused by overexpression, a similar result of JAM-1 localization was reported by Liu et al34 in nontransfected T84 cells. More surprisingly, in transfected MDCK cells, JAM-3 appeared more diffuse and not particularly enriched at a specific subcellular compartment of intercellular junctions. This result argued for the participation of JAM-3 in cell-cell contact in structures distinct from tight junctions, which is in agreement with the diffuse lateral staining observed on the KLN205 cell line.
The function of the 3 JAMs in the intercellular junctions was further addressed using permeability assays with MDCK cells transfected with the different JAM isoforms. When compared with control cells, JAM-1 stable expression did not change FITC dextran diffusion across the MDCK monolayer, whereas JAM-2 increased the paracellular permeability. This was in agreement with the expression of JAM-2 found in permeable endothelial cells such as lymphatic sinuses or HEVs. However, this result contradicted the presumed barrier function of tight junctions and the previously described effect of JAM-1 or JAM-2–EGFP on paracellular permeability of Chinese hamster ovary cells.26,27 Although we cannot exclude that the differential effects of JAM-1 and JAM-2 observed in MDCK cells are attributed to differences in junctional organization between CHO and MDCK intercellular contacts, studies comparing both models remain to be conducted. According to this, the overexpression of JAM-1 in Chinese hamster ovary cells was shown to be sufficient to recruit ZO-1 in cell-cell contacts,29 whereas we did not observe a relocalization of ZO-1 in MDCK cells expressing JAM-1 in basal cell-cell contacts. These results indicate that the specific function of JAM family members may depend on the cell type and the composition of the intercellular junctional complexes.
Therefore, it is not surprising to find heterogeneous expression of JAM family members on endothelial cells linked to each other through adhesive complexes resembling those of epithelial cells but presenting distinct molecular composition and structure. This probably reflects the heterogeneity of interendothelial junctions. The highest vascular expression of JAM-1 was found in adult brain, which is known to develop a network of tight junctional strands unique to the vascular system.50,51 In contrast, endothelial cells of HEVs and lymphatic endothelial cells, which have highly specialized cell-cell junctions,52-54 expressed high levels of JAM-2 and low amounts of JAM-1. Finally, JAM-3 was not expressed in lymphatic sinuses but was found in most endothelial contacts ranging from brain vasculature to HEVs. This argues for a role for JAM-3 in constitutive functions of vascular endothelial cells, consistent with its putative function as an adhesion molecule for circulating leukocytes.33 Taken together, our results suggest that the JAM family members may be involved in different interendothelial junctions reflecting the tissue specificity of vascular beds.
By analogy with the function of JAM-1 as an organizer of occludin clustering in epithelial cells,34,49 one may speculate about a similar function in endothelial cells. Such a regulatory role for JAM-1 on endothelial tight junction is further supported by its expression in brain vasculature and its role in leukocyte recruitment in the cerebrospinal fluid of mice with experimental meningitis.28,31 We cannot exclude a similar regulatory function for JAM-2 and JAM-3, but their participation in endothelial junctional complexes devoid of occludin expression suggests their involvement in different mechanisms. Although the morphologic heterogeneity of endothelial junctions is well established, the specific properties of the different interendothelial junctional complexes are unknown. It has been suggested that interendothelial junctions should be considered as adherens junctions in which gap and tight junctions may be inserted.55 These “mixed junctional complexes” would be more easily regulated than morphologically defined junctions found in epithelial cells, and their molecular composition could regulate their “leakiness.” Therefore, the specific route of leukocyte recirculation in noninflammatory conditions will depend on the establishment and stability of interendothelial junctions. The specific expression of JAM-1, -2, or -3 in different vascular compartments brings new insights in the molecular nature of such junctional complexes. Their structural and molecular characterization is probably the next step in our understanding of specific vascular functions that contribute to the maintenance of homeostasis.
Sequences used in the current study are accessible under the following GenBank accession numbers: CD 2, M16445; PVR (poliovirus receptor),M24406; CD 22, X59350; MCAR (mouse Coxsackie virus receptor), U90715; A33, U79725; CTH, AF061022; CTX, (cortical thymocyte moleculeXenopus), U43330; CTHX (CTX human homologue), hs889n15; CHT1 (cortical thymocyte molecule chicken), Y14063; moJAM-1, U89915; huJAM-1, NM_016 946; moJAM-2, AJ300304; moJAM-3, AJ291757; amalgamDrosophila, M23561.
We thank Dr S. Rosen for kindly providing us with polyclonal antibody against JAM-3. We also thank D. Gay-Ducrest and C. Magnin for their technical expertise in molecular biology and immunohistochemistry.
Supported by Krebsforschung schweiz (KFS 981-02-2000), Foundation (31-49241.96), Foundation Gabriella Giorgi-Cavaglieri, and Human Frontier Science Program Organization (LT 0218/1998-M) (M.A.L.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michel Aurrand-Lions, Department of Pathology, Centre Medical Universitaire, 1 rue Michel-Servet, CH-1211, Geneva, Switzerland; e-mail: michel.aurrand-lions@medecine.unige.ch.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal