The appearance and expansion of donor white blood cells in a recipient after transfusion has many potential biologic ramifications. Although patients with HIV infection are ostensibly at high risk for microchimerism, transfusion-associated graft-versus-host disease (TA-GVHD) is rare. The purpose of this study was to search for sustained microchimerism in such patients. Blood samples were collected from 93 HIV-infected women (a subset from the Viral Activation Transfusion Study, an NHLBI multicenter randomized trial comparing leukoreduced versus unmodified red blood cell [RBC] transfusions) before and after transfusions from male donors. Donor lymphocytes were detected in posttransfusion specimens using a quantitative Y-chromosome–specific polymerase chain reaction (PCR) assay, and donor-specific human leukocyte antigen (HLA) alleles were identified with allele-specific PCR primers and probes. Five of 47 subjects randomized to receive nonleukoreduced RBCs had detectable male lymphocytes 1 to 2 weeks after transfusion, but no subject had detectable male cells more than 4 weeks after a transfusion. In 4 subjects studied, donor-specific HLA haplotypes were detected in posttransfusion specimens, consistent with one or more donors' cells. None of 46 subjects randomized to receive leukoreduced RBCs had detectable male lymphocytes in the month after transfusion. Development of sustained microchimerism after transfusion in HIV-infected patients is rare; HIV-infected patients do not appear to be at risk for TA-GVHD.
Introduction
The appearance and persistence of foreign (allogeneic) white blood cells (WBCs) in a recipient through transplantation, transfusion, or pregnancy has the potential for far-reaching biologic ramifications.1 Transfused WBCs have been associated with febrile transfusion reactions, alloimmunization and subsequent platelet refractoriness,2 transmission of cytomegalovirus (CMV) and other infections,3 up-regulation or reactivation of latent host viruses,4 and immune suppression of the recipient.5 6
For many years it has been observed that red blood cell components from allogeneic donors contain WBCs capable of survival and expansion.7,8 In immunologically intact persons, the life of such cells is usually short (6 or fewer days),9,10 and no clinical consequences result. However, prolonged survival of donor cells, sometimes for years, has occurred after intrauterine transfusion,11,12 and recent studies have demonstrated the asymptomatic persistence of minor populations of donor cells in a proportion of patients after massive transfusion for trauma.13
In immunologically impaired recipients, however, such as children with severe combined immunodeficiency, the consequences of transfusing viable WBCs in blood components can include graft-versus-host disease (GVHD).14,15 This transfusion-associated complication results in expansion of donor WBCs that orchestrate destruction of recipient tissues, including the skin, gastrointestinal tract, and hematopoietic cells. Although rare, transfusion-associated GVHD is almost always lethal because of bleeding and infections associated with profound bone marrow aplasia, and no satisfactory treatment has been discovered. Prevention is critical and requires that, to prevent lymphocyte proliferation, patients at risk be restricted to the transfusion of cellular components that have been gamma-irradiated.16 Because of a small degree of injury inflicted on red blood cells (RBCs) during irradiation17and the expense and administrative complexities associated with the procedure, only a small portion of blood components is usually treated in this fashion. Diagnoses and conditions that put patients at increased risk for transfusion-associated GVHD include severe congenital immunodeficiencies, in utero and neonatal transfusions, Hodgkin and other lymphomas, bone marrow transplantation, and use of drugs with strong immunosuppressant effects, such as 2-deoxycoformycin.18 19
Patients infected with HIV are also immunologically impaired and ostensibly at high risk for long-term persistence and expansion of donor WBCs. However, only one instance of transfusion-associated GVHD has been reported, in a child.20 To better understand this paradox, we designed and carried out a study to assess the survival and quantitation of transfused WBCs given to HIV-infected recipients.
Patients, materials, and methods
Study design
Transfusion recipients in this study were a subset of those enrolled in the multicenter Viral Activation Transfusion Study (VATS) between 1995 and 1998 (see 1 for names of participating institutions and persons).21 The main objective of VATS, a randomized and double-blinded study, was a comparison of the effects of leukoreduced versus unmodified blood transfusions on the clinical and laboratory course of HIV-infected patients. In addition to the primary endpoints of overall mortality and changes in plasma HIV RNA, the investigators also collected data that allowed the evaluation of the survival and persistence of transfused WBCs from male donors in female recipients.
Eligible subjects for VATS were 14 years of age or older, had never received blood products before (except for immune globulin), and were about to be transfused with red blood cells because of symptomatic, nonsurgically induced anemia. Patients with the diagnosis of thrombotic thrombocytopenia purpura and those who received intravenous immune globulin in the past 6 months were excluded. Other eligibility requirements included evidence of previous exposure to, or current infection with, cytomegalovirus, as determined by the presence of antibodies to this virus or a history of CMV end-organ disease. This was to ensure that no enrolled subject was at risk for de novo transfusion-transmitted CMV, as might occur to CMV-naive patients in the unmodified (white-cell replete) study arm.22 Enrolled subjects were randomized by telephone assignment to receive either leukoreduced (less than 5 × 106 residual WBCs per unit) or unmodified RBCs. Leukoreduction of RBCs was achieved through filtration, within 72 hours of blood collection; no specific manufacturer's filter was required. All RBC components were expected to be transfused within 14 days of collection whenever possible. Gamma irradiation was not used in this substudy. If platelet transfusions were required, these were leukoreduced regardless of the study arm assignment.
Patients underwent transfusions as ordered by their personal physicians. A transfusion episode included 1 U (or more) RBCs given over 1 day (or multiple days); each transfusion episode ended once no RBCs were transfused for at least 7 days. Two successive transfusion episodes (T1 and T2) were studied; weekly blood samples were obtained for up to 4 weeks after transfusion and then every 3 months after enrollment until the end of the study (spring 1999).
Subjects included in the donor survival analysis subset reported here were female VATS participants whose study transfusions included at least one component collected from a male donor and whose pre- and posttransfusion blood samples were available for testing.
Cell and DNA preparation and processing
Leukocyte pellets from EDTA-anticoagulated whole blood specimens from donors, and EDTA- and ACD-anticoagulated whole blood specimens from recipients at baseline and after transfusion, were frozen as previously described.10 After thawing, DNA lysates were prepared by adding 50 μL PCR lysis solution (10 mM Tris, pH 8.3; 2.5 mM MgCl2; 1% Tween-20; 1% NP40; 0.4 mL proteinase K) to each pellet and incubating at 60°C for 1.5 hours with vortexing every 20 minutes, followed by 95°C incubation for 2 hours.
Laboratory testing
The adequacy of leukoreduction was assessed in each RBC component using quantitative PCR of a generic human leukocyte antigen (HLA) DQ-alpha sequence in a leukocyte pellet prepared and frozen at the time of transfusion.23 HIV RNA was quantitated from frozen plasma using a reverse transcription—PCR assay with a lower limit of quantification of 200 copies per milliliter (Roche Amplicor assay; Roche Diagnostics; Branchburg, NJ). CD4 cell counts were measured by flow cytometry (FACScan; Becton Dickinson, San Jose, CA) using whole blood samples cryopreserved in dimethyl sulfoxide.23
Donor lymphocyte survival studies
To detect male donor lymphocytes in a female recipient's blood, we used a 125-μL sample of posttransfusion recipient blood for the amplification of a 148-bp region of the sex-determining region of the human Y chromosome (SRY)24 using the allele-specific primer pair SA (5′ CGCATTCATCGTGTGGTCTCGCG 3′) and SD (5′ CTGTGCCTCCTGGAAGAATGGCC 3′), as previously described.13Specific amplified product was detected by oligomer liquid hybridization using a 32P-labeled probe, SB (5′ GAGGCGCAAGATGGCTCTAGAG 3′). Hybridized samples were subjected to 6% polyacrylamide gel electrophoresis (PAGE) at 12.5 V/cm and overnight autoradiography. Testing of all specimens associated with a specific recipient and transfusion episode (through the week 4 posttransfusion samples) was performed as a batch. DNA lysates were tested in duplicate, both neat and at a 1:10 dilution, in a single PCR–hybridization–PAGE autoradiography run. Duplicate standard curves composed of known concentrations (10-fold serial dilutions) of male donor cells in pretransfusion female recipient cells were analyzed in parallel with clinical samples and used to interpolate donor leukocyte concentrations in the latter samples. Autoradiographs were analyzed using the Millipore BioImage Electrophoresis Analyzer application software (Millipore, Ann Arbor, MI) with Whole Band Analyzer application software (Millipore), as detailed elsewhere.13Results were reported as WBC/mL. To avoid false-positive results, we considered male donor cells to be present in a specimen if all replicate results were greater than 0 and at least one of the results showed 25 or more donor cells per milliliter (3 times the theoretical detection limit of the assay).
Quantitative allele-specific PCR for HLA typing of donor cells
In recipients with measurable numbers of male cells after transfusion by Y-chromosome PCR analysis, we pursued identification of a specific donor through the amplification of HLA-DNA. For each transfusion episode, we used primers and probes designed to selectively amplify and detect single-copy HLA class II gene alleles that were unique to any possible male donor target cell populations and that were not present in the recipient. Concentrations of Mg2+, amplification protocols, and reaction buffers were adapted to each specific primer pair to optimize the assay conditions. Sequences of primers and probes and amplification conditions have been previously reported.13
Results
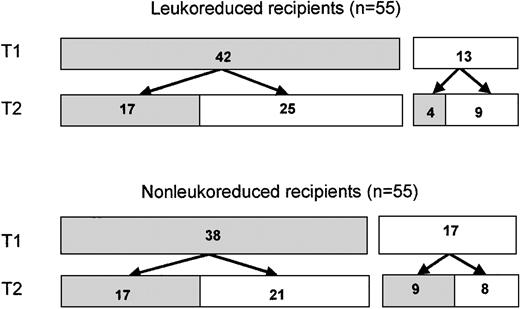
Five hundred thirty-one subjects were enrolled in the VATS trial; 265 were randomized to receive leukoreduced components, and 266 were randomized to receive unmodified blood. Women comprised 21% of enrolled subjects (112 subjects), including 56 in the leukoreduced transfusion arm and 56 in the unmodified arm. Two enrolled women (one in each arm) did not receive transfusions. Of the remaining 110 (Figure1), 80 (73%) received at least one RBC component from a male donor during the first transfusion episode (T1) and provided at least one posttransfusion specimen available for study. Of these 80 subjects, 42 were randomized to receive leukoreduced cells and 38 to receive nonleukoreduced cells. Thirty-four of these women (17 in each arm) underwent a second transfusion of red blood cells and provided posttransfusion samples. Thirteen other women who had received only female cells during a first transfusion (including 4 in the leukoreduced arm and 9 in the nonleukoreduced arm) received male cells in a second transfusion and had posttransfusion samples drawn. Thus, a total of 63 transfusion episodes administered to 46 women in the leukoreduced arm had evaluable posttransfusion samples, as did 64 transfusion episodes to 47 women in the nonleukoreduced arm. These 93 subjects comprise the donor survival study population. Characteristics of these women are shown in Table 1.
Distribution of female blood recipients.
Patients were randomized to receive either leukoreduced or nonleukoreduced red blood cells. ▪, number of subjects who received blood transfusions from one or more male donors, with evaluable posttransfusion blood samples; ■, number of subjects who received blood from only female donors, did not have posttransfusion blood samples for study, or did not undergo a T2 transfusion.
Distribution of female blood recipients.
Patients were randomized to receive either leukoreduced or nonleukoreduced red blood cells. ▪, number of subjects who received blood transfusions from one or more male donors, with evaluable posttransfusion blood samples; ■, number of subjects who received blood from only female donors, did not have posttransfusion blood samples for study, or did not undergo a T2 transfusion.
Characteristics before study transfusions of women with evaluable posttransfusion donor cell survival data
| . | LR arm (T1) . | NLR arm (T1) . | LR arm (T2) . | NLR arm (T2) . |
|---|---|---|---|---|
| Transfused female subjects | 55 | 55 | 24 | 29 |
| Female subjects transfused with ≥ 1 U male donor cells and evaluable data | 42 | 38 | 21 | 26 |
| Days between start of T1 and T2 transfusions* | NA | NA | 54 (27, 144) | 68 (16, 273) |
| Subject age (y)* | 35 (28, 47) | 38 (24, 48) | 35 (27, 44) | 37 (28, 48) |
| Race/ethnicity | ||||
| White (%) | 13 (31) | 16 (42) | 8 (38) | 9 (35) |
| Black (%) | 20 (48) | 17 (45) | 9 (43) | 14 (54) |
| Hispanic (%) | 9 (21) | 2 (5) | 4 (19) | 2 (8) |
| Other (%) | 0 | 3 (8) | 0 | 1 (4) |
| Route of HIV infection | ||||
| Intravenous drug use (%) | 12 (29) | 17 (45) | 8 (38) | 9 (35) |
| Heterosexual contact (%) | 28 (67) | 18 (47) | 12 (57) | 14 (54) |
| Other/unknown (%) | 2 (5) | 3 (8) | 1 (5) | 3 (11) |
| Previous HAART (%)† | 11 (26) | 7 (18) | 5 (24) | 8 (31) |
| Log10 HIV RNA* | 4.7 (2.6, 5.8) | 4.9 (2.7, 5.9) | 5.0 (3.8, 5.9) | 4.7 (2.7, 5.8) |
| CD4 cell count per μL* | 19 (2, 156) | 12 (2, 357) | 14 (2, 84) | 6 (1, 303) |
| . | LR arm (T1) . | NLR arm (T1) . | LR arm (T2) . | NLR arm (T2) . |
|---|---|---|---|---|
| Transfused female subjects | 55 | 55 | 24 | 29 |
| Female subjects transfused with ≥ 1 U male donor cells and evaluable data | 42 | 38 | 21 | 26 |
| Days between start of T1 and T2 transfusions* | NA | NA | 54 (27, 144) | 68 (16, 273) |
| Subject age (y)* | 35 (28, 47) | 38 (24, 48) | 35 (27, 44) | 37 (28, 48) |
| Race/ethnicity | ||||
| White (%) | 13 (31) | 16 (42) | 8 (38) | 9 (35) |
| Black (%) | 20 (48) | 17 (45) | 9 (43) | 14 (54) |
| Hispanic (%) | 9 (21) | 2 (5) | 4 (19) | 2 (8) |
| Other (%) | 0 | 3 (8) | 0 | 1 (4) |
| Route of HIV infection | ||||
| Intravenous drug use (%) | 12 (29) | 17 (45) | 8 (38) | 9 (35) |
| Heterosexual contact (%) | 28 (67) | 18 (47) | 12 (57) | 14 (54) |
| Other/unknown (%) | 2 (5) | 3 (8) | 1 (5) | 3 (11) |
| Previous HAART (%)† | 11 (26) | 7 (18) | 5 (24) | 8 (31) |
| Log10 HIV RNA* | 4.7 (2.6, 5.8) | 4.9 (2.7, 5.9) | 5.0 (3.8, 5.9) | 4.7 (2.7, 5.8) |
| CD4 cell count per μL* | 19 (2, 156) | 12 (2, 357) | 14 (2, 84) | 6 (1, 303) |
Of 110 female subjects transfused during the VATS, 80 had evaluable samples after their first (T1) transfusions, and 47 had evaluable samples after their second (T2) transfusions.
Median values are followed by 10th and 90th percentiles in parentheses.
Highly active antiretroviral therapy exposure before transfusion.
LR, leukoreduced study arm; NLR, nonleukoreduced study arm.
Leukoreduced RBCs had a median of 1.08 × 105 residual WBCs per component, and more than 99% of these components (1854 of 1869) had fewer than 5 × 106 WBCs per unit. The median age of a RBC component at the time of transfusion was 9 days (10th percentile, 3 days; 90th percentile, 14 days). Four percent of RBC components (157 of 3812 U in the full VATS study set) were older than 14 days at the time of transfusion.
Detection of male donor cells using Y-chromosome amplification is outlined in Table 2, and representative amplification gels are shown in Figure 2. Five transfusion episodes (of 64 total episodes) in 5 subjects receiving nonleukoreduced RBCs were associated with evidence of posttransfusion male donor cell survival. Four of these subjects (NLR-1, NLR-2, NLR-3, and NLR-5) had detectable cells at day 7, all after T2 transfusions. In 2 of these subjects (NLR-1 and NLR-2), the cells could no longer be detected 1 week later. NLR-3 had male cells detected on days 4 and 7 (the day 4 specimen was drawn as a quarterly specimen after T1); day 14, day 21, and day 28 samples were unavailable. NLR-5 had male cells on days 7 and 21 (day 14 results showed inconsistent reactivity and were not considered positive); cells were undetectable by day 28. For a T1 transfusion, NLR-4 received 2 U blood older than stipulated by the study protocol (24 days old). On day 14, male donor cells were readily detected, but by day 21 the sample was no longer considered positive. No male donor cells were detected in the nonleukoreduced arm during follow-up quarterly studies.
Detection of male donor WBCs after transfusion of red blood cells to female subjects
| Recipient . | First transfusion episode (T1) . | Transfusion interval (start of T1 to start of T2) . | Second transfusion episode (T2) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor gender: component age (d) . | Cell count (male cells/mL recipient blood) . | Donor gender: component age (d) . | Cell count (male cells/mL recipient blood) . | ||||||||||
| Pre . | D7 . | D14 . | D21 . | D28 . | Pre . | D7 . | D14 . | D21 . | D28 . | ||||
| NLR-1 | M:10; F:7 | 0, 0 | 8, 0 | (NT)† | (NT)† | (NT)† | 13 days | M:12; F:12 | 0, 0 | 64, 71.5 | 0, 0 | 0, 0 | 0, 0 |
| NLR-2 | M:3; M:5 | 0 | NT | 0 | 0 | (NT)† | 22 days | M:3; M:3 | 0, 0; 0 | 25, 55; 20 | 0, 0; 0 | NT | NT |
| NLR-3 | M:7; F:7 | 0 | NT | 0 | 0 | 0 | 66 days | M:7; F:10 | 0, 0 | 23; 49‡ | NT | NT | NT |
| NLR-4 | M:24; F:24 | 0, 0 | 0, 0 | 1830, 2110 | 16, 0 | 0, 0 | 70 days | M:11; F:11 | 0 | 0 | 0 | 0 | NT |
| NLR-5 | M:12; F:12* | 0, 0; 0 | NT | 0, 0; 16, 0 | 17, 5; 15 | 9, 21; 0 | 61 days | M:3; M:4 | 0, 0; 0 | 16.5, 362 | 0, 20; 10 | 8.5, 31 | 0, 0; 0 |
| LR-1 | M:7; M:10; M:13; F:3 | 0 | 0 | 0 | 0 | 02-153 | No second transfusion | ||||||
| Recipient . | First transfusion episode (T1) . | Transfusion interval (start of T1 to start of T2) . | Second transfusion episode (T2) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor gender: component age (d) . | Cell count (male cells/mL recipient blood) . | Donor gender: component age (d) . | Cell count (male cells/mL recipient blood) . | ||||||||||
| Pre . | D7 . | D14 . | D21 . | D28 . | Pre . | D7 . | D14 . | D21 . | D28 . | ||||
| NLR-1 | M:10; F:7 | 0, 0 | 8, 0 | (NT)† | (NT)† | (NT)† | 13 days | M:12; F:12 | 0, 0 | 64, 71.5 | 0, 0 | 0, 0 | 0, 0 |
| NLR-2 | M:3; M:5 | 0 | NT | 0 | 0 | (NT)† | 22 days | M:3; M:3 | 0, 0; 0 | 25, 55; 20 | 0, 0; 0 | NT | NT |
| NLR-3 | M:7; F:7 | 0 | NT | 0 | 0 | 0 | 66 days | M:7; F:10 | 0, 0 | 23; 49‡ | NT | NT | NT |
| NLR-4 | M:24; F:24 | 0, 0 | 0, 0 | 1830, 2110 | 16, 0 | 0, 0 | 70 days | M:11; F:11 | 0 | 0 | 0 | 0 | NT |
| NLR-5 | M:12; F:12* | 0, 0; 0 | NT | 0, 0; 16, 0 | 17, 5; 15 | 9, 21; 0 | 61 days | M:3; M:4 | 0, 0; 0 | 16.5, 362 | 0, 20; 10 | 8.5, 31 | 0, 0; 0 |
| LR-1 | M:7; M:10; M:13; F:3 | 0 | 0 | 0 | 0 | 02-153 | No second transfusion | ||||||
Male donor cells were considered present in a specimen (shaded box) if all replicate test results were >0 and at least one of the results was ≥25 donor cells/mL. Results separated by commas represent replicates of the same specimen tube. Results separated by semicolon represent repeat testing on separate specimen tubes (results are listed in order of testing).
Subject NLR-5 also received 2 U RBCs (M:16; F15) 9 days after the T1 transfusion.
See T2.
Subject NLR-3. T2 day 4 results also available: 90, 241.
Subject LR-1. At 3 months after T1, male cells were detected: 33, 40.
NT, not tested.
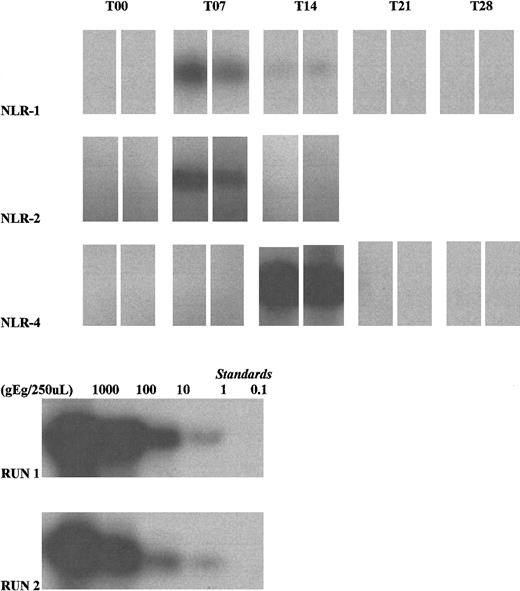
Autoradiographs depicting donor leukocyte survival in 3 HIV-infected female recipients (NLR-1, NLR-2, and NLR-4) (top 3 rows).
Amplification of Y-chromosome DNA was performed using duplicate 125 μL whole blood samples collected at weekly intervals. All T00 (pretransfusion) samples were negative. The T07 blood sample from NLR-1 during the second study transfusion was positive (though a weak band was seen at T14, quantitation was below the definition of positive). NLR-2 was positive on T07, during the second study transfusion. NLR-4 was strongly positive on T14, during the first study transfusion. Lysate standards run with the clinical samples (NLR-1, standard run 2; NLR-2 and NLR-4, standard run 1) demonstrate 10-fold dilutions starting from the leftmost band equivalent to values of 1000, 100, 10, 1 and 0.1 genome equivalents (gEq)/lane.
Autoradiographs depicting donor leukocyte survival in 3 HIV-infected female recipients (NLR-1, NLR-2, and NLR-4) (top 3 rows).
Amplification of Y-chromosome DNA was performed using duplicate 125 μL whole blood samples collected at weekly intervals. All T00 (pretransfusion) samples were negative. The T07 blood sample from NLR-1 during the second study transfusion was positive (though a weak band was seen at T14, quantitation was below the definition of positive). NLR-2 was positive on T07, during the second study transfusion. NLR-4 was strongly positive on T14, during the first study transfusion. Lysate standards run with the clinical samples (NLR-1, standard run 2; NLR-2 and NLR-4, standard run 1) demonstrate 10-fold dilutions starting from the leftmost band equivalent to values of 1000, 100, 10, 1 and 0.1 genome equivalents (gEq)/lane.
In the leukoreduced arm, male donor cells were detected in 1 of 63 transfusion episodes involving leukoreduced RBCs. In this case (LR-1), no cells were detected during any of the 4 weekly samples after T1 transfusion but were found for the first and only time 3 months later. The residual white counts in the 4 filtered units given to LR-1 were all lower than 2.5 × 105 cells per component. Male donor cells were not detected in any other recipient in this arm at any time point.
Pretransfusion characteristics of the 6 women with detectable male donor cells after transfusion are shown in Table3; they were similar to those in the patient population as a whole (Table 1). With the exception of NLR-4, the median age of the transfused male donor components given to these women (6 days; 10th percentile, 3 days; 90th percentile, 14 days) was slightly younger than the median age of male donor components given to the other women (8 days; 10th percentile, 4 days; 90th percentile, 14 days).
Characteristics before study transfusions of women with detectable posttransfusion male donor cells
| Recipient . | Race . | HIV risk . | Age . | Previous HAART use (start day, in relation to T1 transfusion)3-150 . | Log10 HIV viral load . | CD4 cells/μL . | Detection of male cells after transfusion episode . | |
|---|---|---|---|---|---|---|---|---|
| T1 . | T2 . | |||||||
| NLR-1 | Black | Intravenous drug use | 47 | No | 4.7 | 6 | No | Yes |
| NLR-2 | Black | Intravenous drug use | 28 | −27 | 3.3 | 3 | No | Yes |
| NLR-3 | Black | Heterosexual | 34 | −48 | 5.0 | 6 | No | Yes |
| NLR-4 | White | Heterosexual | 45 | No | 4.7 | 10 | Yes | No |
| NLR-5 | Black | Heterosexual | 44 | No | 4.7 | 2 | No | Yes |
| LR-1 | White | Heterosexual | 31 | No | < 2.33-152 | 52 | Yes3-151 | —3-151 |
| Recipient . | Race . | HIV risk . | Age . | Previous HAART use (start day, in relation to T1 transfusion)3-150 . | Log10 HIV viral load . | CD4 cells/μL . | Detection of male cells after transfusion episode . | |
|---|---|---|---|---|---|---|---|---|
| T1 . | T2 . | |||||||
| NLR-1 | Black | Intravenous drug use | 47 | No | 4.7 | 6 | No | Yes |
| NLR-2 | Black | Intravenous drug use | 28 | −27 | 3.3 | 3 | No | Yes |
| NLR-3 | Black | Heterosexual | 34 | −48 | 5.0 | 6 | No | Yes |
| NLR-4 | White | Heterosexual | 45 | No | 4.7 | 10 | Yes | No |
| NLR-5 | Black | Heterosexual | 44 | No | 4.7 | 2 | No | Yes |
| LR-1 | White | Heterosexual | 31 | No | < 2.33-152 | 52 | Yes3-151 | —3-151 |
Highly active antiretroviral therapy exposure before transfusion.
LR-1 did not undergo T2 transfusion; male cells were detected only in the 3-month posttransfusion sample.
Below lower limit of detection.
The detection of specific donor HLA genotypes is described in Tables 4through7. Posttransfusion samples were available for these experiments from 4 subjects (NLR-1, NLR-2, NLR-4, and NLR-5) at time points coinciding with dates when male cells were detected. No relevant samples were available from NLR-3 and LR-1 because all aliquots of frozen whole blood from the implicated time points had been used in other VATS-related research.
Detection of donor HLA class II genotypes after transfusion to recipient NLR-1
| HLA allele . | Recipient NLR-1 . | Donor 1A M, 10 days . | Donor 1B F, 7 days . | Donor 1C M, 12 days . | Donor 1D F, 12 days . | Recipient, posttransfusion analysis day . |
|---|---|---|---|---|---|---|
| Transfusion . | T1 . | T1 . | T2 . | T2 . | T2 day 7 (=T1 day 20) . | |
| DR1 | POS | POS | POS | NT | ||
| DR11 | POS | NT | ||||
| DQB501 | POS | POS | POS | POS | NT | |
| DQB7 | POS | NT | ||||
| DR3 | POS | Detected | ||||
| DQB201 | POS | POS | POS | Detected | ||
| DR7 | POS | POS | POS | NT | ||
| DR4 | POS | NT | ||||
| DQB603 | POS | NT | ||||
| DQB8 | POS | NT |
| HLA allele . | Recipient NLR-1 . | Donor 1A M, 10 days . | Donor 1B F, 7 days . | Donor 1C M, 12 days . | Donor 1D F, 12 days . | Recipient, posttransfusion analysis day . |
|---|---|---|---|---|---|---|
| Transfusion . | T1 . | T1 . | T2 . | T2 . | T2 day 7 (=T1 day 20) . | |
| DR1 | POS | POS | POS | NT | ||
| DR11 | POS | NT | ||||
| DQB501 | POS | POS | POS | POS | NT | |
| DQB7 | POS | NT | ||||
| DR3 | POS | Detected | ||||
| DQB201 | POS | POS | POS | Detected | ||
| DR7 | POS | POS | POS | NT | ||
| DR4 | POS | NT | ||||
| DQB603 | POS | NT | ||||
| DQB8 | POS | NT |
Allele-specific primers and probes were designed to detect HLA genotypes after transfusion that were not present in the recipient before transfusion. Donor gender and age of RBC component at time of transfusion are listed (eg, M, 10 days). Each recipient's genotype is indicated in the second column; corresponding genes found in the transfused blood donors are aligned with the recipient's.
POS, positive for gene; detected (not detected), identified (not identified) by respective primer/probe analysis of recipient's posttransfusion sample; NT, not tested.
Detection of donor HLA class II genotypes after transfusion to recipient NLR-2
| HLA allele . | Recipient NLR-2 . | Donor 2A M, 3 days . | Donor 2B M, 5 days . | Donor 2C M, 3 days . | Donor 2D M, 3 days . | Recipient, posttransfusion analysis day . |
|---|---|---|---|---|---|---|
| Transfusion . | T1 . | T1 . | T2 . | T2 . | T2 day 7 (=T1 day 29) . | |
| DR2 | POS | POS | NT | |||
| DR3 | POS | NT | ||||
| DR15 | POS | POS | NT | |||
| DQB201 | POS | POS | POS | NT | ||
| DQB602 | POS | POS | NT | |||
| DQB603 | POS | POS | NT | |||
| DR7 | POS | POS | Detected | |||
| DQB501 | POS | POS | Detected | |||
| DQB7 | POS | Detected | ||||
| DR13 | POS | Not detected | ||||
| DQB8 | POS | Not detected | ||||
| DR4 | POS | POS | NT | |||
| DR1 | POS | POS | NT |
| HLA allele . | Recipient NLR-2 . | Donor 2A M, 3 days . | Donor 2B M, 5 days . | Donor 2C M, 3 days . | Donor 2D M, 3 days . | Recipient, posttransfusion analysis day . |
|---|---|---|---|---|---|---|
| Transfusion . | T1 . | T1 . | T2 . | T2 . | T2 day 7 (=T1 day 29) . | |
| DR2 | POS | POS | NT | |||
| DR3 | POS | NT | ||||
| DR15 | POS | POS | NT | |||
| DQB201 | POS | POS | POS | NT | ||
| DQB602 | POS | POS | NT | |||
| DQB603 | POS | POS | NT | |||
| DR7 | POS | POS | Detected | |||
| DQB501 | POS | POS | Detected | |||
| DQB7 | POS | Detected | ||||
| DR13 | POS | Not detected | ||||
| DQB8 | POS | Not detected | ||||
| DR4 | POS | POS | NT | |||
| DR1 | POS | POS | NT |
See footnote to Table 4.
Detection of donor HLA class II genotypes after transfusion to recipient NLR-4
| HLA allele . | Recipient NLR-4 . | Donor 4A M, 24 days . | Donor 4B F, 24 days . | Recipient, posttransfusion analysis day . |
|---|---|---|---|---|
| Transfusion . | T1 . | T1 . | T1 day 14 . | |
| DR1 | POS | NT | ||
| DR7 | POS | POS | POS | NT |
| DQB201 | POS | POS | POS | NT |
| DQB501 | POS | NT | ||
| DR15 | POS | Detected | ||
| DR2 | POS | Not detected | ||
| DQB602 | POS | Not detected | ||
| DR4 | POS | NT | ||
| DQB8 | POS | NT |
| HLA allele . | Recipient NLR-4 . | Donor 4A M, 24 days . | Donor 4B F, 24 days . | Recipient, posttransfusion analysis day . |
|---|---|---|---|---|
| Transfusion . | T1 . | T1 . | T1 day 14 . | |
| DR1 | POS | NT | ||
| DR7 | POS | POS | POS | NT |
| DQB201 | POS | POS | POS | NT |
| DQB501 | POS | NT | ||
| DR15 | POS | Detected | ||
| DR2 | POS | Not detected | ||
| DQB602 | POS | Not detected | ||
| DR4 | POS | NT | ||
| DQB8 | POS | NT |
See footnote to Table 4.
Detection of donor HLA class II genotypes after transfusion to recipient NLR-5
| HLA allele . | Recipient NLR-5 . | Donor 5A M, 12 days . | Donor 5B F, 12 days . | Donor 5C F, 15 days . | Donor 5D M, 16 days . | Donor 5E M, 3 days . | Donor 5F M, 4 days . | Recipient, posttransfusion analysis day . | Recipient, posttransfusion analysis day . |
|---|---|---|---|---|---|---|---|---|---|
| Transfusion day . | T1 . | T1 . | T1 (day 9) . | T1 (day 9) . | T2 . | T2 . | T2 day 7 (=T1 day 68) . | T2 day 21 (=T1 day 82) . | |
| DR2 | POS | POS | NT | NT | |||||
| DR15 | POS | POS | NT | NT | |||||
| DQB602 | POS | POS | POS | POS | NT | NT | |||
| DQB603 | POS | POS | POS | NT | NT | ||||
| DR11 | POS | POS | Detected | Detected | |||||
| DR3 | POS | POS | Detected | Detected | |||||
| DQB501 | POS | POS | Detected | Not detected | |||||
| DQB7 | POS | POS | POS | Detected | Not detected | ||||
| DR7 | POS | Detected | Not detected | ||||||
| DR13 | POS | POS | Not detected | Detected | |||||
| DR12 | POS | Not detected | Not detected | ||||||
| DR4 | POS | POS | Not detected | Not detected | |||||
| DQB201 | POS | POS | NT | NT | |||||
| DR1 | POS | NT | NT | ||||||
| DQB8 | POS | POS | NT | NT |
| HLA allele . | Recipient NLR-5 . | Donor 5A M, 12 days . | Donor 5B F, 12 days . | Donor 5C F, 15 days . | Donor 5D M, 16 days . | Donor 5E M, 3 days . | Donor 5F M, 4 days . | Recipient, posttransfusion analysis day . | Recipient, posttransfusion analysis day . |
|---|---|---|---|---|---|---|---|---|---|
| Transfusion day . | T1 . | T1 . | T1 (day 9) . | T1 (day 9) . | T2 . | T2 . | T2 day 7 (=T1 day 68) . | T2 day 21 (=T1 day 82) . | |
| DR2 | POS | POS | NT | NT | |||||
| DR15 | POS | POS | NT | NT | |||||
| DQB602 | POS | POS | POS | POS | NT | NT | |||
| DQB603 | POS | POS | POS | NT | NT | ||||
| DR11 | POS | POS | Detected | Detected | |||||
| DR3 | POS | POS | Detected | Detected | |||||
| DQB501 | POS | POS | Detected | Not detected | |||||
| DQB7 | POS | POS | POS | Detected | Not detected | ||||
| DR7 | POS | Detected | Not detected | ||||||
| DR13 | POS | POS | Not detected | Detected | |||||
| DR12 | POS | Not detected | Not detected | ||||||
| DR4 | POS | POS | Not detected | Not detected | |||||
| DQB201 | POS | POS | NT | NT | |||||
| DR1 | POS | NT | NT | ||||||
| DQB8 | POS | POS | NT | NT |
See footnote to Table 4.
We were able to detect DR3 and DQB201 in a sample of blood from NLR-1 collected 7 days after the T2 transfusion (20 days after the T1 transfusion), when male DNA had been identified by Y-chromosome analysis. The presence of DR3 was consistent only with the genotype of male donor 1A, associated with the T1 transfusion. However, it is possible that we could have been detecting DNA from the other male donor (1C) or from female donor 1B because all 3 donors were DQB201-positive.
We used 5 sets of donor HLA-specific primer pairs and probes to test blood from recipient NLR-2 one week after her T2 transfusion. DR7, DQB501, and DQB7 were found, consistent with cells from T2 donors 2C and 2D. Donor 2B (from the T1 transfusion) was also DR7 positive, but we did not find DQB8, another of his alleles, and we did not find DR13, a distinguishing allele of donor 2A. Thus, at least 2 male donors could have contributed to the posttransfusion DNA detected in this subject.
We studied recipient NLR-4 on day 14 after the T1 transfusion, using 3 sets of primer pairs and probes that would have distinguished donor 4A from the recipient (DR2, DR15, and DQB602). Only DR15 was detected.
Finally we studied recipient NLR-5 at 2 time points after the T2 transfusion (day 7 and day 21). On day 7, at least 3 donors appeared to have contributed to the DNA detected—DR7 from donor 5A; DQB7 and DR11, but not DR13, from either donor 5C or 5F, or both; DQB501 from either donor 5B or 5D, or both; and DR3 from either donor 5D or 5E, or both. On day 21, DNA from donors 5A and 5B was no longer detectable, whereas DNA was still evident consistent with donors 5C, 5D, 5E, and 5F.
Discussion
In the VATS subpopulation of 93 women who received RBC components from one or more male donors, only 5 had detectable male cells in the first month after transfusion. All were in the nonleukoreduced RBC study arm, and this outcome might have been predicted from the comparatively higher WBC content of the unmodified components. Results of DNA studies suggested that more than one donor might have been involved in the microchimerism, though the limitations of this assay make firm donor identification difficult. Four of the 5 recipients demonstrated male chimerism only with the T2 transfusion; whether this was by chance or was related to a cumulative effect of allogeneic transfusions on the likelihood of microchimerism cannot be determined from such small numbers. However, with 47 participants receiving nonleukoreduced RBCs, these 5 who had posttransfusion donor cell survival represented only 11% of subjects; seen another way, this represented 8% of the 64 evaluable transfusion episodes involving nonleukoreduced male donor RBCs. Of key importance, sustained donor leukocyte microchimerism did not develop in any of these recipients. These data provide confirmatory laboratory evidence in support of clinical impressions that HIV-infected patients are not at risk for GVHD.
Which factors might have protected most female transfusion recipients from detectable microchimerism or, conversely, placed 5 others at risk are unclear. In particular, the mean age of the components at transfusion was not meaningfully shorter in the 5 recipients with detectable donor cells than in other recipients in this substudy. On the contrary, the highest detectable number of male cells was found in the recipient of 2 old units (24 days old, NLR-4). Recipient factors such as age, race, HIV risk factor, previous HAART use, baseline viral load, and baseline CD4 cell count were also not uniquely slanted in this small population than in the rest of the sub-study group. In all 4 tested participants, one or more male donors shared some class II genes with the relevant recipient. It is possible that in some of these subjects, the degree of partial HLA matching between donor and recipient facilitated tolerance and allogeneic cell survival.25-27 However, the extent to which this occurred more frequently in these recipients than in other women in the study was not ascertained.
Of the 46 evaluable subjects assigned to the leukoreduced arm (and 63 evaluable transfusion episodes), male cells were detected in only one recipient after transfusion. The data were unusual in that no male cells were identified in her blood at any of the 4 weekly time points in the first month after transfusion. The sole positive sample was drawn 3 months after the transfusion, suggesting that alternative explanations for the presence of male donor cells must be considered in evaluating the results of our study. During pregnancy, 2-way trafficking of cells across the placenta—maternal to fetal circulation, and vice versa—has been documented. Fetal WBCs,28,29 and even free DNA of fetal origin,30 can be identified using DNA-based technology during and immediately after pregnancy. Long-term postpartum persistence of fetal cells also occurs, and the development of systemic sclerosis, with its marked preponderance in women, may be a consequence in some women of prolonged fetal cell survival and a fetal-versus-maternal (graft-versus-host) reaction.31,32In another study, 6 of 8 women with pregnancies more than 6 months earlier had evidence of CD34+ hematopoietic progenitor cells, in one woman as long as 27 years after delivery.33We were unfortunately unable to ascertain whether the subjects in our sub-study had given birth to male children before enrollment in VATS; this information had not been required in the original study database.
The use of male blood recipients could have circumvented the problem of long-term survival of male fetal cells in our female recipients and would have substantially increased our study population size (presumably without affecting the results; we are unaware of published data to suggest that transfusion-associated microchimerism has different features in males than in females). We used Y-chromosome DNA amplification for screening for microchimerism because of its ready applicability to female recipients receiving blood from one or more male donors of otherwise unknown genotype. Historically, the detection of microchimerism capitalized on such sex chromosome differences, primarily through cytogenetic identification of the Y chromosome8,11 or through PCR amplification of Y-chromosome DNA fragments.9,10 More recently, it has become possible to study other polymorphic genetic markers, such as HLA alleles,27,34 or non-HLA, highly polymorphic, or otherwise informative markers such as are found on human globin genes.14,28 However, if multiple donors or donors of unknown genotype are involved, a highly polymorphic system and many different probes must be used to consistently detect microchimerism. Even when a donor's genotype is known, careful choice of probes is required to ensure informative amplifications. In a recent side-by-side comparison of a selection of these approaches, Y-chromosome DNA amplification triumphed as the most sensitive and consistent assay for microchimerism.35
Regardless of the assay used, the detection of rare populations of cells in blood remains challenging. Assays range in sensitivity from 0.1% male DNA in a female DNA background using single primer pairs for the Y chromosome to 0.0001% with nested primers.35Different primers may lead to different amplification efficiencies, and inadvertent mispriming of associated material, such as pseudogenes in the recipient, has led to false-positive results because of misinterpretation of the bands as of donor origin.36 We controlled for this possibility by running coded pre- and posttransfusion samples from each case together. In addition, the testing laboratory was blinded to the clinical assignment of patients to either the leukoreduced or the unmodified arm of the study. To minimize potential PCR carryover and to maximize specificity, we used nonnested assays with radiolabeled probes specific to the amplified product of the minor population under study.
We could also have missed the peak of donor lymphocyte proliferation in our transfusion recipients. In previous studies, the peak increase in concentration of donor lymphocytes has occurred from 3 to 7 days,7,8 10 and our choice of 7 days as the first posttransfusion assay point might have caused earlier transient proliferations to be missed. However, we expected and were looking for prolonged microchimerism that might have clinical relevance; hence, the absence of donor survival beyond 7 days was useful data.
Our subjects all had advanced acquired immunodeficiency syndrome. Given the inherent immunodeficiency of the illness and the severity of their conditions, they comprised a population theoretically likely to be at risk for GVHD. Despite this, measurable numbers of WBCs survived in only a small proportion, and then for only short periods, without amplification of cell numbers. These data are entirely in keeping with clinical observations that transfusion-associated GVHD does not develop in HIV-infected patients, almost without exception.37 In an isolated report in the literature, a child born with HIV received 6 nonirradiated RBC transfusions at age 30 months. Mild and transient GVHD associated with the appearance of additional HLA alleles in blood samples (presumably by serologic methods) linked to at least 4 different donors developed and persisted through 3 months after transfusion, when the symptoms disappeared.20 This single case report stands in stark contrast to statistics on blood usage in HIV infection. HIV infection has been diagnosed in more than 1 000 000 patients in the United States; moderate or severe anemia (hemoglobin level less than 9.4 g/dL) developed in an estimated 20% of them through 1996, and many became transfusion-dependent.38 The rarity of TA-GVHD may stem from infection with HIV, which could protect the infected patient from GVHD either by infecting and destroying transfused CD4+cells or by rendering those cells less able to proliferate in the immunosuppressive environment that is a consequence of HIV-1 replication.39
Of interest is the contrast between these results and those obtained in a small population of apparently immunocompetent women who, after transfusion of multiple units of fresh blood (4 to 18 U, all 15 days old or younger) after trauma, had sustained, asymptomatic, multilineage microchimerism for many months.13 Hemorrhage, trauma, and surgery might separately or synergistically have created a degree of immunosuppression that fostered donor cell survival in these patients. In addition, the exposure to larger numbers of donors should have increased the odds that a partially HLA-matched donor was chosen to whom the recipient was tolerant.
The unexpected survival of, and host perturbation by, transplanted or transfused WBCs can have important repercussions for a recipient. GVHD resulting from the expansion of donor WBCs after solid organ or bone marrow transplantation can cause death or serious morbidity. By contrast, microchimerism, the steady-state survival of smaller numbers of donor cells, may be of critical value after organ transplantation in that these cells may facilitate donor tolerance and graft acceptance.40,41 Transfusion-associated GVHD in the already immunocompromised patient with HIV infection should presumably be a devastating adverse complication. However, our data suggest that donor WBCs are not able to proliferate in the HIV-infected patient. As a result, gamma irradiation or other methods (such as photochemical treatment)42 that prevent transfusion-associated graft-versus-host disease through the inhibition of T-cell proliferation appear unnecessary in this population.
We thank the patients who participated in this study for their efforts, and we thank the transfusion service personnel and nurses at each medical center, without whose assistance the study could not have been accomplished. Please see the for further information about the Viral Activation Transfusion Study Group.
The Viral Activation Transfusion Study Group is the responsibility of the following persons.
Clinical sites
Case Western Reserve University, Cleveland, OH (N01-HB-57115): Michael Lederman, MD; Roslyn Yomtovian, MD; Michael Chance, RN; Donna Hendrix, RN. Georgetown University, Washington, DC (N01-HB-57116): Princy N. Kumar, MD; S. Gerald Sandler, MD; Karyn Hawkins, RN. Miriam Hospital/Brown University, Providence, RI (N01-HB-57117): Timothy P. Flanigan, MD; Joseph Sweeney, MD; Maria D. Mileno, MD; Melissa Di Spigno, RN; Michelle Dupuis, MT (SSB). Mt Sinai School of Medicine, New York, NY (N01-HB-57118): Henry S. Sacks, PhD, MD; Kala Mohandas, MD; Frances R. Wallach, MD; Letty Mintz, ANP. Ohio State University, Columbus (N01-HB-57119): Michael F. Para, MD; Melanie S. Kennedy, MD; Jane Russell, RN; Dave Krugh, MT. University of California, San Diego (N01-HB-57120): Thomas A. Lane, MD; W. Christopher Mathews, MD; Peggy Mollen-Rabwin, RN. University of California, San Francisco (N01-HB-57121): Edward L. Murphy, MD, MPH; Steven G. Deeks, MD; Maurene Viele, MD; Chaolun Han, MD; Joanne Moore, MT (ASCP) SBB. University of North Carolina, Chapel Hill (N01-HB-57122): Charles van der Horst, MD; Meera Kelley, MD; Mark E. Brecher, MD; Linh Ngo, FNP. University of Pittsburgh, PA (N01-HB-57123): John W. Mellors, MD; Darrell J. Triulzi, MD; Deborah K. McMahon, MD; Sharon Riddler, MD. University of Texas Medical Branch, Galveston (N01-HB-57124): David M. Asmuth, MD; Richard B. Pollard, MD; Janice Curry, PAC; Gerald Shulman, MD. University of Washington/Puget Sound Blood Center, Seattle (N01-HB-57125): Ann Collier, MD; Terry Gernsheimer, MD; Dee Townsend-McCall, RN; Jill Corson, RN.
Central laboratory
Blood Centers of the Pacific, San Francisco, CA (N01-HB-57126): Michael P. Busch, MD, PhD; Tzong-Hae Lee, MD, PhD; W. Lawrence Drew, MD, PhD (UCSF Mt Zion Medical Center, San Francisco); Megan Laycock.
Coordinating center
New England Research Institutes, Watertown, MA (N01-HB-57127): Leslie A. Kalish, ScD; Susan F. Assmann, PhD; Jane D. Carrington, RN, BS; Margot S. Kruskall, MD (Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA); Ruth Eisenbud, BA.
Sponsoring agency
National Heart, Lung and Blood Institute: George J. Nemo, PhD, project officer; Paul R. McCurdy, MD; Dean Follmann, PhD.
Steering committee
Paul V. Holland, MD (chair), Sacramento Medical Foundation Blood Centers, CA.
Data Safety Monitoring Board
Jeffrey McCullough, MD (chair), University of Minnesota, Minneapolis; Victor DeGruttola, ScD; Peter Frame, MD; Janice G. McFarland, MD; Ronald T. Mitsuyasu, MD; Elizabeth J. Read, MD; Dorothy E. Vawter, PhD.
Supported by contracts N01-HB-57126, N01-HB-57127, and N01-HB-57115 from the National Heart Lung and Blood Institute (National Institutes of Health).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Margot S. Kruskall, Division of Laboratory and Transfusion Medicine, Beth Israel Deaconess Medical Center, Yamins 309, 330 Brookline Ave, Boston, MA 02215.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal