Fibrinogen Milano XII was detected in an asymptomatic Italian woman, whose routine coagulation test results revealed a prolonged thrombin time. Fibrinogen levels in functional assays were considerably lower than levels in immunologic assays. Polymerization of purified fibrinogen was strongly impaired in the presence of calcium or ethylenediaminetetraacetic acid (EDTA). Two heterozygous structural defects were detected by DNA analysis: Aα R16C and γ G165R. As seen previously with other heterozygous Aα R16C variants, thrombin-catalyzed release of fibrinopeptide A was 50% of normal. Additionally, the release of fibrinopeptide B was delayed. Immunoblotting analysis with antibodies to human serum albumin indicated that albumin is bound to Aα 16 C. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis of plasmin digests of fibrinogen Milano XII in the presence of calcium or EDTA showed both normal and novel D1 and D3 fragments. Further digestion of abnormal D3 fragments by chymotrypsin resulted in degradation products of the same size as the fragments derived from normal fibrinogen. SDS-PAGE analysis under reducing conditions showed no difference between normal fibrinogen and fibrinogen Milano XII or between their plasmic fragments. Circular dichroism analysis revealed a shift in the mean residual ellipticity and a significant reduction of the α-helix content in the variant D3 fragment. It is concluded that the Aα-chain substitution is mainly responsible for the coagulation abnormalities, whereas the substitution in the γ-chain induced a conformational change in the D3 fragment.
Introduction
Fibrinogen, a soluble plasma glycoprotein of 340 kd, is made up of 2 copies of 3 different polypeptide chains (Aα2, Bβ2,γ2), linked together with 29 interchain and intrachain disulfide bridges. The molecule is organized in a dimeric fashion consisting of a central E domain containing the amino termini of all 6 polypeptide chains and 2 outer D domains. In the final stage of blood coagulation, thrombin cleaves the fibrinopeptides A and B in a sequential manner from the amino termini of the Aα- and Bβ-chains. Cleavage occurs between residues R16 (single-letter amino acid abbreviations) and G17 of the Aα-chain and residues R14 and G15 of the Bβ-chain, exposing the A and B polymerization sites. Resultant monomers join together to form 2-stranded, half-staggered protofibrils. It has been shown that protofibrils result from longitudinal D-D interactions and noncovalent contacts between the A and the a sites. The a site is formed by residues 329, 330, 340, and 364 in the carboxy-terminal part of the γ-chain.1 2 Subsequently, the growing fibrils aggregate in a lateral fashion to form fibers that increase progressively in thickness and develop branch points. The evolving network is finally stabilized with covalent bonds by the activated factor XIII, resulting in a clot resistant to mechanical disruption.
Dysfibrinogenemia is a heritable disorder characterized by structural mutations in any of the 3 polypeptide chains of fibrinogen. A repertoire of 191 individual cases in which the structural defects have been elucidated are listed athttp://www.geht.org/pages/database_ang.html (accessed January 2001). Regarding hemostasis, most of the affected patients are asymptomatic, but some suffer from bleeding, thrombosis, or both.3 To date, only one compound heterozygote with 2 mutations in fibrinogen has been described.4 The patient had hypofibrinogenemia but had no polymerization defect.
We report a dysfunctional fibrinogen variant with 2 single heterozygous amino acid substitutions, one at position Aα R16C and the other at position γ G165R. The defect in the Aα-chain is located in the thrombin cleavage site and impairs the exposure of the A site. The amino acid substitution in the γ-chain is novel. Although it is sequentially and structurally remote from all known functional sites, we found that it affects the proper folding of the carboxy-terminal part of the γ-chain. From both in vivo and in vitro data, it is evident that these 2 amino acid substitutions result in a severe polymerization defect.
Materials and methods
Routine coagulation tests
Purification of fibrinogen
Fibrinogen Milano XII was isolated from citrated plasma of the propositus by affinity chromatography using fibrin-monomer-Sepharose CL-2B.7 Because the propositus is a heterozygous carrier of abnormal fibrinogen, the purified fibrinogen preparation contained normal and abnormal Aα- and γ-chains. Normal fibrinogen was isolated from a plasma pool of healthy donors. The purified protein was extensively dialyzed against 0.05 M triethanolamine-HCl, pH 7.4, 0.1 M NaCl (TEA-buffer).
Coagulation profiles of purified fibrinogen
Fibrin polymerization was evaluated turbidimetrically.8 Fibrinogen (540 μL, 0.55 mg/mL) was preincubated with 30 μL 20 mM Ca++ or 20 mM ethylenediaminetetraacetic acid (EDTA) (final concentration 1 mM) in polystyrene cuvettes for 5 minutes at 37°C. After the addition of 30 μL 10 U/mL bovine thrombin (final concentration, 0.5 U/mL; Diagnotec AG, Liestal, Switzerland), the increase in turbidity at 350 nm was measured in a spectrophotometer at 37°C. Each experiment was performed twice.
DNA analysis
Genomic DNA was isolated as previously described.9The entire coding region of all 3 fibrinogen genes was amplified by polymerase chain reaction (PCR), and the PCR products were purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA). Coding and complementary strands of the purified products were sequenced (Automated DNA Sequencing Facility, University of North Carolina at Chapel Hill) using the same primers as for PCR.
Kinetics of fibrinopeptide release
Fibrinogen solutions were diluted with TEA-buffer to a final concentration of 0.2 mg/mL. Human α-thrombin (Enzyme Research Laboratory, South Bend, IN) was added to a final concentration of 0.005 U/mL, and the individual reactions were stopped at designated time points by boiling the incubation mixtures for 15 minutes. To measure the total amount of fibrinopeptide A and B released at an infinity time point, the fibrinogen was incubated with 10 U/mL thrombin for 240 minutes. After centrifugation, the supernatants were immediately analyzed by reverse-phase high-performance liquid chromatography (HPLC) using the Shimadzu HPLC-System (Shimadzu, Columbia, MD) with a Discovery C18 250-mm, 5-μm column (Supelco, Bellefonte, PA). The column was equilibrated with buffer A (25 mM NaH2PO4/Na2HPO4, pH 6.0), and the autosampler loaded 200 μL each sample on the column. Peptides were eluted with a linear gradient from 15% to 36% buffer B (25 mM NaH2PO4/Na2HPO4, pH 6.0, with 50% acetonitrile) and monitored by absorbance reading at 210 nm. Fibrinopeptide peak areas from 2 experiments were determined by the accompanying software class 5.0 VP (Shimadzu). Fibrinopeptide release curves were constructed by plotting the percentage release, assuming the total FpA release from normal fibrinogen was 100%.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblot analysis of purified fibrinogen
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) of nonreduced normal and variant fibrinogen was performed on a 3% to 10% gradient slab polyacrylamide gel with a 3% stacking gel.10 Separated proteins were transferred onto nitrocellulose sheets and incubated overnight at room temperature with either a 1/2000 dilution of peroxidase-conjugated rabbit antibody to human fibrinogen (P0445; Dako A/S, Glostrup, Denmark) or a peroxidase-conjugated rabbit antibody to human serum albumin (P0356; Dako A/S). Bound antibodies were visualized by diaminobenzidine in the presence of H2O2.
To analyze the fibrinogens under reduced conditions, 10 μL reducing solution (10 mg dithiothreitol in 100 μL 0.05 M Tris pH 8.5, 8 M urea) was added to 50 μL fibrinogen (0.8 mg/mL). The mixture was incubated for 20 minutes at 60°C. SDS-PAGE was performed on an 8% slab gel with a 4.5% stacking gel10 and stained with Coomassie blue.
SDS-PAGE and immunoblot analysis of fibrinogen degradation products
Purified normal and variant fibrinogen were proteolytically degraded by the addition of plasminogen (purified from human plasma as described by Deutsch and Mertz11) and streptokinase (American Diagnostica, Greenwich, CT). Digestion was performed at 37°C in the presence of Ca++ or EDTA for 4 hours. The reaction was stopped by heating the samples for 5 minutes in the presence of SDS-containing sample buffer at 95°C. SDS-PAGE of fibrinogen degradation products was performed on 8% and 12% polyacrylamide gels10 under nonreducing and reducing conditions, respectively. Gels were loaded with 20 μg initial fibrinogen preparation and stained with Coomassie blue. The experiment was repeated in the presence of the peptide GPRP (0, 2, and 5 mM) with 1 mM EDTA added to all reactions.12 Immunoblot analysis was performed with nonreduced fibrinogen degradation products. Electrophoresed proteins were transferred onto nitrocellulose sheets and incubated at room temperature with the following antibodies: polyclonal rabbit anti–human fibrinogen (A0080; Dako); polyclonal rabbit anti–human fibrinogen γ-chain prepared by Hazelton Research Products (Denver, PA) using γ-chain purified from inclusion bodies expressed in Escherichia coli as the antigen13; monoclonal anti–human serum albumin (clone HAS-11, A6684; Sigma, St Louis, MO); monoclonal antibody E2F8E5 to fragment E, recognizing the sequence GHRPLDK (β 15-21) (Immunotech, Marseilles, France); peroxidase-conjugated anti–mouse or anti–rabbit IgG (Calbiochem, La Jolla, CA). Bound antibodies were visualized by enhanced chemiluminescence Western blotting detection reagents (Amersham Pharmacia Biotech, Piscataway, NJ). Human serum albumin (A9511; Sigma) was used as a control.
Purification of the fibrinogen degradation fragment D3
The fragment D3 was isolated as described earlier,14 with a slight modification. Briefly, 2 mL normal or variant fibrinogen (3 mg/mL) were proteolytically degraded for 4 hours at 37°C by the addition of 1 U/mL plasminogen (Kabi, Stockholm, Sweden) and 200 U/mL streptokinase (Behring, Marburg, Germany) in the presence 10 mM EDTA. The incubation mixture was dialyzed against the starting buffer (0.01 M TEA, 1 mM CaCl2, pH 7.4) and loaded on a Lysine-Sepharose 4B (Amersham Pharmacia Biotech) column (15 mL) equilibrated with starting buffer. The column was successively rinsed with 20 mL starting buffer, 20 mL elution buffer 1 (0.03 M TEA, 1 mM CaCl2, pH 7.4), and 20 mL elution buffer 2 (0.05 M TEA, 0.1 M NaCl, 1 mM CaCl2, pH 7.4). Fractions of 1 mL were collected and analyzed by SDS-PAGE on 8% polyacrylamide gels. Fragment D3–containing fractions were pooled and dialyzed either against TEA-buffer and frozen or against water and lyophilized.
Digestion of purified fragment D3 with chymotrypsin
Digestion was performed according to Medved et al.15 Lyophilized fragment D3 (500 μg) was dissolved in 100 μL 0.1 M phosphate buffer pH 7.0. α-Chymotrypsin (56 U/mg; type VII, TLCK-treated, bovine pancreas; Sigma) was added in an enzyme-substrate ratio of 1:50. The reaction was stopped at 0, 8, and 23 hours by boiling the samples. Fragments were separated by SDS-PAGE on 10% polyacrylamide gel10 and stained with Coomassie blue.
Circular dichroism analysis of purified fragment D3
Purified fragment D3 was dialyzed against 10 mM phosphate buffer pH 7.4. The concentration of the dialyzed samples was determined at 280 nm in the presence of 1 M urea (E = 20.0).15 Far UV-circular dichroism (CD) spectra from 260 to 190 nm were recorded on a Pistar-180 Circular Dichroism Spectrophotometer (Applied Photophysics, Surrey, United Kingdom). Before taking the CD spectra, the absorbance spectra of the samples were recorded from 320 to 180 nm to verify and adjust the protein concentration to an OD of approximately 0.8. Then a 0.07 mg/mL solution (400 μL) of purified fragment D3 was measured in a quartz cuvette with 0.1-cm path length at room temperature. The CD spectrum of the buffer solution was subtracted from each sample spectrum. CD data were converted to mean residual ellipticity, assuming a mean residue molecular weight of 115.16,17 Protein secondary structures were estimated from CD spectra using Continll18 and Selcon319 software from the CDPro software package (http://lamar.colostate. edu/∼sreeram/CDPro/).
Results
Case report and routine coagulation tests
The propositus of dysfibrinogen Milano XII was born in 1929 in Italy and had no reported history of bleeding or thrombosis. Unfortunately, no other family members were available for testing. Routine coagulation tests with plasma samples from the propositus revealed a significantly prolonged thrombin time; furthermore, her plasma was not clottable with reptilase (Table1). Immunologically determined fibrinogen levels (Laurell) were within normal range, but fibrinogen concentrations measured by a functional method (Clauss) were dramatically lower.
Results of routine coagulation tests in patient's plasma
| . | Milano XII propositus . | Normal range . |
|---|---|---|
| Thrombin time (s) | 106.9 | 11.5-15 |
| Reptilase time (s) | > 180 | 14-17 |
| Fibrinogen (Clauss) (g/L) | 0.14 | 1.5-3.0 |
| Fibrinogen (Laurell) (g/L) | 3.05 | 1.5-3.0 |
| . | Milano XII propositus . | Normal range . |
|---|---|---|
| Thrombin time (s) | 106.9 | 11.5-15 |
| Reptilase time (s) | > 180 | 14-17 |
| Fibrinogen (Clauss) (g/L) | 0.14 | 1.5-3.0 |
| Fibrinogen (Laurell) (g/L) | 3.05 | 1.5-3.0 |
Coagulation profiles of purified fibrinogen
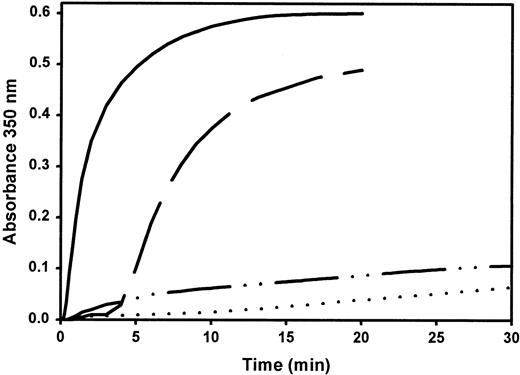
Turbidity curves (Figure 1) represent the kinetics of fibrin formation after the addition of thrombin to purified normal or variant fibrinogen in the presence of 1 mM calcium ions or 1 mM EDTA. In the presence of calcium, the variant showed a prolonged lag time and strongly reduced final turbidity compared to normal fibrinogen. In the presence of EDTA, only a minute turbidity increase occurred over the recording period.
Coagulation profile of purified fibrinogen.
Turbidity increase of normal fibrinogen in the presence of 1 mM Ca++ (——) or 1 mM EDTA (— —) and fibrinogen Milano XII with 1 mM Ca++ (– · · –) or 1 mM EDTA (· · · ·) was recorded at 350 nm after the addition of 0.5 U/mL bovine thrombin to 0.5 mg/mL fibrinogen at 37°C. Curves represent the average of 2 assays.
Coagulation profile of purified fibrinogen.
Turbidity increase of normal fibrinogen in the presence of 1 mM Ca++ (——) or 1 mM EDTA (— —) and fibrinogen Milano XII with 1 mM Ca++ (– · · –) or 1 mM EDTA (· · · ·) was recorded at 350 nm after the addition of 0.5 U/mL bovine thrombin to 0.5 mg/mL fibrinogen at 37°C. Curves represent the average of 2 assays.
DNA analysis
Sequence analysis of the entire coding region of the 3 fibrinogen genes revealed 2 point mutations (data not shown). The first mutation was found in exon 6 of the γ-chain, a substitution of base 4682 guanine to adenine, changing the amino acid glycine to arginine at position 165 of the γ-chain. The second mutation was detected in exon 2 of the Aα-chain. A single base change at position 1202 from cytosine to thymine led to the replacement Aα R16C.
Kinetics of fibrinopeptide release
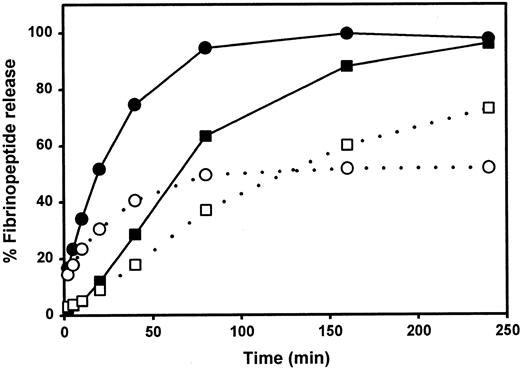
We examined the rate of thrombin-catalyzed fibrinopeptide release by measuring the peak areas of FpA and FpB as detected by reverse-phase HPLC. After a 240-minute incubation, only approximately 50% of the FpA was released from fibrinogen Milano XII (Figure2). Increasing the amount of thrombin, the time of incubation, or both did not change this percentage. The release of FpB from fibrinogen Milano XII was delayed but approached 100% with a higher thrombin concentration (data not shown).
Kinetics of fibrinopeptide release
. ▪, ●, normal fibrinogen; ■, ○, fibrinogen Milano XII; ○, ●, fibrinopeptides A; ■, ▪, fibrinopeptides B. Each curve represents the average of 2 experiments.
Kinetics of fibrinopeptide release
. ▪, ●, normal fibrinogen; ■, ○, fibrinogen Milano XII; ○, ●, fibrinopeptides A; ■, ▪, fibrinopeptides B. Each curve represents the average of 2 experiments.
SDS-PAGE and immunoblot analysis of fibrinogen
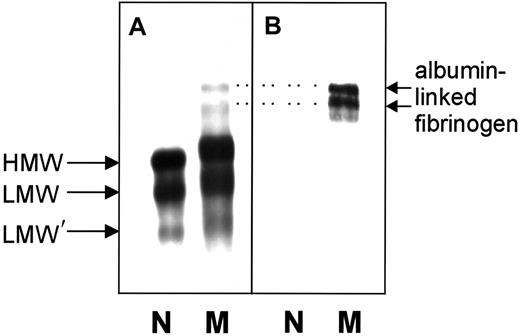
Nonreduced normal and variant fibrinogens were subjected to SDS-PAGE and immunoblot analysis. HMW fibrinogen (Mr340 000), LMW fibrinogen (Mr 305 000), and LMW′ fibrinogen (Mr 270 000) were observed in both fibrinogens (Figure 3A). With the variant fibrinogen, these 3 bands were much broader and consistently migrated more slowly than the comparable bands of normal fibrinogen. Two additional bands with higher molecular weights were detected in the variant sample. Because these 2 bands reacted with an antibody against human albumin, they likely represent HMW and LMW fibrinogen complexed with albumin (Figure 3B), similar to earlier reports for the fibrinogen variants IJmuiden (Bβ R14C) and Nijmegen (Bβ R44C).20SDS-PAGE of reduced samples revealed that the Aα-, Bβ-, and γ-chains from normal and variant fibrinogen were identical and stained with the same intensity. No abnormally migrating chains were visible (data not shown).
Immunoblots of unreduced normal fibrinogen and of fibrinogen Milano XII.
(A) Detection with anti–human fibrinogen antibodies. (B) Detection with anti–human serum albumin antibodies. N, normal fibrinogen; M, fibrinogen Milano XII; HMW, high-molecular-weight fibrinogen (340 kd); LMW, low-molecular-weight fibrinogen (305 kd); LMW′, low-molecular-weight fibrinogen (270 kd). Amount of fibrinogen loaded: panel A, 0.44 μg; panel B, 2.4 μg. Dotted lines connect the albumin-linked bands in panel B with their counterparts in panel A.
Immunoblots of unreduced normal fibrinogen and of fibrinogen Milano XII.
(A) Detection with anti–human fibrinogen antibodies. (B) Detection with anti–human serum albumin antibodies. N, normal fibrinogen; M, fibrinogen Milano XII; HMW, high-molecular-weight fibrinogen (340 kd); LMW, low-molecular-weight fibrinogen (305 kd); LMW′, low-molecular-weight fibrinogen (270 kd). Amount of fibrinogen loaded: panel A, 0.44 μg; panel B, 2.4 μg. Dotted lines connect the albumin-linked bands in panel B with their counterparts in panel A.
SDS-PAGE and immunoblot analysis of fibrinogen degradation products
It is known that the extent of plasmin degradation of fibrinogen is sensitive to calcium bound to the high-affinity site and to the peptide GPRP bound to the a polymerization site. Both sites are located in the D domain, and normal binding of Ca++ or the peptide GPRP protects the fibrinogenolysis fragment D1 against further degradation to D2 and D3. Plasmin digests of normal and Milano XII fibrinogens produced fragment D1 in the presence of calcium and fragment D3 in the presence of EDTA (Figure4A), indicating complete protection of the substrates by bound calcium. Under both conditions, the digests of fibrinogen Milano XII showed an additional band of apparently higher molecular weight, denoted D1* and D3* in Figure 4A. We compared the plasmin digestion of fibrinogen Milano XII to the digestion of fibrinogen St Gallen I (γ G292V),21 a variant that is abnormally digested to give the fragments D2 and D3 because of only partial protection of D1 by Ca++. As shown in Figure 4A, fragment D3* was not the same as fragment D2 and thus did not arise from the incomplete conversion of D1 in D3. When the plasmin digests were examined under reducing conditions, normal and variant chain remnants were indistinguishable (Figure 4B). This indicates that the unusual D1* and D3* fragments arose from disulfide-linked molecules. We also digested the normal and variant fibrinogens in the presence of 2 mM and 5 mM GPRP and saw complete protection in both samples (data not shown). These degradation patterns indicate that both the high-affinity calcium-binding site and the a polymerization site are normal in fibrinogen Milano XII.
SDS-PAGE of fibrinogenolysis fragments digested in the presence of 1 mM EDTA or 1 mM Ca++.
Normal fibrinogen (N), variant fibrinogens Milano XII (M) and St Gallen I (S). Lane S illustrates the size of the intermediate plasmin degradation fragment D2 (see “Results”). (A) Nonreduced samples. (B) Reduced samples. W, molecular weight standard. Fibrinogen (0.4 mg/mL) was digested with plasminogen (10 μg/mL) and streptokinase (40 U/mL) for 4 hours at 37°C.
SDS-PAGE of fibrinogenolysis fragments digested in the presence of 1 mM EDTA or 1 mM Ca++.
Normal fibrinogen (N), variant fibrinogens Milano XII (M) and St Gallen I (S). Lane S illustrates the size of the intermediate plasmin degradation fragment D2 (see “Results”). (A) Nonreduced samples. (B) Reduced samples. W, molecular weight standard. Fibrinogen (0.4 mg/mL) was digested with plasminogen (10 μg/mL) and streptokinase (40 U/mL) for 4 hours at 37°C.
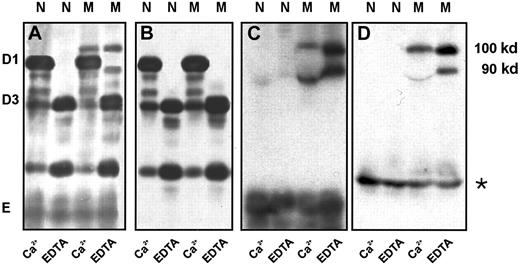
To identify the fragment of fibrinogen Milano XII that bound albumin, we performed immunoblot analysis of nonreduced plasmin digests comparing the patterns obtained with 4 antibodies: polyclonal antibodies against fibrinogen and γ-chain and monoclonal antibodies against β-chain 15-21 (E2F8E5) and human serum albumin (HAS-11). The antifibrinogen blot (Figure 5A) showed 2 bands of 90 kd and 100 kd in the fibrinogen Milano XII samples (lanes M) that were also faintly apparent in the Coomassie-stained gels (Figure 4A, EDTA, lane M), and it showed the bands D1* and D3*. The anti–γ-chain antibody reacted with D1* and D3* but not with the 90- and 100-kd bands or with fragment E (Figure 5B). Because the resolution in these blots was insufficient to separate D1* and D3* species, the bands are not distinct from D1 and D3, appearing rather as single broad bands. The 90-kd and 100-kd bands reacted with antifibrinogen, anti–β-chain and antialbumin antibodies (Figure 5A,C,D), indicating that albumin is linked to fragment E. As expected for fragment E, the same bands were seen in the presence of calcium or EDTA, though the 90-kd bands were distorted by the D1 fragment in the presence of calcium. We were surprised to find that E2F8E5 reacted with the plasmin degradation fragments because it has been reported that Bβ1-42 is removed by plasmin cleavage early in the reaction.22 In control experiments, we found that the E2F8E5 reaction depended on the reaction conditions. We saw only weak reactivity with the 90-kd, 100-kd, and normal E fragments with 10-fold higher concentrations of plasminogen and streptokinase, as expected for the subsequent removal of Bβ1-42 (data not shown). We also showed that E2F8E5 did not cross-react with human albumin (data not shown). Taken together, these results indicate that albumin is bound to the E-domain of fibrinogen Milano XII likely through the neo-cysteine at position 16 in the Aα-chain.
Immunoblot of the plasmic digest of normal fibrinogen and fibrinogen Milano XII obtained in the presence of 5 mM Ca++ or 5 mM EDTA.
Detection antibodies: polyclonal antifibrinogen antibody (A), polyclonal antibody to fibrinogen γ-chain (B), monoclonal antibody to β 15-21(C), monoclonal antibody to human serum albumin (D). Fibrinogen (1.5 mg/mL) was digested with plasminogen (10 μg/mL) and streptokinase (40 U/mL) for 4 hours at 37°C. Amount of fibrinogen loaded: 0.5 μg (A); 1 μg (B); 15 μg (C); 30 μg (D). * indicates bands that resulted from a slight albumin contamination of the purified fibrinogens; N, normal fibrinogen; M, fibrinogen Milano XII.
Immunoblot of the plasmic digest of normal fibrinogen and fibrinogen Milano XII obtained in the presence of 5 mM Ca++ or 5 mM EDTA.
Detection antibodies: polyclonal antifibrinogen antibody (A), polyclonal antibody to fibrinogen γ-chain (B), monoclonal antibody to β 15-21(C), monoclonal antibody to human serum albumin (D). Fibrinogen (1.5 mg/mL) was digested with plasminogen (10 μg/mL) and streptokinase (40 U/mL) for 4 hours at 37°C. Amount of fibrinogen loaded: 0.5 μg (A); 1 μg (B); 15 μg (C); 30 μg (D). * indicates bands that resulted from a slight albumin contamination of the purified fibrinogens; N, normal fibrinogen; M, fibrinogen Milano XII.
Purification of the fibrinogen degradation fragment D3
Using a Lysine Sepharose 4B column and a strategy of different buffers with increasing salt concentration, fragment D3 was eluted in an early peak and was separated from fragment E and plasmin. The D3 and D3* fraction from fibrinogen Milano XII eluted concomitantly and could not be separated from each other (data not shown).
Digestion of purified fragment D3 with chymotrypsin
Fragment D3 (Mr 82 kd) can be further digested by chymotrypsin to a fragment called TSD (thermodynamic stable domain).22 In bovine fibrinogen, a chymotrypsin cleavage site had been identified between γ 151 and γ 152 leading to a digestion fragment of 63 kd. Because of the similarity of bovine and human fibrinogen,23 it is likely that the cleavage site for chymotrypsin in human fibrinogen may be situated close to that found in bovine fibrinogen. Incubation of normal and variant fragment D3 with chymotrypsin led to a 63-kd fragment, and the double band associated with fibrinogen Milano XII disappeared (data not shown). Therefore, removal of the mutation site at position 165 led to a chymotryptic fragment of the same size as the fragment derived from normal fibrinogen.
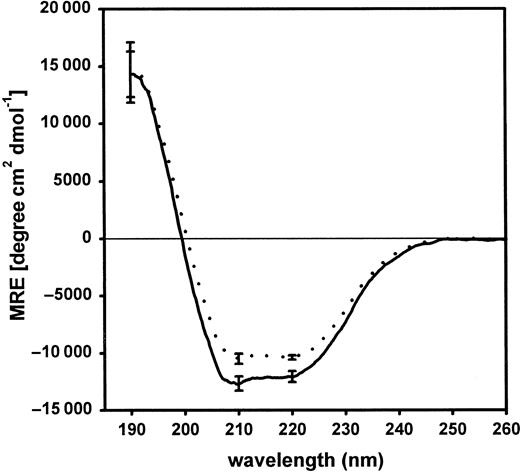
Far UV-CD analysis of purified fragment D3
Figure 6 shows far UV-CD spectra of purified fragments D3 of normal and variant fibrinogen. A slight reduction of the mean residual ellipticity was calculated with fragment D3 derived from fibrinogen Milano XII. The curve representing the normal fragment D3 showed a minimum at 210 nm reflecting the α-helical structure in the protein.24 25 This feature was less prominent in the CD-spectra of variant fragment D3. The α-helical content in the variant fragment D3 (28.2% ± 0.9%) was significantly different from normal D3 (31.8% ± 0.7%;P = .005).
Far UV-CD spectra of purified fragments D3 of normal and variant fibrinogen.
Each curve represents the mean of 3 individual spectra. Each of these 3 spectra is the result of an individual plasmin digestion and subsequent purification of fragment D3. Normal fibrinogen, ——; variant, · · · ·. The standard deviation is given at selected wavelengths.
Far UV-CD spectra of purified fragments D3 of normal and variant fibrinogen.
Each curve represents the mean of 3 individual spectra. Each of these 3 spectra is the result of an individual plasmin digestion and subsequent purification of fragment D3. Normal fibrinogen, ——; variant, · · · ·. The standard deviation is given at selected wavelengths.
Discussion
We describe a dysfunctional fibrinogen variant with 2 mutations, one in the Aα-chain at position 16 (R16C) and the other in the γ-chain at position 165 (G165R). This dysfibrinogen was discovered in a woman of Italian origin, born in 1929. Routine coagulation studies led to a diagnosis of dysfibrinogenemia. Unfortunately, no other family members were available to study the transmission of the mutations. Both defects were present in the fibrinogen that circulated at a normal level in the plasma of the propositus. The mutation Aα R16C inhibited the release of fibrinopeptide A by thrombin. HPLC analysis of fibrinopeptide release revealed that only 50% of fibrinopeptide A was cleaved from the purified fibrinogen Milano XII (Figure 2). This result confirmed the heterozygosity found in DNA analysis. We found that small amounts of abnormal fibrinogen circulate as disulfide-linked complexes with albumin. The disulfide bonds likely form between the neo-cysteine introduced by the mutation Aα R16C and the free sulfhydryl group (C34) present in albumin.26 Covalent binding of albumin to Aα C16 was previously described in fibrinogen Bern V containing the same mutation (Aα R16C).27
The mutation in the γ-chain (G165R) caused abnormal electrophoretic migration on SDS-PAGE of the unreduced variant fibrinogen and of some of its nonreduced degradation fragments. The relative migration rate of a protein in a polyacrylamide gel containing SDS depends on its molecular size, shape,28 and binding affinity for SDS.29 The latter factor was proposed to be responsible for shifts in electrophoretic mobility of the fibrinogen γ-chain that have been observed in several fibrinogen mutants30-35because of the introduction of a more basic or more hydrophobic amino acid.36 Fibrinogen Milano XII showed abnormal migration under nonreducing conditions but normal migration under reducing conditions. We hypothesize that the amino acid exchange from glycine to arginine at position 165 affects the folding pattern of the carboxy-terminal globular domain of the γ-chain. The abnormal shape of the fragment, held together by disulfide bridges, may thus affect the electrophoretic migration of nonreduced fragments but is compatible with the normalization of electrophoretic behavior after the reduction of disulfide bonds. To investigate tentative structural differences, we analyzed purified D3 fragments derived from normal fibrinogen and fibrinogen Milano XII, the latter a mixture of D3 and D3*, using CD spectroscopy. CD analysis has been applied to study the conformational differences among fibrinogen, fragment D, and fragment E16and between the thermodynamically stable domains 1 and 2 of bovine fibrinogen.22 It has also been used to estimate the conformational changes in human fibrinogen after different in vitro phosphorylation procedures.37 Kirschbaum et al17 used this method to visualize the difference between urea-denatured and native D fragments in a platelet-binding study. Furthermore, CD spectroscopy has been a useful tool for detecting structural changes caused by single amino acid substitutions in different enzymes.38 39 We observed a reduction in the mean residual ellipticity (Figure 6) and a significant decrease in the α-helix content between normal and variant fragment D3. These results support our suggestion of a conformational abnormality in the D-domain of fibrinogen Milano XII.
The amino acid substitution Aα R16C is common and accounts for 18% of all reported dysfunctional fibrinogen variants. This mutation leads to abnormal coagulation test results in plasma and to impaired polymerization of purified fibrinogen. Delayed clotting of fibrinogen Milano XII is in agreement with reports on functional behavior of other Aα R16C variants.27 40 Because the polymerization defect attributed to the Aα-chain mutation is so dramatic, it may obscure any additional disturbance of fibrin clot formation resulting from the γ G165R mutation. Our plasmin digestion experiments in the presence of GPRP suggest that the a sites in the carboxy-terminal domains of fibrinogen Milano XII are intact. Therefore, if the mutation in the γ-chain has an effect on polymerization, it is unlikely to have been caused by impaired A-a interactions.
Fibrinogen Milano XII is the first dysfunctional fibrinogen variant with 2 amino acid substitutions associated with a strong polymerization defect. Brennan et al4 reported earlier a fibrinogen variant with 2 mutations in the coding region of fibrinogen. Both structural defects were detectable in the circulating fibrinogen but did not affect fibrin polymerization. Neither the substitution γ G165R nor the unusual migration of plasmic fragments D on SDS-PAGE has previously been reported. The mutation is located in a loop of the γ-chain formed by a cysteine bridge between residues 153 and 182 in the globular domain of the γ-chain (Figure7). It affects the structure of the D-domain and may contribute to the observed polymerization defect. We intend to study the functional properties of the homozygous γ G165R substitution using a recombinant fibrinogen variant with the same mutation.
Position of the amino acid substitution in the γ-chain of fibrinogen Milano XII.
The mutation site (γ G165R) is situated in a loop formed by an intrachain disulfide bridge between γ 153 C and γ 182 C. The figure shows the γ-chain of fibrinogen and is adapted from the crystal structure (IFZB from the Brookhaven Data Bank) of fragment D reported by Spraggon et al.1
Position of the amino acid substitution in the γ-chain of fibrinogen Milano XII.
The mutation site (γ G165R) is situated in a loop formed by an intrachain disulfide bridge between γ 153 C and γ 182 C. The figure shows the γ-chain of fibrinogen and is adapted from the crystal structure (IFZB from the Brookhaven Data Bank) of fragment D reported by Spraggon et al.1
We thank Drs F. Baudo and R. Redaelli (Ospedale Niguarda, Milan, Italy) for providing the patient's plasma; Dr A. Tripathy (University of North Carolina, Chapel Hill) for help with CD analysis; and Dr Kelly A. Hogan for critical review of the manuscript.
Supported by grants from the Swiss National Science Foundation (32-47033.96) and the National Institutes of Health (R01 HL 31048).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Bettina Bolliger-Stucki, Department of Pathology and Laboratory Medicine, University of North Carolina at Chapel Hill, CB #7525, 605 Brinkhous-Bullitt Bldg, Chapel Hill, NC 27599-7525; e-mail: bettina_bolliger@med.unc.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal