The human leukemia cell lines K562, CEM, CEM/VLB100, human leukemic blasts, and the bladder cancer J82 cell line have different sensitivities to UV light–induced apoptosis. It is reported that resistance to UV light–induced apoptosis occurs at a point in the apoptotic pathway upstream of caspase-3 but downstream of mitochondrial cytochrome c release. It is demonstrated that the block is due to deficiency of Apaf-1, a critical member of the apoptosome. Sensitivity to apoptosis was independent of caspase-9b or XIAP (inhibitors of apoptosis proteins) expression or levels of procaspase-9. Transfection of Apaf-1 conferred sensitivity to apoptosis in resistant cells. Apaf-1 deficiency may constitute a significant mode of resistance to apoptosis in human leukemia.
Introduction
The apoptosome, which includes the protein Apaf-1, cytochrome c/dATP, and procaspase-9, plays a central role in the apoptotic process.1-8 Cytochrome c is released from mitochondria into the cytosol after the induction of apoptosis by many different stimuli, including CD95, tumor necrosis factor-α, UV irradiation, and chemotherapeutic and DNA-damaging agents.1,9,10 Released cytochrome c associates with Apaf-1 in the presence of dATP or ATP and induces the oligomerization of Apaf-1.4,7,11,12 This recognizes inactive procaspase-9 and forms the apoptosome, triggering autocatalytic processing of procaspase-9.4,6,13,14Activated caspase-9 then processes effector caspases (caspase-3, -6, and -7), which in turn cause cell collapse by cleaving a specific set of substrates. By contrast, granzyme B-induced processing of procaspase-3 does not rely on the apoptosome.15
Cells from Apaf-1 or procaspase-9 knock-out mice are resistant to several apoptotic stimuli.8,16-19 Deletions of Apaf-1 or caspase-9 in ras- and myc-transformed murine cells can replicate the tumorigenic effects of p53 deletion.8 Oncogene (such as E1A)-transformed cells have increased levels of Apaf-1 and procaspase-9 protein expression and are sensitized to etoposide-induced apoptosis.5 Inactivation of Apaf-1 and caspase-9 substantially reduces the number of cells required to form tumors.8 Transfection of theApaf-1 gene into leukemic cells increases the sensitivity of cells to etoposide-induced apoptosis.20
An endogenous, alternatively spliced isoform of caspase-9 (named caspase-9b), which lacks the central large subunit caspase domain, can interact with the caspase recruitment domain of Apaf-1 and inhibit the apoptotic process.21 Even in the presence of functional Apaf-1 and procaspase-9, inhibitors of apoptosis, such as XIAP (inhibitors of apoptosis proteins), can prevent the proteolytic processing of procaspase-3 by blocking the cytochromec–induced activation of procaspase-9.22IAPs, in turn, can be regulated by the mitochondrial protein, Smac.23 24 These data provide strong evidence that appropriate and functional levels of Apaf-1 or procaspase-9 proteins are crucial for the inhibition of tumor progression and for maintaining the sensitivity of tumor cells to apoptosis. However, it is unknown whether variations in constitutive levels of Apaf-1 have implications for human leukemia.
Human leukemic cells differ widely in their susceptibility to apoptosis. This is exemplified by the human leukemic cell lines, K562, CEM, and CEM/VLB100, which have different sensitivities to tumor necrosis factor (TNF)-α–induced apoptosis. However, we found the block conferring resistance to TNF-α–induced apoptosis is at the mitochondrial level,10 25-27 whereas the block to UV light–induced apoptosis appeared to be downstream of the mitochondria. Using UV light, which can pass through the mitochondrial barrier, as a stimulus, we demonstrated that cytochrome c release from mitochondria occurred in all tumor cells studied and that the differential susceptibility to cytochrome c–dependent apoptosis was associated with the protein level of Apaf-1 in these cells. We report for the first time that constitutive variations in relative levels of the Apaf-1 protein in human leukemic cells can determine sensitivity to apoptosis downstream of mitochondrial involvement.
Materials and methods
Cell lines, acute myeloid leukemia blasts, andApaf-1 gene transfection
Human myeloid leukemic K562 cell line, T-lymphoblastic cell line CEM and its vinblastine-resistant subclone CEM/VLB100, and human bladder cancer J82 cell line were used in this study. The sensitivity of the CEM and CEM/VLB100 cell lines differed by approximately a factor of 3 in their sensitivity to TNF-α–induced apoptosis.26 27 Normal human CD34+ cells were obtained by immunomagnetic bead separation (Minimacs; Miltenyi Biotech, Sunnyvale, CA) after leukopheresis of granulocyte–colony-stimulating factor–treated donors. Human acute myeloid leukemia (AML) blasts were obtained from patients with AML and were separated over a Ficoll-Hypaque gradient (Amersham Pharmacia Biotech, Uppsala, Sweden). The percentage of blasts in each sample was greater than 80% by morphologic assessment.
Cell lines were cultured in RPMI-1640 medium as described previously.25 Flag-tagged Apaf-1L (GenBank accession no.AF134397) pCMV2 plasmid4 14 was grown in Escherichia coli DH5α strain and was purified using Plasmid DNA MiniPrep Kit (QIAGEN, Valencia, CA). For leukemic cells, 4 μg plasmid DNA was transiently transfected into 2 × 106 leukemic cells using DMRIE-C reagent (Gibco BRL, West Sussex, United Kingdom) in serum-free OPTI-MEM medium (Gibco BRL). For the adherent J82 cells, 2 μg plasmid DNA was transfected using Lipofectamine Plus reagent (Gibco BRL). Transfection efficiency was 80% to 100%, as measured by the cotransfection of 2 μg pRSC-GFP plasmid and as observed by fluorescence microscopy. Cell viability was 95% to 100% after transfection, examined by trypan blue staining. Cells were harvested after 24 hours of transfection.
Determination of DNA damage–alkaline unwinding assay
The extent of bulk DNA breakage was assessed by the enhanced fluorescence alkaline unwinding method. The assay is based on the differential binding and fluorescence of the indicator, Hoechst 33258 (Calbiochem, Nottingham, United Kingdom) to single-stranded and double-stranded DNA after a fixed period of alkaline denaturation. DNA fluorescence was determined by a TD-700 fluorometer (Turner Designs, Sunnyvale, CA) with excitation at 350 nm and emission detection at 450 nm. The degree of damaged DNA was expressed by the reduction in the ratio of duplex DNA to total DNA.28
Western blotting
Proteins were subjected to standard SDS-PAGE at 20 to 40 mA/gel and were transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hertfordshire, United Kingdom) for 1 to 2 hours at 100 V after they were probed for various proteins using monoclonal or polyclonal antibodies (as described individually in the figure legends). Bound antibodies were detected using appropriate horseradish peroxidase–conjugated secondary antibodies, followed by detection using SuperSignal enhanced chemiluminescence (ECL; Pierce, Rockford, IL). The density of each band was analyzed using an AlphaImager 2000 Densitometer (Alpha Innotech, San Jose, CA).
Measurement of apoptosis by flow cytometry
To induce apoptosis in intact cells, leukemic cells (5 × 105/mL) were exposed to UV irradiation (120 mJ/cm2) (model TM-20; Chromato-UV-E Transilluminator) for 2 minutes and then further cultured for up to 12 hours. Irradiation was performed with a γ-ray source (cesium Cs 137; Gamma Cell 1000, Atomic Energy of Canada, Ontario) at a dose of 100 Gy. Cells were permeabilized with 70% ethanol, stained with 100 μg/mL propidium iodide (Sigma), and measured by flow cytometry (FACScan, Becton Dickinson).26
Preparation of subcellular fractions
Cells were suspended in 1 mL Buffer A (250 mM sucrose, 10 mM HEPES-KOH, pH 7.4, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 20 μM cytochalasin B) and incubated for 20 minutes on ice. Cells were then broken with a glass Dounce homogenizer (Jencons, Leighton Buzzard, United Kingdom). Mitochondrial purification was described previously.10 S-100 fraction was obtained by ultracentrifuge cytosol at 100 000g for 1 hour.
Measurement of activity of caspase-3 and caspase-9
Fifteen microliters S-100 (50 μg protein) was diluted to 95 μL with Buffer A. The reaction was initiated by the addition of 5 μL 400 μM (final concentration was 20 μM) fluorescent substrates, Ac-Leu-Glu-His-Asp-AFC (Ac-LEHD-AFC; Calbiochem) for caspase-9 or Ac-Asp-Glu-Val-Asp-AMC (Ac-DEVD-AMC; Calbiochem) for caspase-3. After incubation at 30°C for 15 minutes, the reaction was stopped by the addition of 50 μL 1% sodium acetate trihydrate in 175 mM acetic acid. Fluorescence at 400/505 nm for caspase-9 or at 380/460 nm for caspase-3 was measured with a TD-700 fluorometer. Measurements were calibrated against a standard linear regression curve of AFC or AMC. Caspase activity was defined as μM AFC or AMC release per milligram protein per hour (μM/h per milligram protein).
Coupled in vitro transcription/translation
[35S] Methionine-labeled caspase-9 was in vitro transcribed and translated using the T7 polymerase TNT kit (Promega, Southampton, United Kingdom). One microgram pRSC-LacZ-caspase-9 plasmid4 was used in a 50 μL transcription/translation reaction mixture containing 1 μL [35S] methionine. Translation was completed at 30°C for 2 hours.
Immunodepletion of Apaf-1
One microliter monoclonal anti–Apaf-1 antibody was added to 100 μL S-100 (300 μg protein) and incubated for 2 hours at 4°C on a rotor. Thirty microliters protein G Sepharose (Pharmacia Biotech, Amersham, United Kingdom) was then added into S-100 to bind Apaf-1 and antibody complex, and the incubation was continued for 2 hours. After spinning down, the supernatant was used for the cell-free reactions.
Cell-free reactions
Cell-free reactions were set up in 20 μL reaction volumes. For cytochrome c/dATP–induced activation of caspases, 50 μg S-100 in Buffer A was incubated with or without bovine heart cytochrome c (50 ng)/dATP (10 nm) at 30°C up to 1 hour. To detect cytochrome c/dATP–induced activation of exogenous procaspase-9, 0.5 μL [35S]-labeled procaspase-9 was added to the reaction system before cytochromec/dATP. The protein was subjected to 10% SDS-PAGE. The gel was dried and exposed to x-ray film. For granzyme B–induced caspase-3 and caspase-9 activation, 2 U granzyme B (Calbiochem) was added to each reaction and incubated at 30°C for 1 hour. Activities of caspase-3 and -9 were measured by fluorogenic assay as described above. Cleavage of the caspase proteins was confirmed by Western blot analysis. Monoclonal anticaspase-9 antibody5 was used at a 1:1000 dilution for the detection of the procaspase-9 cleaved into 37-kd and 35-kd fragments. Monoclonal anti–caspase-3 antibody (Transduction Laboratories, Lexington, KY) was used in 1:1000 dilution to detect the cleavage of procaspase-3. A reduction in density or disappearance of procaspase-3 bands defined the cleavage of caspase-3. Monoclonal anti–β-actin antibody (Sigma) (1:10 000) was used as a control protein.
RT-PCR detection for caspase-9b/procasapse-9
Total RNA was extracted with the RNeasy kit (QIAGEN). Two micrograms RNA was used for reverse transcription, which was performed using Retroscript first-strand synthesis kit for RT-PCR (Ambion, TX). Reverse-transcribed products were amplified by PCR using 2 caspase-9–specific primers, Mch6-ATG (ATGGACGAAGCGGATCGG) and Mch6-TAA (CCCTGGCCTTATGATGTT). Samples were subjected to an initial 5-minute denaturation at 95°C, followed by 35 cycles of denaturation at 94°C for 45 seconds, annealing at 56°C for 45 seconds, and extension at 72°C for 1.5 minutes. Additional extension was at 72°C for 10 minutes. Twenty-five cycles were previously determined to be on the linear part of the curve for caspase-9b cDNA amplification under these conditions (data not shown); 18S served as the housekeeper gene. Mch6 (procaspase-9)-pRSC-LacZ and Mch6b (caspase-9b)-pcDNA3 plasmids served as positive controls for procaspase-9 and caspase-9b.21
Results
Sensitivity of leukemic cells to UV light–induced apoptosis is not correlated with DNA damage or cytochrome c release
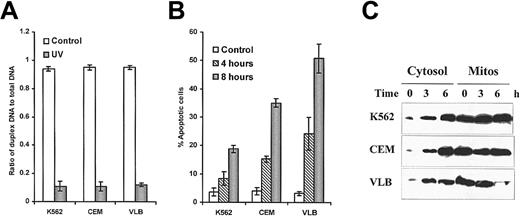
UV light–induced DNA damage after 1 hour was assessed by the alkaline unwinding assay.28 There was no difference in the sensitivity of these cell lines to UV light–induced double-strand breaks (Figure 1A). Leukemic cells started to undergo apoptosis after 4 hours of exposure to UV light. As shown in Figure 1B, leukemic cells had different susceptibilities to UV light–induced apoptosis. The K562 cell line was remarkably resistant to UV light–induced apoptosis. However, the CEM/VLB100cell line showed greater sensitivity to UV light–induced apoptosis than either K562 or its parental CEM cells. The sensitivity of leukemic cells to UV light–induced apoptosis did not correlate with the degree of DNA double-strand breakage (correlation, P > .5).
UV light–induced DNA damage, apoptosis, and cytochrome
c release. (A) UV light–induced DNA damage. DNA double-strand breaks were assessed by alkaline unwinding assay. (B) UV light–induced apoptosis was assessed by flow cytometry. (C) Detection of cytochrome c release by Western blotting. 50 μg cytosolic protein or 25 μg mitochondrial protein was analyzed by immunoblotting with denatured anticytochrome c antibody (7H8.2C12) at 1:1000 dilution.
UV light–induced DNA damage, apoptosis, and cytochrome
c release. (A) UV light–induced DNA damage. DNA double-strand breaks were assessed by alkaline unwinding assay. (B) UV light–induced apoptosis was assessed by flow cytometry. (C) Detection of cytochrome c release by Western blotting. 50 μg cytosolic protein or 25 μg mitochondrial protein was analyzed by immunoblotting with denatured anticytochrome c antibody (7H8.2C12) at 1:1000 dilution.
Cytochrome c translocation from mitochondria to cytosol was convincingly demonstrated in all cell lines after UV light stimulation for 3 hours, as detected by Western blot analysis (Figure 1C) and confirmed by fluorescence microscopy (10 and data not shown). Again, however, we observed that the amount of released cytochrome c in each cell line did not correlate with the extent of apoptosis in response to UV light. These results suggest that the resistance to UV light–induced apoptosis does not occur at the point of release of cytochrome c.
Block conferring resistance to UV light–induced apoptosis is upstream of caspase-3 activation
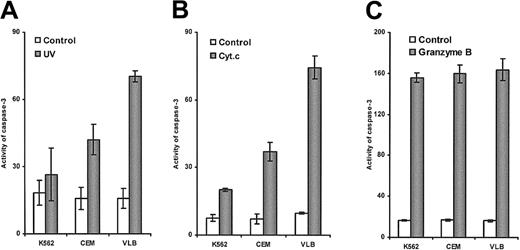
Differential sensitivity of leukemic cells to UV light–induced caspase-3 activation was shown by fluorescent AMC release (Figure2A). The CEM/VLB100 cell line showed significantly increased susceptibility to UV light–induced caspase-3 activation compared with the parental CEM cell line. The K562 cell line was remarkably resistant to UV light–induced caspase-3 activation. The degree of UV light–induced apoptosis was proportional to the activity of caspase-3 (P < .001), suggesting that UV light–induced apoptosis in these cell lines was associated with caspase-3 activity. This result also suggests that the block conferring the resistance is upstream of caspase-3 activation.
Caspase-3 activation induced by UV light, cytochrome
c, and granzyme B. (A) UV light–induced caspase-3 activation. S-100 was extracted from cells after exposure to UV light for 3 hours. (B) Cytochrome c–induced casapase-3 activation. S-100 from the 3 cell lines was incubated with cytochromec and dATP for 1 hour. (C) Granzyme B–induced caspase-3 activation. S-100 from the 3 cell lines was incubated with 2 U granzyme B for 1 hour.
Caspase-3 activation induced by UV light, cytochrome
c, and granzyme B. (A) UV light–induced caspase-3 activation. S-100 was extracted from cells after exposure to UV light for 3 hours. (B) Cytochrome c–induced casapase-3 activation. S-100 from the 3 cell lines was incubated with cytochromec and dATP for 1 hour. (C) Granzyme B–induced caspase-3 activation. S-100 from the 3 cell lines was incubated with 2 U granzyme B for 1 hour.
Leukemic cells displayed differential sensitivity to exogenous cytochrome c but not granzyme B
We were, therefore, interested in whether the cytosol from the different cell lines had a similar response to exogenous cytochromec because they expressed similar levels of caspase-3 protein (data not shown). Caspase-3 activation in cytosolic extraction from K562, CEM, and CEM/VLB100 cells in response to exogenous cytochrome c was almost identical to that induced by UV light stimulation (P < .0001) (Figure2A,B). However, caspase-3 activation in the 3 cell lines showed similar sensitivity to granzyme B–induced activation (Figure 2C), and there was no correlation with cytochromec–induced caspase-3 activation (P > .5). The results strongly suggest that the susceptibility of leukemic cells to apoptosis is cytochrome c–dependent and is delayed or blocked downstream of cytochrome c release and upstream of caspase-3 in these apoptosis-resistant cell lines.
Resistance to cytochrome c-induced caspase-3 activation is due to reduced Apaf-1 protein expression in resistant leukemic cells
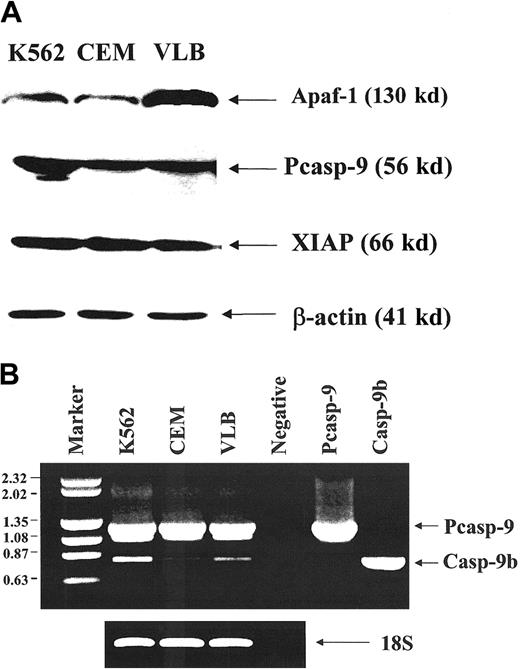
The K562 and CEM cell lines expressed less Apaf-1 than the CEM/VLB100 cell line (Figure3A) as analyzed by densitometry. CEM and CEM/VLB100 cells expressed similar levels of procaspase-9; however, the K562 cell line, which is most resistant to cytochromec, expressed more procaspase-9 than the CEM and CEM/VLB100 cell lines. There was no difference in XIAP expression among the 3 cell lines. Caspase-9b mRNA expression was assessed by semiquantitative RT-PCR (Figure 3B). Although the K562 cell line expressed more caspase-9b than the CEM and CEM/VLB100cell lines, the CEM cell line expressed less caspase-9b than the CEM/VLB100 cell line. This suggests that overexpression of the potential inhibitory caspase-9b was not associated with sensitivity to cytochrome c–induced apoptosis.
Expression of Apaf-1, procaspase-9, and XIAP in leukemic cell lines.
(A) Western blot analysis of the expression of Apaf-1, procaspase-9, and XIAP protein. Two hundred micrograms of protein was used for Apaf-1 expression. Monoclonal anti–Apaf-1 antibody5 was used at 1:1000 dilution. The ratio K562:CEM:CEM/VLB100 for Apaf-1 expression was 1:1:3. 50 μg protein was used for procaspase-9 expression. Monoclonal anti–caspase-9 antibody5 was used at 1:1000 dilution. The ratio K562:CEM:CEM/VLB100 for procaspase-9 expression was 2:1:1. 25 μg protein was used for XIAP expression and detected by polyclonal anti-XIAP antibody and was used at 1:1000 dilution. The ratio K562:CEM:CEM/VLB100 for XIAP expression was 1:1.2:1. (B) RT-PCR analysis of the expression of procaspase-9 and caspase-9b. (left) DNA size markers (kb); right, 2 caspase-9 isoforms (procaspase-9 1.2 kb; caspase-9b, 0.8 kb).
Expression of Apaf-1, procaspase-9, and XIAP in leukemic cell lines.
(A) Western blot analysis of the expression of Apaf-1, procaspase-9, and XIAP protein. Two hundred micrograms of protein was used for Apaf-1 expression. Monoclonal anti–Apaf-1 antibody5 was used at 1:1000 dilution. The ratio K562:CEM:CEM/VLB100 for Apaf-1 expression was 1:1:3. 50 μg protein was used for procaspase-9 expression. Monoclonal anti–caspase-9 antibody5 was used at 1:1000 dilution. The ratio K562:CEM:CEM/VLB100 for procaspase-9 expression was 2:1:1. 25 μg protein was used for XIAP expression and detected by polyclonal anti-XIAP antibody and was used at 1:1000 dilution. The ratio K562:CEM:CEM/VLB100 for XIAP expression was 1:1.2:1. (B) RT-PCR analysis of the expression of procaspase-9 and caspase-9b. (left) DNA size markers (kb); right, 2 caspase-9 isoforms (procaspase-9 1.2 kb; caspase-9b, 0.8 kb).
In vitro translated procaspase-9 showed different sensitivity to cytochrome c–induced cleavage in the presence of S-100 extracted from the 3 cell lines (Figure4A). Cytochrome c–induced cleavage of procaspase-9, in the presence of cytosol from the CEM/VLB100 cell line, was observed after 5 minutes but was less evident in the CEM and K562 cell lines. At 1 hour, complete degradation of exogenous procaspase-9 was observed in the CEM/VLB100 cells. This is further confirmation that the sensitivity of these cell lines to cytochrome c was not associated with endogenous procaspase-9 expression.
Effect of Apaf-1 protein levels on cytochrome
c–mediated cleavage of exogenous procaspase-9.(A) Cytochrome c–induced procaspase-9 cleavage in the presence of S-100 from different cell lines. In vitro translated [35S] procaspase-9 cleavage was initiated by the addition of cytochrome c and dATP in the presence of S-100 from cell lines. (B) Cytochrome c–mediated procaspase-9 cleavage in the absence of Apaf-1. The S-100 in which Apaf-1 was depleted has been indicated as ID. Failure to cleave procaspase-9 by cytochromec is shown by the absence of the cleavage bands (P35 and P37) compared with untreated S-100. (C) Purified Apaf-1 protein enhanced cytochrome c–induced procaspase-9 activation in K562 and CEM cell lines. 1 μL Apaf-1 protein was added to the S-100 (50 μg protein) lysates from K562 and CEM cells. Cytochromec–mediated procaspase-9 cleavage was allowed to proceed for 1 hour. Enhanced procaspase-9 cleavage is shown as disappearance of the procaspase-9 and P37 bands.
Effect of Apaf-1 protein levels on cytochrome
c–mediated cleavage of exogenous procaspase-9.(A) Cytochrome c–induced procaspase-9 cleavage in the presence of S-100 from different cell lines. In vitro translated [35S] procaspase-9 cleavage was initiated by the addition of cytochrome c and dATP in the presence of S-100 from cell lines. (B) Cytochrome c–mediated procaspase-9 cleavage in the absence of Apaf-1. The S-100 in which Apaf-1 was depleted has been indicated as ID. Failure to cleave procaspase-9 by cytochromec is shown by the absence of the cleavage bands (P35 and P37) compared with untreated S-100. (C) Purified Apaf-1 protein enhanced cytochrome c–induced procaspase-9 activation in K562 and CEM cell lines. 1 μL Apaf-1 protein was added to the S-100 (50 μg protein) lysates from K562 and CEM cells. Cytochromec–mediated procaspase-9 cleavage was allowed to proceed for 1 hour. Enhanced procaspase-9 cleavage is shown as disappearance of the procaspase-9 and P37 bands.
After immunodepletion of Apaf-1 protein from S-100, the effect of cytochrome c–induced exogenous procaspase-9 cleavage completely vanished (Figure 4B). In contrast, the addition of purified Apaf-1 protein14 to K562 and CEM cytosol restored sensitivity to cytochrome c–mediated procaspase-9 activation (Figure 4C). These results demonstrate that the resistance of K562 and CEM cell lines to cytochrome c–dependent activation of procaspases was due to insufficient Apaf-1 protein levels in their cytosol.
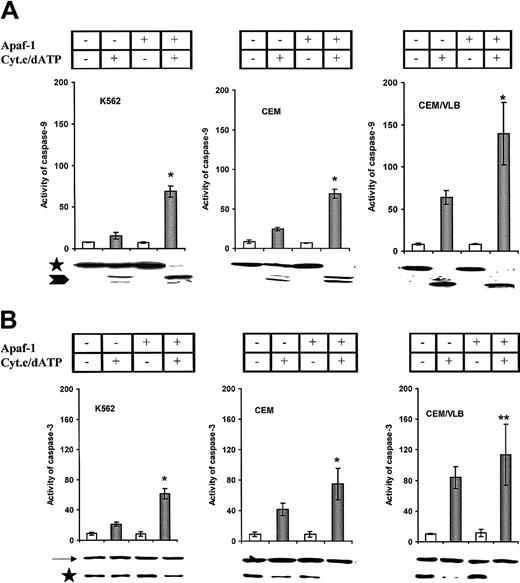
Apaf-1 gene transfection increased the sensitivity of leukemic cells to cytochrome c–mediated activation of procaspases and apoptosis
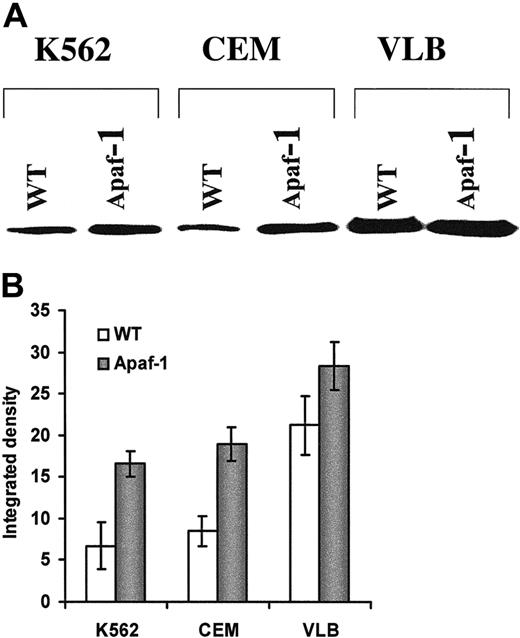
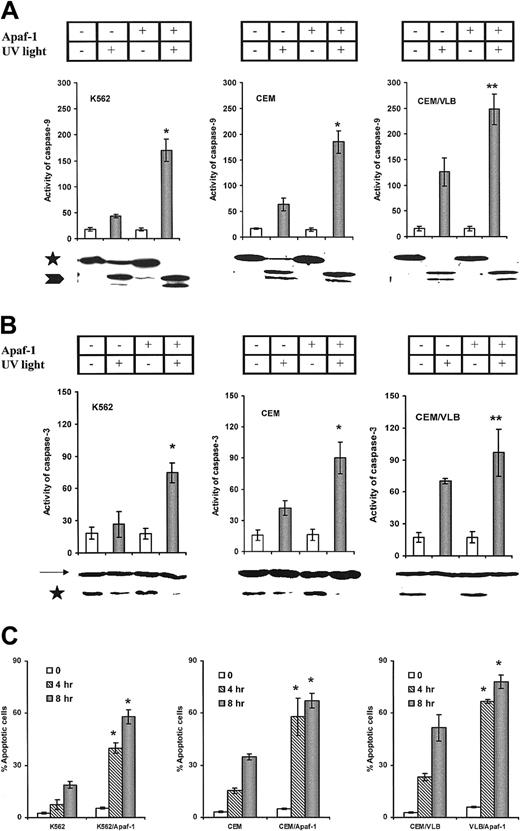
We next transfected the Apaf-1 gene into leukemic cells. Increased Apaf-1 protein expression was detected after transfection for 24 hours in all 3 cell lines (Figure5A); however, the differential expression of Apaf-1 still existed after transfection (Figure 5B). S-100 prepared from all Apaf-1–transfected cells significantly increased cytochromec–mediated activation of procaspase-9 and procaspase-3 in a cell-free system (Figure 6A,B). The enhanced cleavage of procaspase-9 and -3 proteins in Apaf-1–transfected cells was observed by Western blot as a disappearance or reduction in procaspase bands in response to cytochrome c stimulation. As with the response to cytochromec, Apaf-1–transfected cells were sensitized to the activation of caspase-9 (Figure 7A) and caspase-3 (Figure 7B) with UV light irradiation. In addition, the percentage of apoptotic Apaf-1–transfected cells was significantly greater than wild-type cell lines after exposure to UV light (Figure7C). The sensitivity of cells to UV light or cytochromec–induced activation of caspases and apoptosis was dependent on the expression of Apaf-1 in both the wild-type and Apaf-1–transfected cells (P < .001). Mock transfection with vector alone or DMRIE-C, Lipofectamine Plus (Gibco BRL) did not affect Apaf-1 expression or caspase activation (data not shown). Taken together, these data suggest that the restoration of Apaf-1 protein levels can sensitize leukemic cells to cytochromec–dependent apoptosis.
Increased Apaf-1 expression in leukemic cells after
Apaf-1 gene transfection. (A) Western blot detection of Apaf-1 expression in the wild-type (WT) and transfected leukemic cells (Apaf-1). Leukemic cells were transfected with Apaf-1/pCMV2 plasmid DNA. (B) Relative intensity of Apaf-1 expression. The immunoblot shown in (A) was analyzed with a densitometer, and the intensities of Apaf-1 bands were indicated as percentage of integrated density.
Increased Apaf-1 expression in leukemic cells after
Apaf-1 gene transfection. (A) Western blot detection of Apaf-1 expression in the wild-type (WT) and transfected leukemic cells (Apaf-1). Leukemic cells were transfected with Apaf-1/pCMV2 plasmid DNA. (B) Relative intensity of Apaf-1 expression. The immunoblot shown in (A) was analyzed with a densitometer, and the intensities of Apaf-1 bands were indicated as percentage of integrated density.
Cytochrome
c–mediated activation of caspases. S-100 was prepared from the wild-type and Apaf-1–transfected cells. Wild-type cells are indicated as Apaf-1 (−), and Apaf-1–transfected cells are indicated as Apaf-1 (+). (A) Cytochrome c–mediated procaspase-9 activation. The increased activity of caspase-9 in Apaf-1–transfected cells was compared with their wild type (*P < .0001). Processing of procaspase-9 was confirmed by immunoblotting. Caspase-9 precursors are indicated by an asterisk, and the processed caspase-9 are indicated by an arrowhead. (B) Cytochromec–mediated procaspase-3 activation. *P < .0001; **P < .01. Processing of procaspase-3 protein was indicated by immunoblotting. Caspase-9 precursors are indicated by an asterisk, and β-actin is indicated by an arrow.
Cytochrome
c–mediated activation of caspases. S-100 was prepared from the wild-type and Apaf-1–transfected cells. Wild-type cells are indicated as Apaf-1 (−), and Apaf-1–transfected cells are indicated as Apaf-1 (+). (A) Cytochrome c–mediated procaspase-9 activation. The increased activity of caspase-9 in Apaf-1–transfected cells was compared with their wild type (*P < .0001). Processing of procaspase-9 was confirmed by immunoblotting. Caspase-9 precursors are indicated by an asterisk, and the processed caspase-9 are indicated by an arrowhead. (B) Cytochromec–mediated procaspase-3 activation. *P < .0001; **P < .01. Processing of procaspase-3 protein was indicated by immunoblotting. Caspase-9 precursors are indicated by an asterisk, and β-actin is indicated by an arrow.
UV light–induced activation of caspases and apoptosis in leukemic cells.
(A) UV light–induced procaspase-9 activation. Increased activity of caspase-9 in Apaf-1–transfected cells was compared with their wild type (*P < .0001; **P < .001). Processing of procaspase-9 was confirmed by immunoblotting. Caspase-9 precursors are indicated by an asterisk, and the processed caspase-9 is indicated by an arrowhead. (B) UV light–induced procaspase-3 activation. *P < .0001; **P < .01. Processing of procaspase-3 protein was indicated by immunoblotting. Caspase-9 precursors are indicated by an asterisk, and β-actin is indicated by an arrow. (C) UV light–induced apoptosis. Apaf-1–transfected cells were statistically significantly more sensitive than wild-type cells (*P < .0001).
UV light–induced activation of caspases and apoptosis in leukemic cells.
(A) UV light–induced procaspase-9 activation. Increased activity of caspase-9 in Apaf-1–transfected cells was compared with their wild type (*P < .0001; **P < .001). Processing of procaspase-9 was confirmed by immunoblotting. Caspase-9 precursors are indicated by an asterisk, and the processed caspase-9 is indicated by an arrowhead. (B) UV light–induced procaspase-3 activation. *P < .0001; **P < .01. Processing of procaspase-3 protein was indicated by immunoblotting. Caspase-9 precursors are indicated by an asterisk, and β-actin is indicated by an arrow. (C) UV light–induced apoptosis. Apaf-1–transfected cells were statistically significantly more sensitive than wild-type cells (*P < .0001).
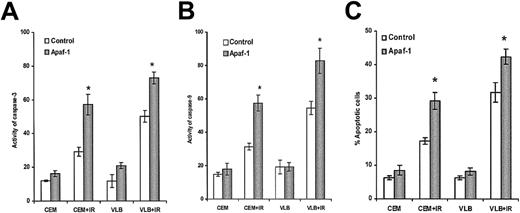
We also tested whether differential Apaf-1 levels could determine the sensitivity of cell lines to γ-irradiation (IR), another DNA-damaging agent. The CEM cell line was more resistant to irradiation-induced activation of caspase-3, caspase-9, and apoptosis than the CEM/VLB100 cell line (Figure8A-C) (P < .001). Apaf-1 transfection restored the sensitivity of CEM cells to irradiation-induced activation of caspases and apoptosis (Figure 8A-C).
Irradiation-induced activation of caspases and apoptosis.
Cells were exposed to irradiation (100 Gy) and further cultured for 6 hours. (A) Activity of caspase-3. (B) Activity of caspase-9. (C) Percentage of apoptosis. Apaf-1–transfected cells were statistically significantly more sensitive than wild-type cells (*P < .001).
Irradiation-induced activation of caspases and apoptosis.
Cells were exposed to irradiation (100 Gy) and further cultured for 6 hours. (A) Activity of caspase-3. (B) Activity of caspase-9. (C) Percentage of apoptosis. Apaf-1–transfected cells were statistically significantly more sensitive than wild-type cells (*P < .001).
Apaf-1 protein deficiency found in human AML blasts and human solid tumor cells
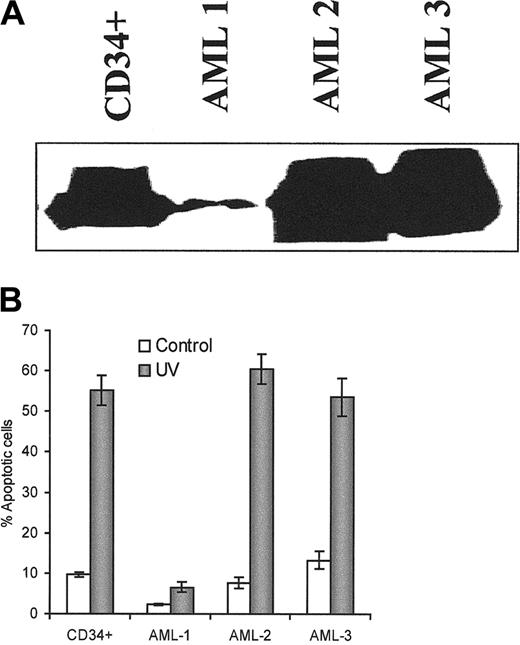
Finally, we were interested in whether Apaf-1 deficiency was associated with resistance to apoptosis in primary leukemic cells and other solid tumor cells. We tested the association of Apaf-1 expression and the sensitivity to UV light–induced apoptosis in some human AML blasts and compared them with the normal human CD34+hemapoietic cells. One of 3 human AML blast populations showed reduced levels of Apaf-1 protein and was resistant to UV light–induced apoptosis (Figure 9A,B; Table1). Ten hours after UV light exposure, the percentage of apoptosis in this resistant AML blast population was 6.6% compared with 55.2% in nonleukemic CD34+ cells and with 60.2% and 58% in other 2 AML blast populations with higher Apaf-1 protein levels. Even after 24 hours, apoptosis in the AML blast population with reduced Apaf-1 level was less than 10%. This suggests that AML-1 blast samples showed lower levels of Apaf-1 protein and more resistance to UV light–induced apoptosis. Using a cell-free system, we also confirmed that AML-1 blasts were more resistant to cytochromec–induced activation of caspases than AML-2 and AML-3 samples. There was a significant correlation (P < .05) between Apaf-1 levels and the sensitivity to cytochromec–dependent activation of caspases/apoptosis (Table 1).
Apaf-1 expression and the sensitivity to UV light–induced apoptosis in the human AML blasts.
(A) Western blot analysis of the expression of Apaf-1 in human normal CD34+ cells and AML blasts. 50 μg protein was used for Apaf-1 expression. Monoclonal anti–Apaf-1 antibody was used at 1:500 dilution. (B) UV light–induced apoptosis. After exposure to UV light for 5 minutes and further culture for 10 hours, cells were fixed with 4% paraformaldehyde and permeabilized by 0.05% saponin for 20 minutes. Slides were stained with 50 ng/mL DAPI. Apoptotic cells were counted under a fluorescence microscope.
Apaf-1 expression and the sensitivity to UV light–induced apoptosis in the human AML blasts.
(A) Western blot analysis of the expression of Apaf-1 in human normal CD34+ cells and AML blasts. 50 μg protein was used for Apaf-1 expression. Monoclonal anti–Apaf-1 antibody was used at 1:500 dilution. (B) UV light–induced apoptosis. After exposure to UV light for 5 minutes and further culture for 10 hours, cells were fixed with 4% paraformaldehyde and permeabilized by 0.05% saponin for 20 minutes. Slides were stained with 50 ng/mL DAPI. Apoptotic cells were counted under a fluorescence microscope.
Correlation between Apaf-1 expression and sensitivity of leukemia cells to cytochrome c-dependent apoptosis
| . | Apaf-1* . | Apoptosis† . | Caspase-3‡ . | Caspase-9‡ . |
|---|---|---|---|---|
| K562 | 1 | 8 | 20 | 15 |
| CEM | 1 | 15 | 37 | 24 |
| CEM/VLB | 3 | 24 | 74 | 64 |
| AML-1 | 2 | 6 | 9 | 5 |
| AML-2 | 49 | 60 | 466 | 202 |
| AML-3 | 49 | 58 | 477 | 230 |
| Correlation (r) | 0.97 | 1.0 | 0.98 | |
| P | <.05 | <.05 | <.05 |
| . | Apaf-1* . | Apoptosis† . | Caspase-3‡ . | Caspase-9‡ . |
|---|---|---|---|---|
| K562 | 1 | 8 | 20 | 15 |
| CEM | 1 | 15 | 37 | 24 |
| CEM/VLB | 3 | 24 | 74 | 64 |
| AML-1 | 2 | 6 | 9 | 5 |
| AML-2 | 49 | 60 | 466 | 202 |
| AML-3 | 49 | 58 | 477 | 230 |
| Correlation (r) | 0.97 | 1.0 | 0.98 | |
| P | <.05 | <.05 | <.05 |
Apaf-1 protein expression analyzed by Western blotting in leukemia cell lines (Figure 3A) and AML blasts, AML-1, AML-2, and AML-3 (Figure 9). Ratios of Apaf-1 expression were analyzed in two groups: (1) K562:CEM:CEM/VLB100 and (2) AML-1:AML-2:AML-3 by densitometry.
UV light–induced apoptosis. Data presented for leukemic cell lines are from Figure 1B. Percentage apoptotic cells were shown at 4 hours. Percentages of apoptosis in AML blasts are shown as Figure 9.
Cytochrome c/dATP induced activation of caspase-3 and -9. S-100 containing 50 μg/mL protein was incubated with cytochrome c and dATP at 30°C for 1 hour. Caspase activity was measured by fluorogenic substrates and expressed as μmol/L per hr/mg protein). Correlations of Apaf-1 protein levels with sensitivity to UV light–induced apoptosis and cytochromec–induced activation of caspases were analyzed by correlation matrices (Statistica 5.0).
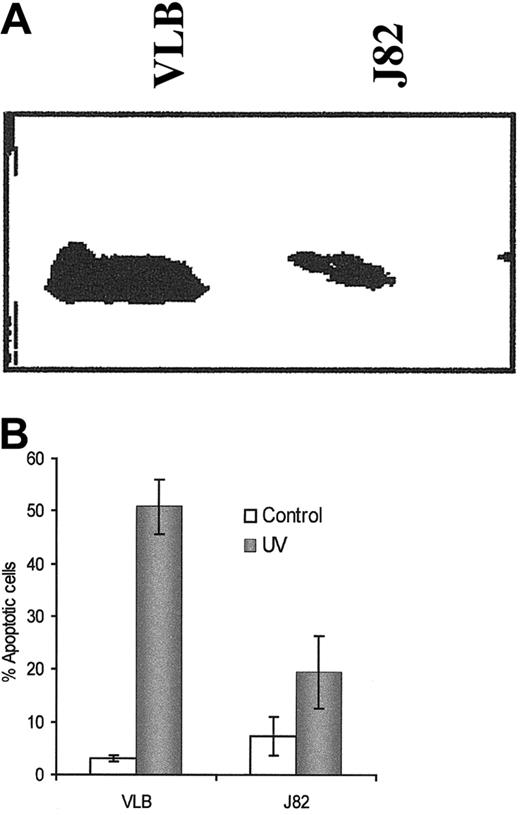
The bladder cancer J82 cell line expressed less Apaf-1 protein than the CEM/VLB100 cell line, and the sensitivity of J82 cells to UV light–induced apoptosis was considerably lower than in CEM/VLB100 cells (Figure10A,B). The percentage of apoptosis and the activation of caspases-3 and -9 were significantly increased after transfection with the Apaf-1 gene (data not shown). In addition, decreased colony formation was correlated with increased levels of Apaf-1 expression as determined by clonogenic assay in 0.3% soft agar (data not shown). Moreover, Apaf-1 transfection suppressed colony formation in J82 cells after UV light stimulation.
Apaf-1 expression and the sensitivity to UV light–induced apoptosis in the human bladder cancer J82 cells.
(A) Apaf-1 expression in J82 cells and compared with CEM/VLB100 cells. (B) UV light–induced apoptosis in the J82 cell line and compared with the CEM/VLB100 cell line. Apoptosis was measured by flow cytometer using propidium iodide–stained cells after exposure to UV light for 8 hours.
Apaf-1 expression and the sensitivity to UV light–induced apoptosis in the human bladder cancer J82 cells.
(A) Apaf-1 expression in J82 cells and compared with CEM/VLB100 cells. (B) UV light–induced apoptosis in the J82 cell line and compared with the CEM/VLB100 cell line. Apoptosis was measured by flow cytometer using propidium iodide–stained cells after exposure to UV light for 8 hours.
Discussion
Previous studies have suggested that Apaf-1 is required for mitochondria-dependent apoptosis5,8,18,19 and that overexpression of Apaf-1 can increase sensitivity to apoptosis induced by chemotherapeutic drugs.20 We suggest that we have provided, for the first time, direct evidence that Apaf-1 protein levels vary constitutively in human leukemic cells. In addition, we have shown that a relative deficiency of Apaf-1 can result in resistance to apoptosis in human leukemic cells and tumor cells. Restoration of Apaf-1 protein levels after gene transfection is able to resensitize previously resistant cells to UV light–induced killing.
Previous studies on the mechanism of leukemic cell resistance to apoptosis have focused on mitochondrial components, such as Bcl-2 or Bcl-XL overexpression,9,29,30 the ratio of Bax to Bcl-2 protein at the mitochondrial membrane,10functional deficiency of the mitochondrial electron transport chain,25,31 or factors upstream of the mitochondrial level, such as NF-κB, IAPs,32 and Bcr-Abl.33 34
We have previously shown that the leukemic cell lines K562, CEM, and CEM/VLB100, have differential sensitivities to TNF-α–induced apoptosis. The block, which confers resistance to TNF-α, is at, or may be upstream of, the level of the mitochondria.10 25 Similar to TNF-α, these leukemic cells showed differential susceptibility to UV light–induced apoptosis; however, UV light–induced DNA damage was similar in the 3 cell lines. Unlike TNF-α, UV light can induce cytochrome crelease from mitochondria in all the tested cell lines. This suggests that the level at which sensitivity to UV-induced apoptosis is determined is downstream of the mitochondria.
UV light induces apoptosis through cytochrome c release and activation of caspase-3.9,19 In our cells, UV light–induced activation of caspase-3 was proportional to the degree of apoptosis. Using a cell-free system, we showed that exogenous cytochrome c was able to activate procaspase-3 to the same extent as UV light. Procaspase-3 itself was not defective because granzyme B, which directly processes caspase-3,15 induced similar levels of caspase-3 activity in all 3 cell lines. Thus, we suggest that the block conferring resistance is between cytochrome c and caspase-3.
Tumors in Apaf-1 or caspase-9 knock-out mice are resistant to apoptosis induced by c-myc.8 T cells from Apaf-1 knock-out mice exhibited resistance to apoptosis induced by DNA-damaging agents, such as etoposide or UV light, but were sensitive to Fas-mediated killing.19 Apaf-1 mRNA is expressed ubiquitously in human tissue, and normal leukocytes express high levels of Apaf-1.3 In our study, the leukemic cell lines K562 and CEM, some human AML blasts, and the tumor cell line J82—all of which are relatively resistant to UV light—expressed lower levels of Apaf-1 protein in the cytosol than did the UV- and irradiation-sensitive cell line CEM/VLB100. In addition, Apaf-1 levels correlated with the sensitivity of all 4 cell lines and AML blasts to cytochromec–dependent activation of caspases and apoptosis. Depletion of Apaf-1 from S-100 cytosol abrogated cytochromec–mediated cleavage of procaspase-9. By contrast, adding purified Apaf-1 to the cytosol of K562 and CEM cells restored the sensitivity to cytochrome c–mediated cleavage of procaspase-3. Transfection of the Apaf-1 gene into leukemic cells increased protein levels of Apaf-1 in all 3 cell lines. As expected, Apaf-1 gene transfection sensitized leukemic cells to cytochrome c–mediated activation of caspase-9 and caspase-3. It also increased the sensitivity of Apaf-1–transfected cells to UV light and IR-induced activation of caspases and consequent apoptosis, and it accelerated the response compared with wild-type cells. Thus we provide direct evidence that Apaf-1 level is an important determinant of cytochrome c–dependent apoptosis in leukemic and tumor cells, even when an alternative mechanism for regulating apoptosis, such as overexpression of Bcr-Abl,20,33,34 exists. We and others33 have found that the Bcr-Abl–positive K562 cell line was resistant to apoptosis induced by the DNA damaging agent etoposide. The blocks are located upstream and downstream of mitochondria. Apaf-1 can sensitize cells to etoposide-induced apoptosis only after the upstream block was removed (Jia et al, manuscript submitted).
The K562 cell line expressed more inhibitory caspase-9b than the CEM/ VLB100 cell line, which can compete with procaspase-9 for binding to Apaf-1.21 However, CEM cells expressed less caspase-9b than CEM/ VLB100 cells. The expression of caspase-9b does not explain the differential sensitivity of the cell lines to cytochrome c-mediated procaspase activation. XIAP inhibitory protein levels were similar among the 3 cell lines. In addition, the ability of exogenous cytochromec to cleave exogenous procaspase-9 in the presence of cytosol from each of the 3 cell lines studied was identical to the cleavage of endogenous procaspase-3, suggesting that the differential sensitivity was not due to differences in endogenous procaspase-9 levels.
In summary, we suggest that Apaf-1 deficiency may be an important mechanism of resistance to apoptosis in human leukemic cells. Further studies are required to assess the overall importance of the mechanism in human leukemia and to understand the mechanism by which Apaf-1 expression is regulated.
We thank Dr Y. A. Lazebnik (Cold Spring Harbor Laboratory, New York) for the gifts of monoclonal anti–Apaf-1 and anti–caspase-9 antibodies, and we thank Dr Michael Neat and Ms Claire Wiggins for clinical samples.
Supported by the Leukemia Research Fund (S.M.K., L.J.) and by Elimination of Leukemia Training and Traveling Fellowship, the Pathology Society of Great Britain and Ireland Traveling Fellowship, and the Cancer Research Committee of the St Bartholomew's Hospital of London (L.J.).
Submitted September 27, 2000; accepted March 20, 2001.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Li Jia, Dept of Haematology, St Bartholomew's and The Royal London School of Medicine and Dentistry, Turner Street, London E1 2AD, United Kingdom; e-mail: l.jia@mds.qmw.ac.uk.

![Fig. 4. Effect of Apaf-1 protein levels on cytochrome. / c–mediated cleavage of exogenous procaspase-9.(A) Cytochrome c–induced procaspase-9 cleavage in the presence of S-100 from different cell lines. In vitro translated [35S] procaspase-9 cleavage was initiated by the addition of cytochrome c and dATP in the presence of S-100 from cell lines. (B) Cytochrome c–mediated procaspase-9 cleavage in the absence of Apaf-1. The S-100 in which Apaf-1 was depleted has been indicated as ID. Failure to cleave procaspase-9 by cytochromec is shown by the absence of the cleavage bands (P35 and P37) compared with untreated S-100. (C) Purified Apaf-1 protein enhanced cytochrome c–induced procaspase-9 activation in K562 and CEM cell lines. 1 μL Apaf-1 protein was added to the S-100 (50 μg protein) lysates from K562 and CEM cells. Cytochromec–mediated procaspase-9 cleavage was allowed to proceed for 1 hour. Enhanced procaspase-9 cleavage is shown as disappearance of the procaspase-9 and P37 bands.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/2/10.1182_blood.v98.2.414/5/m_h81411263004.jpeg?Expires=1767754980&Signature=jC1NKX36B8~3~8wpTk2-KXdRE2kcaLAzLAcW1JmfQiiSdeecWDLHKah~A14IoGCqXrKTd0YWRtIXKVYZdaUb~Oo6yrEvt8o4TDCyrYjim8JGE~JLHLrMgJls9T6-ZPSWfftovAEgzymv~6Czsh5HlPpWTMFvX1m-Kd0XsVPYgHzrR5kMAknuwHfhpC9hWX-yKiuV2e~g7MDPteT67uaqOPtvv0QlR-buigmQBxE7VZz1hM7rPL7vV9IhgyXK7hSqRjLKomZp58xKg8U2zImo-CTUDEYe3LoppbqlP4GfKI5WOaQPSZyTpy~t53VJv0J2zMHfONGDQrNeBUIPcmBcDQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal