Abstract
The p12I protein, encoded by the pX open reading frame I of the human T-lymphotropic virus type 1 (HTLV-1), is a hydrophobic protein that localizes to the endoplasmic reticulum and the Golgi. Although p12I contains 4 minimal proline-rich, src homology 3–binding motifs (PXXP), a characteristic commonly found in proteins involved in signaling pathways, it has not been known whether p12I has a role in modulating intracellular signaling pathways. This study demonstrated that p12I binds to the cytoplasmic domain of the interleukin-2 receptor (IL-2R) β chain that is involved in the recruitment of the Jak1 and Jak3 kinases. As a result of this interaction, p12I increases signal transducers and activators of transcription 5 (STAT5) DNA binding and transcriptional activity and this effect depends on the presence of both IL-2R β and γc chains and Jak3. Transduction of primary human peripheral blood mononuclear cells (PBMCs) with a human immunodeficiency virus type 1–based retroviral vector expressing p12I also resulted in increased STAT5 phosphorylation and DNA binding. However, p12I could increase proliferation of human PBMCs only after stimulation of T-cell receptors by treatment of cells with low concentrations of αCD3 and αCD28 antibodies. In addition, the proliferative advantage of p12I-transduced PBMCs was evident mainly at low concentrations of IL-2. Together, these data indicate that p12I may confer a proliferative advantage on HTLV-1–infected cells in the presence of suboptimal antigen stimulation and that this event may account for the clonal proliferation of infected T cells in vivo.
Introduction
Human T-lymphotropic virus type 1 (HTLV-1) causes adult T-cell leukemia/lymphoma (ATLL),1 and its genome carries genetic information for the structural and enzymatic proteins, the regulatory proteins Tax and Rex, and other open reading frames (orfs) encoding small proteins with largely unknown functions.2-5 HTLV-1 infects and immortalizes primary human T cells in vitro, and after several months, these cells acquire the ability to grow in the absence of interleukin-2 (IL-2).1 The switch to IL-2 independence correlates in most cases with acquisition of a constitutive activation of the Jak/signal transducers and activators of transcription (STAT) pathway6-8 and decreased expression of the src homology 2–containing tyrosine phosphatase 1 protein,9 which regulates signaling from several hematopoietic surface receptors.10 HTLV-1 also confers longevity on T cells in vivo, since expansion of T cells with identical integrations for HTLV-1 can be found at several-year intervals in the same infected individuals.11 These findings raise the question of how CD4+ T cells carrying the HTLV-1 provirus can expand and survive for a long time in vivo.
The HTLV-1 p12I protein, a hydrophobic protein resident in the endoplasmic reticulum (ER) and Golgi58 that is encoded by the 3′ end orf I of the viral genome,12 forms dimers,13 has weak oncogenic properties, and binds to the p16 subunit of the vacuolar hydrogen adenosine diphosphatase (H+ ATPase).14 Expression of p12Iin infected cells is suggested by the presence of transcripts in cultured3,5,15 and ex vivo cells from individuals infected with HTLV-1.5 The orf I is likely expressed in vivo because antibodies (Abs) and cytotoxic T lymphocytes to peptides from the orf I protein have been detected.16,17 Importantly, ablation of the splice acceptor site for the singly spliced p12I messenger RNA from a molecular clone of HTLV-1 impaired viral infectivity in a rabbit model in vivo.18This may be partly related to the finding that p12Iinterferes with major histocompatibility complex class I (MHC I) heavy-chain trafficking and may facilitate escape of HTLV-1–infected cells from the host's immune surveillance.58
We previously reported that p12I also binds the IL-2 receptor (IL-2R) β and γc chains and affects their expression on the cell surface.19 IL-2 is an essential cytokine for the growth and survival of T cells.20 Binding of IL-2 to the IL-2R β and γc chains results in activation and recruitment of Jak1 and Jak3 and phosphorylation and nuclear translocation of STAT5a and STAT5b. STAT proteins have been found to be activated with a high frequency in a wide variety of human cancers,21 and STAT1, STAT3, and STAT5 were shown to be activated in 70% of ATLL primary cells.22 In fact, there is good evidence that constitutively activated STAT signaling participates in oncogenesis by simultaneously promoting cell proliferation and preventing apoptosis.23
Because p12I interacts with the IL-2R β and IL-2R γc chains, we hypothesized that p12I may interfere with or augment signaling pathways and contribute to proliferation of T cells infected with HTLV-1. In this study, we found that p12I, through its binding to the IL-2R β chain, increased STAT5b transcriptional activation and simultaneously decreased the T-cell requirement for IL-2 for proliferation.
Materials and methods
Expression plasmids
The p12I complementary DNA tagged with the AU1 or HA1 epitopes, as well as the wild-type IL-2R β and IL-2R γc chains and mutants for both chains, were cloned in the pME18S plasmid. IL-2R β-chain mutants β330, β350, ΔH, ΔS, ΔA, and ΔST were described previously.20 The remaining IL-2R β-chain mutants were generated by polymerase chain reaction using the ΔST plasmid and resulted in deletion of amino acid residues 264 to 296 and 350 to 525, 264 to 350 and 432 to 525, and 264 to 390, indicated by subscripts in the following: ΔST+296-350, ΔST+350-432, and ΔST+390-525. All plasmids were verified by sequencing. The βCD4β and the ααβ chimeric receptors, which were described previously,24 contain the β chain in which the transmembrane region has been exchanged with the equivalent region of the CD4 receptor (βCD4β) or the extracellular and transmembrane region of the α chain has been joined to the β-chain cytoplasmic portion. Lentiviral human immunodeficiency virus (HIV)–based retroviral vector HR′cytomegalovirus (CMV)-Luc, CMV-driven HIV helper virus deleted for the env and nefgenes (pDNL6), and the HIV long terminal repeat–vesicular stomatitis virus G (VSV-G) envelope were described previously.25HA-tagged p12I was amplified by polymerase chain reaction using Turbo PFU polymerase (Promega, Madison, WI), sequenced, and cloned between BamH1 and Xho1 restriction sites (HR′CMV p12I HA).
DNA transfections, immunoprecipitations, Western blotting, and immunofluorescence
One million COS-7 or 1.2 × 106 293T cells were plated in a 100-mm dish and transfected the next day with 10 μg of each plasmid by using the calcium phosphate method.26Cells were then collected, washed in phosphate-buffered saline (PBS), lysed in lysis buffer by freeze-thawing, and assayed for luciferase activity (Promega).
Abs against the AU1 (Berkeley Antibody, Richmond, CA) and HA1 (12CA5; Boehringer Mannheim, Indianapolis, IN) epitopes were used to immunoprecipitate the wild-type p12I mutants. Monoclonal Ab anti–β chain (R&D Systems, Minneapolis, MN) was used to immunoprecipitate the IL-2R β chain in association with protein glycine agarose (Boehringer Mannheim). Polyclonal rabbit serum was used to detect β-chain protein with antirabbit horseradish peroxidase conjugated as secondary Ab. Immunoprecipitations were done in radioimmunoprecipitation assay (RIPA) buffer overnight at 4°C by using 2 μg/mL Ab. Protein glycine agarose was then added. After an additional 2 hours at 4°C, immunocomplexes were washed in RIPA buffer containing Tween 20 0.1%, resolved on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels (Novex, San Diego, CA), and transferred to nitrocellulose membranes (Hybond; Amersham, Piscataway, NJ). Detection was done with a chemiluminescent system (Amersham, Arlington Heights, IL). Labeled proteins were analyzed in 15% SDS-PAGE gel and visualized on X-Omat film (Kodak, Rochester, NY). Western blotting for STAT5 was done by using Ab Y694 and Y699 (Upstate, Lake Placid, NY).
Pseudotype virus production and concentration
The 293T cells (2 × 106) were seeded on a 10-cm dish and transfected the following day with VSV-G (2 μg), pDNL6 (4 μg), and HR′CMV-Luc or HR′CMV p12I (4 μg) by using an Effectene reagent kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Supernatant from 20 dishes was collected every 12 hours from 24 hours to 72 hours after transfection, cleared of cellular debris by centrifugation at 8000g for 10 minutes at room temperature, filtered through a 0.45 μm filter, and stored at −80°C. Pseudotype viruses were formed into pellets by ultracentrifugation at 50 000g (28 000 rpm with a SW41 rotor) for 1.75 hours at 4°C. Virus was then resuspended in PBS by means of a 4-hour incubation on ice, collected, divided into aliquots, and stored at −80°C. HIV gag p24 was measured by using an antigen capture assay (Coulter, Miami, FL). Primary peripheral blood mononuclear cells (PBMCs) were infected by using comparable amounts of viral particles for both HR′CMV-Luc and HR′CMV p12I. Infectivity and expression were verified by immunofluorescence and immunoprecipitation Western blot assays.
Primary T-cell infection and proliferation assay
Mononuclear cells from peripheral blood of healthy volunteers were purified by Ficoll-Hypaque methods (Hyclone, Logan, UT), washed in PBS, and either used as such or after suboptimal stimulation obtained by phytohemagglutinin (PHA; 1 ng/mL) or CD3/CD28 (0.5 ng/mL) for 24 hours before infection with HR′CMV p12I or HR′CMV-Luc VSV pseudotyped viruses. Infected cells were plated in duplicate in a 96-well plate (3 × 104 cells/well) in 100 μL RPMI 1640 supplemented with 20% fetal-calf serum (FCS) in the absence or presence of 2.5, 5, or 10 U/mL IL-2 (Boehringer Mannheim). Cellular proliferation assays were done on days 2, 3, 4, and 6 after infection by using a Colorimetric Cell Proliferation Kit II (Roche Molecular, Indianapolis, IN) as described by the manufacturer. Spectrophotometric absorbance was measured with a mean filter at 492 nm and a reference filter at 690 nm. SDs were calculated from values obtained from 2 independent infections of each donor cell, and results were representative of 3 independent experiments done using cells from 3 human volunteers.
Electrophoretic mobility shift assays
PBMCs from healthy volunteers were purified with the Ficoll-Hypaque method, washed in PBS, activated with 5 μg/mL PHA for 3 days, washed again in PBS, and cultured for 5 days in RPMI 10% FCS and 10 U/mL IL-2. Lymphocytes were infected with HR′CMV p12I or HR′CMV-Luc pseudotype viruses. After 18 hours, PBMCs were washed twice in medium free of serum and IL-2 and deprived overnight of serum (0.1%) and stimulation with IL-2. The next day, infected PBMCs were divided into 6 batches of 2 × 106cells and pulsed with 0.1, 1, 10, 100, or 1000 U/mL IL-2 for 10 minutes at 37°C. Unstimulated cells were used as controls for basal levels of STAT5 activation. Cells were collected by centrifugation and washed and lysed in 20 μL lysis buffer.22 Binding reactions were done with 2 μg protein extract, as previously reported,22 with an increase in incubation time from 30 minutes to 1 hour. STAT5 DNA binding activity was measured by using phosphorus 32 (32P)–labeled β-casein γ-interferon activated sequence (GAS) elements. The DNA-protein complexes were separated on prerun 5% native polyacrylamide gels in 0.5 × Tris borate EDTA buffer at 200 V for 2 hours. Gels were dried and exposed to x-ray film at −80°C. Gel-shift analysis using the β-casein GAS element was done as described previously.27 COS-7 cells were transfected and pulsed with the selected amounts of IL-2, nuclear extracts were made, and 10 μg of nuclear extracts were incubated with 20 000 cpm 32P-labeled β-casein GAS element and run on 5% native polyacrylamide gels.
Results
The 20–amino acid stretch of IL-2R β involved in Jak1 and Jak3 recruitment binds p12I
To identify the site of p12I interaction within the β chain, the βCD4β and ααβ chimeric molecules were used.24 Both chimeric molecules coprecipitated with p12I, indicating that neither the transmembrane nor the extracellular domains of β were the primary sites of p12Iinteraction (data not shown). Mutants lacking different regions of the β-chain cytoplasmic domain were coexpressed with p12I, and the cell lysate was divided in half and subjected to immunoprecipitation and Western blot analysis with Ab to the receptor (R&D Systems; goat antiserum against human IL-2R β [RDβ]) and to p12I (αHA1) (Figure 1). The βΔST mutant lacking most of the cytoplasmic tail (Δ267) failed to bind p12I (Figure 1, bottom panel, lane 5). Although the β330 mutant also failed to bind p12I, the β350 mutant (lane 7) did bind, indicating that the region encompassing amino acids 330 to 350 of the receptor was required for p12I binding (Figure 1, bottom panel, lanes 6 and 7). The βΔA mutant, which lacks amino acids 313 to 382, also did not bind p12I, whereas the βΔS mutant (Δ267-322) did (Figure1, bottom panels, lanes 13 and 14), further supporting this idea.
The p12I protein binds the acidic region of IL-2R β between residues 330 and 350.
(A) Schematic representation of the IL-2R β-chain truncation and deletion mutants. Middle panels show controls for expression of the IL-2R β truncation mutants and p12I. (B) Coprecipitation of p12I with the IL-2R β mutants. IP indicates immunoprecipitation; WB, Western blot.
The p12I protein binds the acidic region of IL-2R β between residues 330 and 350.
(A) Schematic representation of the IL-2R β-chain truncation and deletion mutants. Middle panels show controls for expression of the IL-2R β truncation mutants and p12I. (B) Coprecipitation of p12I with the IL-2R β mutants. IP indicates immunoprecipitation; WB, Western blot.
Because the βΔH mutant coprecipitated more p12I when increased than the β350 mutant (Figure 1, bottom panel, lanes 7 and 8), we investigated whether the binding site of p12I on the IL-2R β chain extended beyond amino acid 350, which is present in the βΔH mutant (Figure 1, top panel). To do this, 3 additional deletion mutants were constructed and coexpressed with p12I (Figure 1, top panel). Coprecipitation of p12I was observed with the βΔST+296-350mutant, whereas neither βΔST+350-432 nor βΔST+390-525 coprecipitated p12I (Figure 1, bottom panel, lanes 18-20). Therefore, the primary binding site of p12I to the IL-2R β chain appears to be proximal to amino acid 350, although there could be additional contacts beyond this region. Importantly, this region was previously shown to be critical for the recruitment of Jak1 and Jak3 after IL-2 signaling.10 27
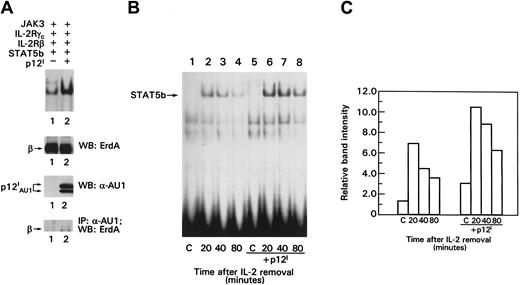
p12I increases STAT5b DNA binding activity
Because p12I binds the IL-2R β chain in a region involved in transduction of the IL-2 signal, we hypothesized that p12I could modulate activation of the nuclear effector of IL-2 signaling, STAT5. To investigate this hypothesis, the IL-2R signaling pathway was reconstituted in a transient transfection system27 in which IL-2R β, γc, Jak3, and STAT5b proteins were coexpressed in the absence or presence of p12I. In this experimental system, the phosphorylation and DNA binding of STAT5 can be assessed before and after IL-2 triggering of cells. COS-7 cells were pulsed with IL-2 for 20 minutes, and nuclear lysates were evaluated for the presence of activated STAT5b by measuring STAT5 binding to the β-casein probe in electrophoretic mobility shift assays (EMSAs) as described previously.27 A 2- to 4-fold increase in the basal level of STAT5 binding to the β-casein probe was observed in the presence of p12I(Figure 2A, top panel, lanes 1 and 2). Results with controls for equivalent expression of IL-2R β and p12I and the association of p12I with the IL-2R β chain are also shown in Figure 2A. Together, these findings suggest that p12I increases STAT5 DNA binding in the absence of IL-2 and that less IL-2 is required to promote STAT5b binding activity in the presence of p12I.
The p12I protein increases STAT5b DNA binding and does not interfere with STAT5b degradation.
(A) EMSA of lysates from COS-7 cells. Top panel shows control for the amount of IL-2R β expression on Western blot analysis, analysis of p12I expression by Western blotting, and p12Iand IL-2R β chain coprecipitation. (B) Transfected cells were pulsed with 10 nM IL-2 for 20 minutes, washed, incubated in medium without IL-2 for the indicated times, lysed, and processed for EMSA as in Figure 2A. (C) Graph shows quantitation of the data in Figure 2B, obtained by using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). C indicates control; IP, immunoprecipitation; WB, Western blot.
The p12I protein increases STAT5b DNA binding and does not interfere with STAT5b degradation.
(A) EMSA of lysates from COS-7 cells. Top panel shows control for the amount of IL-2R β expression on Western blot analysis, analysis of p12I expression by Western blotting, and p12Iand IL-2R β chain coprecipitation. (B) Transfected cells were pulsed with 10 nM IL-2 for 20 minutes, washed, incubated in medium without IL-2 for the indicated times, lysed, and processed for EMSA as in Figure 2A. (C) Graph shows quantitation of the data in Figure 2B, obtained by using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). C indicates control; IP, immunoprecipitation; WB, Western blot.
p12I does not influence degradation of activated STAT5b
IL-2–induced STAT5 activation is tightly regulated by dephosphorylation and protein degradation.28 To investigate whether the increase in p12I-induced STAT5 activation resulted from a delay in its degradation, we studied the decay of STAT5 binding activity after IL-2 pulsing and removal of the ligand. Higher levels of STAT5 binding activity were detected in the presence of p12I at all time points after the IL-2 washout (Figure 2B). However, the rate of decay of the STAT5 binding activity did not appear to be affected by p12I, since the amount of STAT5b binding to the β-casein probe was reduced by about 50% at 80 minutes (Figure 2C). Thus, p12I does not appear to increase IL-2R signaling by delaying STAT5 inactivation or degradation.
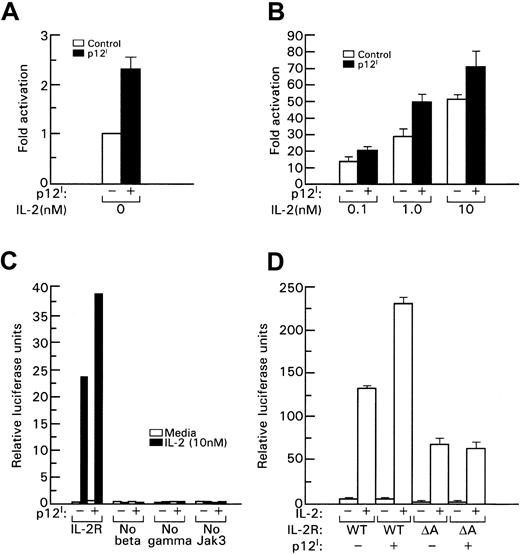
p12I binding to the IL-2R β chain is necessary to increase STAT5b transcriptional activity
To assess whether the increased STAT5b DNA binding induced by p12I also resulted in increased transcriptional activity of STAT5b, the IL-2R signaling pathway was reconstituted in 293T cells and a β-casein promoter luciferase reporter gene was used to measure STAT5b transcriptional activation in the presence of p12I. We found that p12I increased STAT5b transcriptional activity by approximately 2-fold to 3-fold (Figure3A); this increase was also evident in the presence of increasing amounts of IL-2 (Figure 3B). Approximately 10-fold less IL-2 was required to obtain an equivalent level of STAT5b transcriptional activity, as indicated, for example, by the fact that luciferase activity in the presence of p12I with 1 nM IL-2 was equivalent to that obtained in the absence of p12I with 10 nM IL-2 (Figure 3B).
STAT5 transcriptional activation induced by p12I depends on binding to the IL-2R β chain.
In this experiment, 293T cells were transfected with plasmids expressing IL-2R β, IL-2R γc, Jak3, and STAT5b with or without p12I. IL-2 was added 20 hours later; after 24 hours, activation of the β-casein luciferase reporter was determined by using a luminometer. A Renilla luciferase reporter (Promega, Madison, WI) was used to control transfection efficiency. (A) Basal transcriptional activation of STAT5b in the absence of IL-2. (B) STAT5b activation with increasing amounts of IL-2 (nM). The error bars in panels A, B, and D indicate the SE from 3 independent experiments. Panel C shows a representative experiment. At least 3 independent experiments were done to obtain the results shown in each panel. WT indicates wild-type IL-2R β, and A indicates the βΔA mutant.
STAT5 transcriptional activation induced by p12I depends on binding to the IL-2R β chain.
In this experiment, 293T cells were transfected with plasmids expressing IL-2R β, IL-2R γc, Jak3, and STAT5b with or without p12I. IL-2 was added 20 hours later; after 24 hours, activation of the β-casein luciferase reporter was determined by using a luminometer. A Renilla luciferase reporter (Promega, Madison, WI) was used to control transfection efficiency. (A) Basal transcriptional activation of STAT5b in the absence of IL-2. (B) STAT5b activation with increasing amounts of IL-2 (nM). The error bars in panels A, B, and D indicate the SE from 3 independent experiments. Panel C shows a representative experiment. At least 3 independent experiments were done to obtain the results shown in each panel. WT indicates wild-type IL-2R β, and A indicates the βΔA mutant.
The increase in STAT5b transcriptional activity induced by p12I required the presence of all components of the IL-2R pathway, since the absence of β, γc, or Jak3 resulted in loss of STAT5 activation despite the presence of p12I(Figure 3C). In addition, p12I binding to the β chain at amino acids 330 to 350 was also essential for p12Iinduction of STAT5b activity because in the presence of the βΔA mutant, which maintains its ability to transduce the IL-2 signal29 (although at a lower level) but lacks the 330– to 350–amino acid stretch that binds p12I, no increase in STAT5b activity was observed (Figure 3D). Increased transcriptional activation was also observed when STAT5a was used instead of STAT5b (data not shown). Together, these results show that p12Idecreases the threshold of IL-2 signaling and that this effect depends on binding to the IL-2R β chain. Because the β and γcchains are also part of the IL-15R, it would be interesting to determine whether p12I could also affect IL-15R signaling.59
p12I increases STAT5 phosphorylation and DNA binding in primary human PBMCs
HTLV-1 infects and transforms human primary PBMCs both in vivo and in vitro. Therefore, we investigated whether p12I would increase STAT5 activation in its natural target-cell population. An HIV-1–based retroviral vector expressing p12I was constructed (HR′CMV p12I). Expression of HA1-tagged p12I was assessed first in transfected 293T cells by Western blotting, and the characteristic doublet of p12Iwas readily detected (Figure 4A). Transduction of HR′CMV p12I in primary human PBMCs was properly localized in the ER-Golgi compartment, as demonstrated by confocal immunofluorescence (Figure 4B).
HR′CMV p12Itransduction in primary PBMCs increases STAT5 activation.
(A) The HR′CMV p12I and HR′CMV-Luc constructs were transfected in 293T cells and protein extracts subjected to immunoprecipitation and Western blot analysis using αHA1 Ab. (B) Confocal immunofluorescence of PBMCs after infection with pseudotype viruses expressing either Luc or p12I. (C) Increased STAT5 DNA binding was indicated by EMSA. STAT5 DNA binding activity was measured by using a 32P-labeled β-casein GAS element. B indicates basal STAT5 activity; S, starved for serum and IL-2 overnight; minus sign, no Ab to STAT5 after pulse with 1000 U IL-2; plus sign, Ab to STAT5 was added; and 1000, pulse with 1000 IU IL-2. Cells were pulsed with various amounts of IL-2 (0, 0.1, 1, 10, and 100 IU/mL) as indicated. (D) Western blot analysis using a phospho-STAT5–specific Ab was done on the same cellular extracts used in the gel-shift assay.
HR′CMV p12Itransduction in primary PBMCs increases STAT5 activation.
(A) The HR′CMV p12I and HR′CMV-Luc constructs were transfected in 293T cells and protein extracts subjected to immunoprecipitation and Western blot analysis using αHA1 Ab. (B) Confocal immunofluorescence of PBMCs after infection with pseudotype viruses expressing either Luc or p12I. (C) Increased STAT5 DNA binding was indicated by EMSA. STAT5 DNA binding activity was measured by using a 32P-labeled β-casein GAS element. B indicates basal STAT5 activity; S, starved for serum and IL-2 overnight; minus sign, no Ab to STAT5 after pulse with 1000 U IL-2; plus sign, Ab to STAT5 was added; and 1000, pulse with 1000 IU IL-2. Cells were pulsed with various amounts of IL-2 (0, 0.1, 1, 10, and 100 IU/mL) as indicated. (D) Western blot analysis using a phospho-STAT5–specific Ab was done on the same cellular extracts used in the gel-shift assay.
To assess the effect of p12I on PBMCs, peripheral blood lymphocytes from healthy volunteers were infected with pseudotype viruses carrying p12I or the luciferase genes. After 18 hours, PBMCs were deprived of serum and IL-2 to lower the STAT5 basal level of DNA binding activity. Cells were then pulsed for 10 minutes with various concentrations of IL-2, and STAT5 phosphorylation and activation was assessed by either gel-shift assay or Western blot using Ab to phosphorylated STAT5. The expected DNA-protein complex in nuclear extracts of cells pulsed with IL-2 was readily observed by using the β-casein probe (Figure 4C), and the identity of STAT5 in the DNA complex was demonstrated by using a specific STAT5 Ab to supershift the complex (Figure 4C). At 10 U and 100 U IL-2, the level of STAT5-DNA complex was a few-fold higher in cells infected with HR′CMV p12I than in cells infected with the control HR′CMV-Luc virus, suggesting that p12I may enhance STAT5 phosphorylation and DNA binding. To confirm this finding, the same nuclear lysates were used in Western blot analysis using a mouse monoclonal Ab to the phosphorylated STAT5. This experiment confirmed the presence of a higher level of phosphorylated STAT5 in cells infected with HR′CMV p12I than in those infected with HR′CMV-Luc after pulsing with IL-2 (Figure 4D). Thus, as observed in COS-7 cells, p12I increased STAT5 activation in primary human PBMCs.
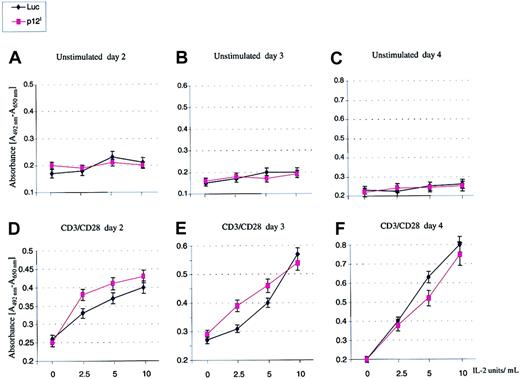
p12I decreases the IL-2 requirement for T-cell proliferation
An expected consequence of an increased basal level of STAT5 activation is that cell proliferation may occur with a lower concentration of exogenous IL-2. To investigate this hypothesis, PBMCs from healthy volunteers were isolated, washed in PBS, and either used as such or after suboptimal stimulation with either PHA (1 ng/mL) or CD3/CD28 (0.5 ng/mL) for 24 hours before infection with HR′CMV p12I or HR′CMV-Luc pseudotype viruses. Infected cells were then divided into 4 aliquots and plated in duplicate in 96-well plates in the absence or presence of 2.5, 5, and 10 or 100 U IL-2. Cellular proliferation was monitored as described.30 The p12I-transduced PBMCs stimulated in suboptimal conditions proliferated faster than the Luc-transduced PBMC control (Figure5) and, although this effect was evident with 2.5 U and 5 U IL-2 (Figure 5D-E), it was lost at higher concentrations10 of the ligand (Figure 5E). Furthermore, this effect was evident only within the first few days after infection, it became undetectable 4 days after infection (Figure 5F), and it was not observed at all in unstimulated resting PBMCs (Figure 5A-C). Other studies suggested that p12I may affect HTLV-1 infectivity by promoting activation of quiescent primary T lymphocytes.31 Our results, obtained from experiments with samples from 3 different donors, do not seem to support this conclusion, since p12I did not stimulate T cells to proliferate in the absence of an exogenous activating signal. Together, these data suggest that p12I expression alone is not sufficient to activate T cells but that it enables T cells, activated in suboptimal conditions, to proliferate even in the presence of a low level of IL-2.
The p12I protein increased T-cell proliferation after suboptimal antigen stimulation.
PBMCs from healthy volunteers were purified by the Ficoll method, washed in PBS, and either used as such or after suboptimal stimulation with 0.5 ng/mL CD3/CD28 Ab for 24 hours before infection with HR′CMV p12I or HR′CMV-Luc pseudotype viruses. Cellular proliferation was measured by using a Colorimetric Cell Proliferation Kit II. SDs were calculated from 2 independent infections of each donor sample, and values are representative of results from 3 separate donors. Similar results were obtained after stimulation with 1 ng/mL PHA (data not shown).
The p12I protein increased T-cell proliferation after suboptimal antigen stimulation.
PBMCs from healthy volunteers were purified by the Ficoll method, washed in PBS, and either used as such or after suboptimal stimulation with 0.5 ng/mL CD3/CD28 Ab for 24 hours before infection with HR′CMV p12I or HR′CMV-Luc pseudotype viruses. Cellular proliferation was measured by using a Colorimetric Cell Proliferation Kit II. SDs were calculated from 2 independent infections of each donor sample, and values are representative of results from 3 separate donors. Similar results were obtained after stimulation with 1 ng/mL PHA (data not shown).
Discussion
HTLV-1 has persisted in nonhuman and human primates for thousands of years, as shown by the finding of proviral DNA in Andean mummies.32 HTLV-1 virions, in contrast to HIV-1 virions, are poorly infectious in vitro, and HTLV-1 is thought to be transmitted efficiently mainly through cell-to-cell contact.1Therefore, maintenance of a large pool of long-lasting infected T cells carrying the provirus in infected individuals11 has likely been instrumental in preventing extinction of HTLV-1. However, the expansion of infected T cells does not occur without risk. In fact, in 1% to 2% of individuals infected with HTLV-1, ATLL, a fatal clonal disease, develops, presumably by means of accidental accumulation of somatic mutations of genes that regulate T-cell growth.4The ways in which HTLV-1 immortalizes T cells in vitro likely mirror the mechanisms used by the virus in vivo. In vitro, the viral transactivator Tax overrides normal mechanisms for controlling cell growth by affecting the activity of regulators of cell-cycle progression, such as p53,33-35p16INK4A,36-38 the Rb protein,39and MAD1,40 and by either suppressing expression of the cell-cycle regulators c-Myb41 and B-Myb42 or increasing expression of antiapoptotic proteins, including Bcl-X and Bcl-2. In addition, Tax appears to have a costimulatory effect on T cells by inducing dephosphorylation of nuclear factor of activated T cells 1,44 resulting in activation of CD28-responsive elements in the IL-2 promoter45 and the IL-2R α chain,46 which together with the β and γc chains, forms the high-affinity receptor for IL-2.
However, although Tax is important in preventing T-cell apoptosis and cell-cycle arrest, its immunogenicity in vivo47 may interfere with the ability of infected cells to survive immune recognition. The HTLV-1 p12I protein may play a crucial role in this regard. First, the ability of p12I to target the MHC I for degradation58 may markedly decrease the density of the MHC I/viral-peptide complexes (including Tax peptides) on the cell surface and render the infected cells invisible to recognition by cytotoxic T lymphocytes. Second, as demonstrated in this study, the ability of p12I to increase the basal level of STAT5 activation and T-cell responsiveness to IL-2, coupled with the costimulatory effect of Tax, may ultimately result in a limited burst of virus as well as limited replication of HTLV-1–infected T cells. Both the Tax costimulatory effect and the increased STAT5 activation induced by p12I appear to depend on antigen or mitogen stimulation, suggesting that cell-cycle entry of HTLV-1–infected T cells may be promoted in the presence of antigen stimulation in the microenvironment. In support of this hypothesis, epidemiologic studies have indicated that patients with concomitantStrongyloides stercoralis infection have a higher frequency of ATLL.48-51
Interestingly, HTLV-1 and HIV-1 have independently evolved 2 proteins (p12I and Nef) with similar cellular targets. Both p12I and Nef target subunits (although different) of the vacuolar H+ATPase14,52; p12I interferes with trafficking of MHC I heavy chain to the cell surface,58 whereas Nef down-regulates the MHC I that has already reached the cell surface, and both proteins increase IL-2 responsiveness (though by different mechanisms).9,53-55 In addition, ablation of Nef and p12I from molecular clones of simian immunodeficiency virus and HTLV-1, respectively, results in a similar phenotype, ie, their absence impairs viral infectivity in vivo, but their presence is not essential for viral infectivity or replication in tissue culture.56 57 Thus, the 3′ region of both human retroviruses encodes proteins (p12I and Nef) that help to preserve life-long infection of humans while increasing the likelihood of viral transmission.
We thank Steven Snodgrass for editorial assistance.
C.N. and J.C.M. contributed equally.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Genoveffa Franchini, National Cancer Institute, Basic Research Laboratory, 41/D804, Bethesda, MD 20892; e-mail:veffa@helix.nih.gov.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal