The D816V mutation of c-kit has been detected in patients with mastocytosis. This mutation leads to constitutive tyrosine kinase activation of Kit. Because stem cell factor (SCF), the ligand for Kit (CD117+), is a chemoattractant for CD117+ cells and one feature of mastocytosis is an abnormal collection of mast cells in tissues derived from CD34+CD117+ mast cell precursors, the hypothesis was considered that the D816V mutation would enhance chemotaxis of these precursor cells. Constructs encoding wild-type Kit or Kit bearing the D816V mutation were transfected into Jurkat cells, labeled with Calcein-am, and migration to SCF assessed in the presence or absence of tyrosine kinase inhibitors. Chemotaxis to SCF was enhanced in D816V transfectants compared to wild-type Kit transfectants (P < .002). Migration of both transfectants was inhibited by tyrosine kinase inhibitors, although D816V transfectants were more sensitive. Chemotaxis was next performed on CD34+CD117+ circulating mast cell precursors obtained from patients with mastocytosis. Analysis of prechemotaxis and migrated cells showed that whereas less than 10% in the prechemotaxis sample had the D816V mutation, 40% to 80% of migrated cells had this mutation. These results demonstrate that the D816V Kit mutation enhances chemotaxis of CD117+ cells, offering one explanation for increased mast cells observed in tissues of patients with mastocytosis.
Introduction
Signaling through Kit, the receptor for stem cell factor (SCF), is crucial for the proliferation of CD34+CD117+ mast cell precursors as well as their subsequent migration into tissues where they differentiate into mast cells.1-8 Activating mutations in Kit (D816V and V560G; single-letter amino acid codes) have been described in the human mast cell leukemia cell line HMC-1.9 In addition, activating mutations corresponding to D816V in the murine mastocytoma cell line p815 (D814Y)10 and in the rat RBL-2H3 mast cell line (D817Y)11 have been identified. These mutations are associated with the constitutive tyrosine kinase activation and phosphorylation of Kit as well as cell proliferation in the absence of the Kit ligand, SCF.9 12-14
The D816V mutation first reported in the peripheral blood mononuclear cells of patients with mastocytosis with an associated hematologic disorder15,16 has now been found in the urticaria pigmentosa lesional skin of all adult patients and a subset of children with severe disease.17,18 Because mastocytosis is a disease characterized by increased mast cell numbers in the skin, spleen, liver, and bone marrow, and because the D816V mutation is associated with increased cell proliferation, it has been concluded that one effect of the mutation is to increase mast cell proliferation in patients with mastocytosis who have this mutation in hematopoietic lineage cells.15-18
However, increased proliferation of mast cell precursors alone would not appear to account for increased tissue mast cell numbers, particularly in the skin at sites of lesions of urticaria pigmentosa where the D816V mutation may be identified even in circumstances where it cannot be identified in marrow or peripheral blood. One alternative explanation would be a preferential accumulation of cells bearing the D816V mutation at these skin sites.19 We thus hypothesized that the D816V mutation in Kit enhances cell migration of CD34+CD117+ circulating mast cell precursors over that observed dependent on wild-type Kit.
As will be shown, the activating Kit mutation D816V does, as hypothesized, enhance cell migration to the Kit ligand, SCF. This enhanced migration was demonstrated both in transfected Jurkat cells encoding the D816V mutation and in CD34+CD117+circulating mast cell precursors isolated from patients with mastocytosis. These findings thus provide a companion explanation for the excessive numbers of mast cells in lesional tissues of patients with mastocytosis and suggests that mastocytosis is, in part, a disease of disordered cell trafficking.
Materials and methods
Reagents and antibodies
Calcein-am was purchased from Molecular Probes (Portland, OR); Amplitaq DNA polymerase and dNTPs from PerkinElmer (San Francisco, CA); Taqstart antibody from Clontech (Palo Alto, CA); andHinfI restriction enzyme from New England Biolabs (Beverly, MA); recombinant human SCF (rhSCF), stromal cell–derived factor-1α (SDF-1α), recombinant human interleukin (rhIL)-3 and rhIL-6 from Peprotech (Rocky Hill, NJ); phycoerythrin (PE) antihuman CD117 (clone 5745) and an IgG1 isotype-matched control (clone MOPC 31C) from Ancell (Bayport, MN); and antihuman CD34 antibody (clone 561) from Dynal (Oslo, Norway).
Construction of plasmid complementary DNA3-kitD816V
The vectors plasmid complementary DNA (pcDNA3)-kitwt and pAlter-kitwt were a kind gift from Dr L. Rönnstrand (Ludwig Institute for Cancer Research, Uppsala, Sweden). Construction of pAlter-kitD816Vwas achieved by site-specific mutagenesis of the pAlter-kitwt vector using the oligonucleotide: 5′-CTA GCC AGA GTC ATC AAG AAT G-3′ (mutated base is underlined) according to manufacturer's instructions (Altered Sites II in vitro Mutagenesis System, Promega, Madison, WI). The mutation was confirmed by sequencing of the pAlter-kitD816V vector using an ABI 310 Genetic Analyzer (PerkinElmer, Branchburg, NJ). A 443-bpAccIII, BglII fragment from the pAlter-kitD816V vector was subcloned into pcDNA3-kitwt creating pcDNA3-kitD816V.
Transient transfection of Jurkat cells
The human T-cell leukemia cell line, Jurkat, was maintained in complete medium comprising RPMI 1640 medium supplemented withl-glutamine (2 mM), penicillin (100 U/mL), streptomycin (100 μg/mL), and 10% heat-inactivated fetal calf serum (FCS) prior to transfection. Jurkat cells (4.0 × 107/0.4 mL RPMI) were transiently transfected with 50 μg pcDNA3-kitwt, pcDNA3-kitD816V, and the vector pcDNA-3 by electroporation (250 V, 960 μF capacitance, pulse time 29-30 ms) using a gene pulser (Bio-Rad, Richmond, CA). Following electroporation, the transfected cells were transferred to complete medium and incubated overnight at 37°C in a 5% CO2 incubator. Viabilities of transfectants ranged from 60% to 80%.
Determination of transfection efficiency by flow cytometric analysis
Transfection efficiency of the D816V and wild-type Kit transfectants was assessed by measuring Kit (CD117+) surface expression by flow cytometry. Briefly, immunofluorescence staining was performed by incubating transfected cells (1 × 106) with either PE-conjugated antihuman CD117 or an IgG1 isotype–matched control for 30 minutes at 4°C. After washing and resuspension in cold phosphate-buffered saline (PBS)/0.1% bovine serum albumin (BSA), the cells were analyzed on a FACScan (Becton Dickinson, San Jose, CA). CD117+ expression on transfectants was compared to a CD117+-expressing human mast cell leukemia cell line, HMC-1.2.
Chemotaxis of Jurkat transfectants
Jurkat transfectants were labeled with the intracellular fluorescent dye, calcein-am, at 1 μM for 45 minutes at 37°C in a 5% CO2 incubator. Following labeling, cells were washed twice with PBS. A concentration of 2 × 105cells in 50 μL migration buffer (RPMI without phenol red, supplemented with 0.5% BSA) were loaded on the top filter of the chemotaxis chamber (96-well Chemotx, 5-μm pore size, 6-mm diameter, Neuroprobe, Gaithersburg, MD). Increasing concentrations of rhSCF (1-400 ng/mL) or SDF-1α (1-400 ng/mL; ligand for the CXCR4 chemokine receptor, which is endogenously expressed on Jurkat cells) were placed in the bottom wells of the chemotaxis chamber. After incubation for 3 hours at 37°C in a 5% CO2 incubator, fluorescence of cells migrating into the bottom chamber was measured using a fluorescence reader (excitation filter 485 nm and emission filter 535 nm) (HTS 7000 Bio Assay Reader, PerkinElmer, San Francisco, CA). Percent migration was calculated by expressing the fluorescence of migrated cells as a percentage of the fluorescence of the total number of cells loaded on the top filter of the chemotaxis chamber.
Chemokinesis was assessed by standard analysis using the optimal SCF concentration for migration (10 ng/mL). In selected experiments, Jurkat cells labeled with calcein-am were incubated with the tyrosine kinase inhibitors Genistein and Tyrphostin AG1296 (Calbiochem, San Diego, CA) for 30 minutes prior to performing the chemotaxis assay.
Isolation and culture of CD34+CD117+ mast cell precursors from patients with mastocytosis
Peripheral blood mononuclear cells from 2 patients with indolent mastocytosis16 (collected by leukapheresis) and known to have the D816V mutation were obtained and processed after informed consent. Progenitor CD34+CD117+ cells from which mast cells are derived were enriched by immunomagnetic positive selection (Miltenyi Biotec, Auburn, CA).20 Approximately 45% of the enriched cell population consisted of CD34+/CD117+ cells as assessed by FACS analysis (Becton Dickinson). CD34+CD117+cells were then cultured in media consisting of Stempro-34 with nutrient supplement (Life Technologies, Grand Island, NY), l-glutamine (2 mM), penicillin (100 U/mL), streptomycin (100 μg/mL), rhIL-3 (30 ng/mL), rhIL-6 (100 ng/mL), and rhSCF (100 ng/mL). CD34+CD117+ cells were harvested for the chemotaxis assay after 1 and 3 days of culture.21
Chemotaxis of CD34+CD117+ cells
The CD34+CD117+ cells were washed twice with Stempro-34 media without IL-3, IL-6, and SCF. The cells were then suspended in Stempro-34 media supplemented with 0.5% BSA (migration buffer). A total of 2.5 × 104 cells in 25 μL migration buffer was loaded on the top filter of the chemotaxis chamber (96-well Chemotx, 5-μm pore size, 3.2-mm diameter, Neuroprobe). The optimal concentration for SCF (10 ng/mL) in 30 μL was added to the bottom wells of the chemotaxis chamber. Cells were left to migrate for 3 and 6 hours at 37°C in a 5% CO2 incubator. The migrated cells were then collected from the bottom wells of the chemotaxis chamber. Nested polymerase chain reaction (PCR) followed by HinfI restriction digestion was performed on the migrated cells as described below.
Nested PCR, restriction digestion, and quantitation of D816V mutation by gel fluorescence imaging
A 111-bp region of c-kit DNA spanning D816V was amplified by nested PCR. The primers Kit-1-F (TCCTCCAACCTAATAG TGTATTCACAG) and Kit–1-R (TTTGCAGGACTGTCAAGCAGAGAATG) were used for the first round and Kit–2-F (TATCCTCCTTACTCATGGTCGG) and Kit-2-R (AGAGAATGGGTACTCACGTTTCC) for the second round of amplification. The PCR was performed in a final volume of 50 μL containing 50 mM KCl, 10 mM Tris-HCl, 200 μM of each dNTP, 5 U Amplitaq DNA polymerase and 0.5 μM of each primer. One microliter of the PCR product from the first round was used as the template for the second round of PCR. The rest of the components in the reaction mixture were the same as described above for the first round. The same temperature conditions were used for the first and second round touchdown PCR. There was an initial 95°C 1-minute denaturation step followed by denaturation at 94°C for 1 minute and an extension at 72°C for 2 minutes; for the first cycle, annealing was at 68°C for 30 seconds, which dropped down to 58°C at the end of the fifth cycle at the rate of 2°C/cycle. The annealing temperature for the remaining 23 cycles was 58°C. The PCR ended with a 5-minute extension at 72°C. The PCR was performed on a PTC-100 programmable thermal cycler (MJ Research, Watertown, MA).
To detect the mutation that creates an additional HinfI site, the second-round PCR product was digested with the restriction enzyme HinfI for 2 hours at 37°C. The digested product was electrophoresed on a 20% tris-borate EDTA (TBE) polyacrylamide gel (Novex, San Diego, CA). Predicted sizes for the wild-typec-kit sequence were 68 and 43 bp and for the heterozygous mutated sequence were 68, 54, 43, and 14 bp. Because the mutation is found on only one of the 2 alleles, cells with mutations also have the wild-type fragment of 68 and 43 bp generated from the wild-type copy of the c-kit gene. Control cell lines, HMC-1.1 (without the Kit D816V mutation) and HMC-1.2 (heterozygous for the Kit D816V mutation) were analyzed in parallel.
The proportion of cells bearing the D816V mutation in both the starting and migrated cells was calculated following gel fluorescence imaging (Gel Expert, Nucleotech, San Carlos, CA) of the restriction enzyme digestion products. A ratio was obtained by dividing the fluorescence of the 54-bp band with that of the 68-bp band. This value was then expressed as a percentage of the ratio of fluorescence of the positive control HMC-1.2.
Statistical analysis
Values are presented as mean ± SEM. Comparative analysis was achieved using the unpaired 2-tailed t test using statistical software (Statview, Abacus Concepts, Berkeley, CA). AP value of .05 or less was considered significant.
Results
Assessment of transfection efficiency by flow cytometric analysis
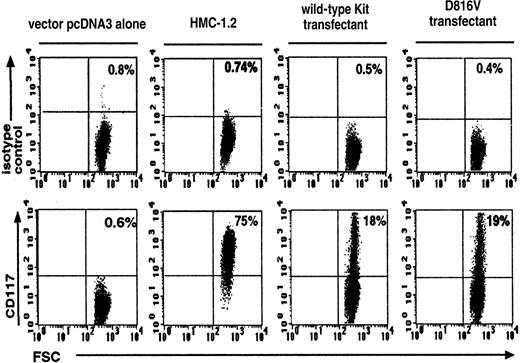
Efficiency of transfection was assessed by measuring CD117+ expression on wild-type Kit– and D816V-transfected Jurkat cells. There were similar percentages of CD117+expression on both the wild-type Kit (18% ± 1.8%, n = 3) and the D816V (19% ± 1.3%, n = 3) transfectants (Figure1). As expected, Jurkat cells transfected with the vector pcDNA-3 alone expressed background levels of CD117+ (0.6% ± 0.01%, n = 3). The corresponding value for the HMC-1.2 cell line was 75% ± 2.2% (n = 3).
Flow cytometric analysis of Kit expression on wild-type Kit– and D816V-transfected Jurkat cells.
Transfection efficiency of D816V and wild-type Kit transfectants were compared with Jurkat cells transfected with the vector pcDNA-3 alone and the Kit-expressing cell line HMC-1.2. The upper panel represents cells stained with the IgG1 isotype control and the lower panel, cells stained with Kit monoclonal antibody. FSC indicates forward scatter. Numbers shown in each quadrant refer to the percentages of cells bearing surface Kit. Data are representative of 3 separate experiments.
Flow cytometric analysis of Kit expression on wild-type Kit– and D816V-transfected Jurkat cells.
Transfection efficiency of D816V and wild-type Kit transfectants were compared with Jurkat cells transfected with the vector pcDNA-3 alone and the Kit-expressing cell line HMC-1.2. The upper panel represents cells stained with the IgG1 isotype control and the lower panel, cells stained with Kit monoclonal antibody. FSC indicates forward scatter. Numbers shown in each quadrant refer to the percentages of cells bearing surface Kit. Data are representative of 3 separate experiments.
SCF-dependent chemotaxis of transfected cells
We next determined the chemotaxis responses of the wild-type Kit and D816V transfectants to SCF compared to SDF-1α (Figure2). There was a significant enhancement of migration to SCF of the D816V transfectants, peaking at 18.2% ± 1.9% (n = 3; 10 ng/mL) compared to the wild-type Kit transfectants (9.3% ± 1.0%, n = 3; P < .002; Figure 2A). As expected, there was minimal migration to SCF by Jurkat cells transfected with the pcDNA-3 vector alone (1.4% ± 0.18%; n = 3). In contrast, migration of Jurkat cells to SDF-1α was comparable in the wild-type Kit and D816V transfectants, as well as in Jurkat cells transfected with the pcDNA-3 vector alone (Figure 2B). To determine whether the differences in migration to SCF observed between the D816V and wild-type Kit transfectants were due to random migration, we performed control chemokinesis experiments. Briefly, standard analysis using the optimal SCF concentration for cell migration (10 ng/mL) was performed. The results confirmed that the enhanced migration observed in D816V transfectants was due mainly to chemotaxis and not chemokinesis, because significant random migration was not demonstrated when SCF was added either to the top well or to both the top and bottom wells of the chemotaxis chamber (Figure 2C). However, there was more chemokinesis in Jurkat cells expressing the mutant receptor compared to the wild-type Kit as would be expected when comparing a receptor with an activating mutation to the wild-type receptor.
Chemotaxis responses of transfectants.
The cell migration of wild-type kit (●), D816V (♦), and pcDNA-3 (vector alone) transfectants (▪) to (A) stem cell factor (SCF) and (B) stromal cell derived factor 1-α (SDF-1α). Data are presented as mean ± SEM (n = 3). Asterisks in panel A represent values that were significant at P ≤ .05 when migration of D816V and wild-type Kit transfectants were compared. (C) Comparison of chemokinesis in D816V (▪), wild-type Kit (■), and pcDNA-3 transfectants (▧). Data are presented as mean ± SEM (n = 2).
Chemotaxis responses of transfectants.
The cell migration of wild-type kit (●), D816V (♦), and pcDNA-3 (vector alone) transfectants (▪) to (A) stem cell factor (SCF) and (B) stromal cell derived factor 1-α (SDF-1α). Data are presented as mean ± SEM (n = 3). Asterisks in panel A represent values that were significant at P ≤ .05 when migration of D816V and wild-type Kit transfectants were compared. (C) Comparison of chemokinesis in D816V (▪), wild-type Kit (■), and pcDNA-3 transfectants (▧). Data are presented as mean ± SEM (n = 2).
Effect of tyrosine kinase inhibitors on migration to SCF of transfected cells
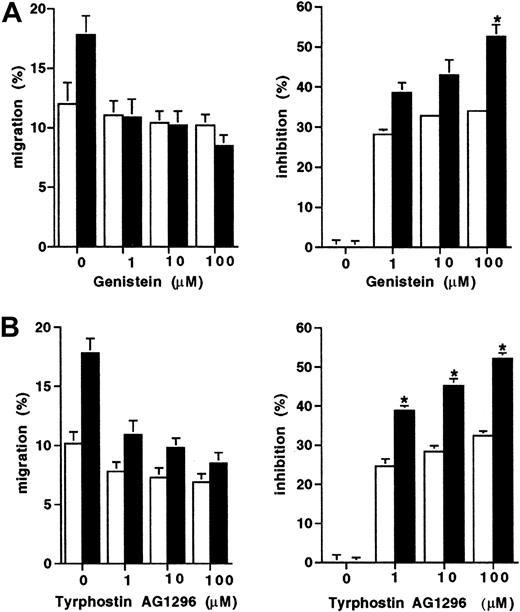
Because the D816V mutation results in constitutive activation of the tyrosine kinase activity of Kit, we next assessed whether the enhanced migration displayed by D816V transfectants would be differentially effected by tyrosine kinase inhibitors. Migration to SCF of both the D816V and wild-type Kit transfectants was partially inhibited by both tyrosine kinase inhibitors Genistein and Tyrphostin AG 1296 (Figure 3). However, migration of the D816V transfectants was inhibited to a greater degree by both Genistein (100 μM; 52% versus 33%; Figure 3A;P < .009) and Tyrphostin AG 1296 (100 μM; 53% versus 31%; Figure 3B; P < .007) compared to the wild-type Kit transfectants (n = 2).
Inhibition of migration to SCF of transfectants.
The effect of tyrosine kinase inhibitors expressed both as percent migration and percent inhibition (A) Genistein (1-100 μM) and (B) Tyrphostin AG1296 (1-100 μM) on cell migration to SCF (10 ng/mL) in D816V (▪), and wild-type Kit transfectants (■). Data are presented as mean ± SEM (n = 2). When error bars are not shown, the SEM was too small to be diagrammed. Asterisks represent values that were significant at P ≤ .05.
Inhibition of migration to SCF of transfectants.
The effect of tyrosine kinase inhibitors expressed both as percent migration and percent inhibition (A) Genistein (1-100 μM) and (B) Tyrphostin AG1296 (1-100 μM) on cell migration to SCF (10 ng/mL) in D816V (▪), and wild-type Kit transfectants (■). Data are presented as mean ± SEM (n = 2). When error bars are not shown, the SEM was too small to be diagrammed. Asterisks represent values that were significant at P ≤ .05.
Preferential migration to SCF of mutant CD34+CD117+ cells from patients with mastocytosis
To determine whether these observations are clinically relevant in mastocytosis, we performed chemotaxis of CD34+CD117+ cells from which mast cells are derived. Samples taken from 1- and 3-day cultures of CD34+CD117+ cells from patients with mastocytosis were analyzed for the presence of the D816V mutation both in the starting prechemotaxis sample as well as 3 and 6 hours after chemotaxis. A schematic diagram of the HinfI restriction sites in the 111-bp PCR product from HMC-1.1 and HMC-1.2 as well as the predicted sizes for the HinfI restriction products from the positive control cell line HMC-1.2 (68, 54, 43, and 14 bp) and wild-type control cell line HMC-1.1 (68 and 43 bp) are shown in Figure4A. HinfI restriction digestion of the PCR product from the positive control HMC-1.2 (heterozygous for Kit D816V mutation) revealed 3 bands at 68, 54, and 43 bp (Figure 4B; the 14-bp band is not shown in this figure) confirming the presence of the mutation, whereas 2 bands were detected in the wild-type control cell line HMC-1.1 (without the Kit D816V mutation) at 68 and 43 bp. Mutant cells were below the limits of detection in the prechemotaxis day 1 and 3 cultured CD34+CD117+-enriched cells, consistent with published data on the presence of the D816V mutation in this cell population.22 However, analysis of PCR products from migrated cells at both 3 and 6 hours following HinfI digestion of CD34+CD117+ cells revealed the presence of the mutation after chemotaxis in both 1- and 3-day cultures. Moreover, mutated cells were not detected in the upper chamber after chemotaxis (data not shown).
Preferential migration of CD34+CD117+ cells with the D816V mutation.
(A) A schematic diagram of the HinfI restriction sites in the 111-bp PCR product from HMC-1.1 and HMC-1.2 control cell lines. Predicted sizes for the sequence from the wild-type control cell line HMC-1.1 (without the kit D816V mutation) are 68 and 43 bp and for the positive control cell line HMC-1.2 (heterozygous for the Kit D816V mutation) are 68, 54, 43, and 14 bp. (B) RepresentativeHinfI restriction digestion and quantification by gel fluorescence imaging of the D816V mutation in CD34+CD117+ cells obtained from a patient with mastocytosis compared to control cell lines HMC-1.1 (wt) and HMC-1.2 (D816V) (the 14-bp band is not shown in this figure). CD34+CD117+ cells were harvested after 1 and 3 days of culture and analyzed for the D816V mutation in the starting population and in the bottom chamber at 3 and 6 hours following chemotaxis to SCF (10 ng/mL). An experiment using cells from a second patient yielded similar results. The percent total cell migration for each population was HMC1.2, 7.5%; HMC1.1, 5.7%; CD34+CD117+, day 1, 3 hours, 6.5%; CD34+CD117+, day 1, 6 hours, 8%; CD34+CD117+, day 3, 3 hours, 6.3%; CD34+CD117+, day 3, 6 hours, 13.8%.
Preferential migration of CD34+CD117+ cells with the D816V mutation.
(A) A schematic diagram of the HinfI restriction sites in the 111-bp PCR product from HMC-1.1 and HMC-1.2 control cell lines. Predicted sizes for the sequence from the wild-type control cell line HMC-1.1 (without the kit D816V mutation) are 68 and 43 bp and for the positive control cell line HMC-1.2 (heterozygous for the Kit D816V mutation) are 68, 54, 43, and 14 bp. (B) RepresentativeHinfI restriction digestion and quantification by gel fluorescence imaging of the D816V mutation in CD34+CD117+ cells obtained from a patient with mastocytosis compared to control cell lines HMC-1.1 (wt) and HMC-1.2 (D816V) (the 14-bp band is not shown in this figure). CD34+CD117+ cells were harvested after 1 and 3 days of culture and analyzed for the D816V mutation in the starting population and in the bottom chamber at 3 and 6 hours following chemotaxis to SCF (10 ng/mL). An experiment using cells from a second patient yielded similar results. The percent total cell migration for each population was HMC1.2, 7.5%; HMC1.1, 5.7%; CD34+CD117+, day 1, 3 hours, 6.5%; CD34+CD117+, day 1, 6 hours, 8%; CD34+CD117+, day 3, 3 hours, 6.3%; CD34+CD117+, day 3, 6 hours, 13.8%.
The proportion of mutated cells both in the starting population (prechemotaxis) as well as the migrated cells (after chemotaxis) from both day 1 and 3 cultures was estimated following gel fluorescence imaging. The ratio of the fluorescence intensity of the 54-bp band and the 68-bp band was calculated and expressed as a percentage of the ratio obtained for HMC-1.2, the positive control. We concluded that whereas less than 10% (< ratio of 0.05) mutated cells were detectable in the prechemotaxis sample (both day 1 and 3 cultures), the sample after chemotaxis contained 40% to 80% (ratio of 0.18-0.42) of mutated cells (both day 1 and 3 cultures; Figure 4B), demonstrating preferential migration to SCF of cells bearing mutated Kit.
Discussion
The D816V mutation is the most commonly detected activating Kit mutation in patients with mastocytosis, a disease characterized by abnormal proliferation and subsequent accumulation of mast cells in various tissues. In the present study, we have demonstrated that this activating mutation enhances cell migration to its ligand, SCF. This was first demonstrated in Jurkat cells (Figure 1) transfected with Kit bearing the D816V mutation. SCF induced a standard bell-shaped dose-dependent chemotactic response in both D816V and wild-type Kit transfectants (Figure 2A). However, SCF induced an enhanced chemotactic response in the D816V compared to the wild-type Kit transfectants. In contrast, there was comparable migration to SDF-1α (Figure 2B), the ligand for the endogenously expressed CXCR4 chemokine receptor in Jurkat cells transfected with the D816V, wild-type Kit, or pcDNA-3 vector. Moreover, control experiments confirmed that the differences in cell migration observed between the D816V and wild-type Kit transfectants were due mainly to chemotaxis and not chemokinesis although chemokinesis was enhanced to some degree in Jurkat cells expressing the mutant compared to the wild-type Kit as would be expected (Figure 2C).
To assess whether the enhanced tyrosine kinase activity conferred by the D816V mutation is responsible for the observed effects on chemotaxis, we compared the effect of tyrosine kinase inhibition on cell migration in both D816V and wild-type Kit transfectants (Figure3). Although both transfectants were partially inhibited by the tyrosine kinase inhibitors Genistein and Tyrphostin AG1296, suggesting that both the wild-type and mutant transfectant use the same signal transduction pathway, the D816V transfectants were more sensitive to the inhibitory effect. This is consistent with evidence from murine studies that activating Kit mutations similarly enhance tyrosine kinase signaling.23-25 Previous work from our laboratory has similarly demonstrated that CD34+ mast cell precursors from patients with mastocytosis are hypersensitive to SCF, in that CD34+ precursors from patients with mastocytosis (when cultured in SCF) give rise to more mast cells per CD34+ cell plated than when CD34+precursors are obtained from normal donors.26
To extend the observation of enhanced cell migration to SCF by the D816V transfectants to patients with mastocytosis, chemotaxis was performed on CD34+CD117+ cells enriched from the peripheral blood of these patients. Although the mutated cultured CD34+CD117+ cells were minimally detected (< 10%) in the starting population, between 40% and 80% of the migrated cells carried the mutation at both 3 and 6 hours following cell migration to SCF (Figure 4). Moreover, mutated cells were not detected in the upper chamber following chemotaxis (data not shown). These results demonstrate preferential migration of CD34+CD117+ mast cell precursors bearing the activating Kit mutation D816V to SCF and support our findings in transfected cells. Furthermore, the short time frame in which these experiments were performed (3-6 hours) precludes local proliferation as a plausible explanation for the increased numbers of CD34+CD117+ mast cell precursors found in the bottom wells of the chemotaxis chamber.
In view of these data, we speculate that in patients with mastocytosis, CD34+CD117+ mast cell precursor cells bearing the D816V mutation preferentially migrate to SCF produced by stroma cells, endothelial cells, fibroblasts, and keratinocytes found in tissues such as the skin. Differentiation of the mutant CD34+CD117+ progenitor cells into mature mast cells then occurs as a result of the microenvironment. Thus, enhanced cell migration in addition to aberrant proliferation, the other known effect of the D816V mutation, may contribute to the extensive mast cell hyperplasia observed in these tissues.
In conclusion, our studies demonstrate enhanced migration of cells bearing the activating Kit mutation D816V both in a transfected cell model and in human hematopoietic mast cell progenitor cells isolated from patients with mastocytosis. These findings offer a plausible mechanism in addition to aberrant proliferation for excessive mast cell numbers at tissue sites that produce SCF.
Supported by Fogarty Fellowship, National Institutes of Health, and the Swedish Cancer Society. M.S. and G.N. are supported by the Swedish Cancer Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Marcia L. Taylor, Laboratory of Allergic Diseases, NIAID, NIH, Bldg 10, Rm 11C207, 10 Center Dr, MSC1881, Bethesda, MD 20892-1881; e-mail: mtaylor@niaid.nih.gov.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal