Development of inhibitory antibodies is a serious complication of treatment with repeated factor IX infusions in a minority of patients with hemophilia B. Such antibodies detected in 8 patients have been characterized. Typing studies revealed that patients' immune response toward factor IX is highly heterogeneous and involves immunoglobulin G (IgG) antibodies, preferentially IgG1 and IgG4. The preservation of the sequence and the 3-dimensional orientation of the amino acids constituting one epitope are highly important for the assembly of an antibody-antigen complex. To localize the epitopes on the factor IX molecule, an original approach was designed using a set of factor X chimeras carrying regions of factor IX. Results showed that some patients' antibodies were directed against both the domain containing the γ-carboxy glutamic acid residues (Gla domain) and the protease domain of factor IX. In contrast, no binding was observed to the epidermal growth factor–like domains or to the activation peptide. Functional characterization showed that the purified IgG from patients' serum inhibited the factor VIIIa-dependent activation of factor X. Moreover, patients' IgG directed against the Gla domain inhibited the binding of factor IX to phospholipids as well as the binding of factor VIII light chain to factor IXa. These data demonstrate that inhibitors appearing in patients with severe hemophilia B display specificity against restricted functional domains of factor IX.
Introduction
Blood coagulation is a response to vascular injury leading to platelet activation and sequential activation of coagulation factors, including factor IX (FIX), to their enzyme forms.1 The blood clotting reactions result in the formation of a fibrin plug. During blood coagulation, activated FIX (FIXa) associates with its activated cofactor, factor VIIIa (FVIIIa), converts its specific substrate factor X (FX) into its activated derivative, FXa. This reaction occurs in the presence of calcium ions at the surface of an appropriate membrane, principally provided by activated platelets.
FIX belongs to the vitamin K-dependent proteins, including factor VII, FX, and protein C. These proteins share a common gene organization in which exons encode distinct domain structures, indicating a typical divergent evolution involving a number of mutations after exon-shuffling events.2-4 Therefore, FIXa, FXa, FVIIa, and activated protein C are highly homologous, both in domain organization and 3-dimensional structure.5
At the amino-terminal end, FIX contains the Gla domain that encompasses Ca++-binding γ-carboxy glutamic acid (Gla) residues. FIX binds to activated platelets via the amino terminal residues 3-11 in the Gla domain.6 Two epidermal growth factor (EGF)-like domains follow the Gla domain, the first of which contains a high affinity Ca++-binding site.7 The second EGF-like domain is connected to an activation peptide that is released during zymogen activation. Finally, the carboxy-terminal serine protease domain contains a single metal ion-binding site.8
Patients with the X-linked bleeding disorder hemophilia B show qualitative and/or quantitative defects in FIX. The deficiency affects 1 in 30 000 males and is sporadic in a third of the cases. Patients affected with the severe form of the disease suffer from bleeding in joints, muscles, and soft tissues. They are treated for bleeding episodes by infusion of purified plasma or recombinant FIX concentrates. A serious complication of the FIX replacement therapy in multitransfused patients with hemophilia B is the development of antibodies that inhibit FIX coagulant activity. It has been established that such inhibitory antibodies (inhibitors) arise in 1% to 4% of the treated patients with severe hemophilia B (FIX activity < 0.01 IU/mL).9-12
The mechanism involved in inhibitor development in hemophilia B remains poorly documented and understood compared with hemophilia A particularly with regard to inhibitor recognition sites and inhibitory properties. The present study was directed to gain more insight into these aspects of FIX inhibitors. Exon shuffling between FIX and FX complementary DNA (cDNA) to obtain recombinant FX chimeric proteins carrying specific domains of FIX was used to study the epitopes recognized by FIX antibodies in greater detail. These chimeras allowed the identification of 2 regions recognized by patients' immunoglobulins G (IgGs), the Gla and protease domains. Functional analysis of patients' inhibitor showed that these antibodies inhibit the interaction of FIX with its cofactors, FVIIIa and procoagulant phospholipids. This study combined with the known structure-function relationships of FIX allows a better understanding of the action of inhibitors at the molecular level.
Patients, materials, and methods
Materials
Protein G Sepharose 4 Fast Flow and CNBr Sepharose CL-4B were from Amersham Pharmacia Biotech AB (Uppsala, Sweden). Polystyrene microtiter plates were from Nunc A/S (Rosklide, Denmark). Restriction enzymes, geneticin, and other antibiotics were obtained from Life Technologies (Cergy Pontoise, France). Dulbecco modified Eagle medium (DMEM), Fungizone, L-α-phosphatidyl-L-serine, and L-α-phosphatidylcholine were purchased from Sigma (St Louis, MO). Fetal calf serum (FCS) was from Bio-Whittaker Europe (Emerainville, France). Pfu polymerase was obtained from Stratagene (Cambridge, United Kingdom). Oligonucleotide primers were from Isogen Bioscience BV (Maarssen, The Netherlands).
Patients' characteristics
Eight unrelated multitransfused patients with severe hemophilia B who developed inhibitors against FIX were analyzed (Table1). FIX inhibitor titers were measured by the Bethesda assay.13 One Bethesda unit (BU) is defined as the amount of inhibitory activity of patient plasma that produces 50% inhibition of FIX activity in a one-stage assay. All patients were high responders with more than 5 BU/mL.
Characteristics of patients
| Patient no. . | Gene analysis . | Inhibiter titer (BU/mL) . | Allergic reactions . | |
|---|---|---|---|---|
| Peak . | Present study . | |||
| 1 | Arg248Stop | 64 | 44 | No |
| 2 | Gene deletion exon4-exon8 | > 100 | 26-40 | Yes |
| 3 | Not determined | 170 | 100 | No |
| 4 | Leu57Stop | 140 | 42 | No |
| 5 | Complete gene deletion | 19 | 7 | No |
| 6 | Asp152Stop | > 100 | 26 | Unknown |
| 7 | Not determined | 103 | 17 | Yes (lethal) |
| 8 | Complete gene deletion | 130 | 12 | Yes |
| Patient no. . | Gene analysis . | Inhibiter titer (BU/mL) . | Allergic reactions . | |
|---|---|---|---|---|
| Peak . | Present study . | |||
| 1 | Arg248Stop | 64 | 44 | No |
| 2 | Gene deletion exon4-exon8 | > 100 | 26-40 | Yes |
| 3 | Not determined | 170 | 100 | No |
| 4 | Leu57Stop | 140 | 42 | No |
| 5 | Complete gene deletion | 19 | 7 | No |
| 6 | Asp152Stop | > 100 | 26 | Unknown |
| 7 | Not determined | 103 | 17 | Yes (lethal) |
| 8 | Complete gene deletion | 130 | 12 | Yes |
Preparation of serum and purification of patients' IgG
Plasma was obtained from patients and normal volunteers, 15 unrelated healthy individuals, in accordance with the Declaration of Helsinki. Informed consent of the patients or family to perform research studies was obtained. Plasma was recalcified by adding 25 mM CaCl2 at room temperature. After adding 5 U/mL of purified thrombin and incubation at 37°C for 30 minutes to induce clotting, the serum was isolated by centrifugation at 12 000g for 15 minutes at room temperature, and IgG was isolated by affinity chromatography on protein G Sepharose 4 Fast Flow equilibrated with 25 mM Tris-HCl, pH 7.4, 150 mM NaCl. After extensive washes, immobilized IgG was eluted with 0.1 M Glycine-HCl, pH 2.5, and collected by 1-mL fractions in tubes containing 100 μL 1M Tris-HCl, pH 9.0. IgG was dialyzed against 25 mM Tris-HCl, pH 7.4, 150 mM NaCl and stored at −20°C, and its concentration was calculated from the absorbance at 280 nm by using an extinction coefficient of 1.4, and its purity was analyzed by sodium dodecyl sulfate 10% polyacrylamide gel electrophoresis.
Recombinant human wild-type FX
Human FX cDNA was isolated from a human liver cDNA library from Clontech (Leyden, The Netherlands) by polymerase chain reaction (PCR) using 2 oligonucleotides, one complementary to the 5′ end of FX cDNA, and the other complementary to the 3′ end. BamHI and HindIII restriction sites were appended to the PCR product by the 2 oligonucleotides to allow the insertion of the amplification product into a vector. The codon corresponding to the −2 residue of FX was changed (ACG → AGG, Thr → Arg) to allow correct cleavage of the propeptide as previously described.14 The entire wild-type cDNA sequence was verified using the ABI PRISM Dye Terminator Cycle Sequencing Reaction Kit (PE Biosystems, Courtaboeuf, France) on an ABI PRISM 373 DNA sequencer according to the manufacturer's specifications. Madin-Darby canine kidney (MDCK) cells were grown in DMEM supplemented with 10% FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Transfection was performed by using the calcium phosphate precipitation method. After selection of transfected cells with medium containing geneticin at a concentration of 800 μg/mL, single clones were picked and propagated in selective medium to obtain stable cell lines. Production of FX antigen was assayed by enzyme-linked immunosorbent assay (ELISA) and cell lines producing FX were selected.
Recombinant FX chimera constructions
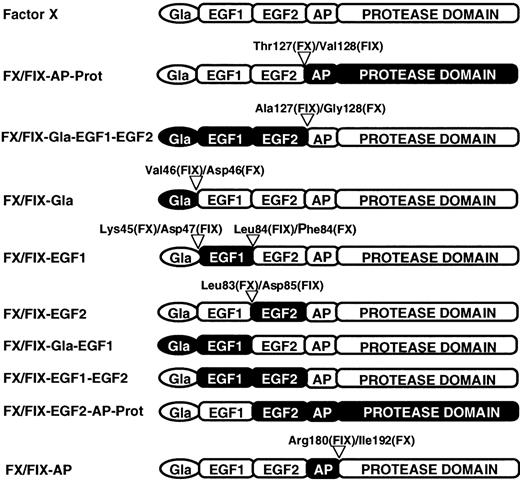
The mammalian expression plasmid pKG5 containing either human FIX cDNA15 or human FX cDNA (see above) was used as a template to construct full-length cDNAs encoding FX/FIX chimeras. Mutagenesis was performed by a PCR-based method,16,17using partially overlapping primers consisting of hybrid oligonucleotides of FX and FIX cDNAs. Primer sequences were selected on the basis of the highly conserved intron-exon boundaries amongFX and FIX genes and on the basis that exons of FIX and FX genes roughly correspond to specific functional domains of the proteins.2 3 Exon shuffling between FIX and FX cDNA by this approach lead to recombinant FX/FIX chimeric proteins carrying specific domains of FIX. The obtained chimeras possess domain borders as specified in Figure1. The FIX and FX domains are expressed by the chimera names, ie, FX/FIX-Gla-EGF1-EGF2 contains the Gla domain, and the 2 EGF-like domains of FIX or FX/FIX-AP-Prot, its counterpart, contain the activation peptide and the protease domain of FIX (Figure 1).
Schematic representation of FX chimera constructs.
There are 8 recombinant chimeric proteins. The protein domains are as follows: γ-carboxy glutamic acid rich (Gla), epidermal growth factor-like (EGF1 and EGF2), activation peptide (AP), and protease. FX and domains of FX are in white, whereas FIX domains are in black. Within the chimeras, numbers indicate the specific adjacent residues from FIX and FX that comprise the exchange sites.
Schematic representation of FX chimera constructs.
There are 8 recombinant chimeric proteins. The protein domains are as follows: γ-carboxy glutamic acid rich (Gla), epidermal growth factor-like (EGF1 and EGF2), activation peptide (AP), and protease. FX and domains of FX are in white, whereas FIX domains are in black. Within the chimeras, numbers indicate the specific adjacent residues from FIX and FX that comprise the exchange sites.
Proteins
Mouse monoclonal anti-FIX antibodies CLB-FIX 11 and CLB-FIX 14 have been described previously18 as well as anti-FVIII light chain antibodies CLB-CAg-12 and CLB-CAg-117.19Polyclonal antibodies against FIX were obtained as described.20 Antibodies were conjugated with horseradish peroxidase. Polyclonal antibodies against FX conjugated or not with horseradish peroxidase were obtained from DAKO (Dakopatts, Glostrup, Denmark). FXIa and antithrombin were obtained from Enzyme Research Laboratories (South Bend, IN). Bovine serum albumin (BSA) is from Sigma. Human FVIII and its constituent subunits were purified as previously outlined.20 Thrombin-cleaved light chain of FVIII was prepared by incubating isolated FVIII light chain (310 nM) with α-thrombin (20 nM) for 1 hour at 37°C in 100 mM NaCl, 10 mM CaCl2, 50 mM Tris (pH 7.4). After incubation, thrombin was inhibited by the addition of PPACK (30 nM), and the cleaved light chain was purified by immunoaffinity chromatography using the monoclonal antibody CLB-CAg-117 as previously described.20 FX was purified as previously described.20 Normal plasma-derived FIX (pd-FIX) was prepared and was converted into FIXa by FXIa as described.18
Protein concentrations
FIX and FX proteins were assayed by ELISA employing anti-FIX and anti-FX polyclonal antibodies, respectively. FIX and FX were expressed in units, where 1 unit represents the amount in 1 mL of normal human plasma. Proteins were quantified by the method of Bradford,21 using BSA as a standard. FVIII activity was measured by spectrophotometric assay using bovine coagulation factors (Coatest FVIII; Chromogenix AB, Mölndal, Sweden). The amount of FVIII in 1 mL of normal human plasma (1 U/mL) was assumed to correspond to 0.35 nM. FVIII light chain was quantified in an immunologic assay employing the anti-FVIII light chain antibodies CLB-CAg-12 and CLB-CAg-117.20 Molar concentrations of FIXa, FXa, and α-thrombin were determined by active site titration.20
Radiolabeling of proteins
Antibodies and FIX were labeled with 125I (Amersham) using Iodo Gen (Pierce Chemical, Rockford, IL) as described.22 Specific radioactivities varied from 74 to 222 kBq/μg.
Identification of immunoglobulin class and subclass of anti-FIX antibodies
Polystyrene microtiter plates were coated with 100 μL purified human plasma FIX (final concentration 2 μg/mL) in carbonate buffer (50 mM NaHCO3, pH 9.5) overnight at 4°C. Residual binding sites were blocked with 200 μL Tris-buffered saline (TBS; 25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.01% [vol/vol] Tween 20) containing 3% BSA for 1 hour at room temperature. Immunoglobulin class of the inhibitor was determined by using serial dilutions of serum (100 μL, ranging from 1/5 to 1/12 800) from patients and control individuals in TBS-0.1% BSA. After washing with TBS, the class of the bound immunoglobulin was identified using horseradish peroxidase (HRP)-conjugated IgG specific for human IgA or IgG raised in rabbits (Dakopatts) or HRP affinity-isolated antibodies specific for human IgM raised in goats (Sigma). Normal serum and purified IgG from healthy volunteers obtained as described earlier were used as controls. The IgG subclass of the anti-FIX antibodies was determined by using monoclonal HRP IgG raised in mice and specifically directed against either human IgG1, 2, 3, or 4 (Tebu SA, Le Perray en Yvelines, France). These monoclonal antibodies were found to be specific in ELISA.23-26 The normal absorbance range for each IgG subclass was obtained by analyzing normal serum from healthy volunteers. Ranges of absorbance (average ± SD) of normal individuals tested at 20-fold plasma dilution for each subclass were 0.071 ± 0.021 for IgG1, 0.068 ± 0.019 for IgG2, 0.073 ± 0.022 for IgG3, and 0.065 ± 0.023 for IgG4. Samples with an absorbance between 0.1 and 0.3 were expressed as (+), between 0.3 and 0.7 as (++), and higher than 0.7 as (+++). Results higher than normal ranges were checked by 3 control studies. First, a competitive blocking test was used by addition of soluble FIX (50 μg/mL). Second, the patients' serum IgGs were absorbed by protein G Sepharose and the supernatant was assayed. Third, polystyrene microtiter plates coated with purified plasma human von Willebrand factor (generous gift from Dr J. P. Girma) were used.
Localization of epitopes recognized by anti-FIX IgG
The assay was performed by using polystyrene microtiter plates coated with polyclonal anti-FX antibody (2.5 μg/mL). Residual binding sites were blocked with 200 μL TBS containing 3% BSA. Media from cells expressing FX/FIX chimeras were diluted (1:2) in TBS containing 0.1% BSA and then added to the plates for 2 hours of incubation at room temperature. Various concentrations of patients' purified IgGs (from 50 μg/mL to 0.25 μg/mL) diluted in TBS-0.1% BSA containing 5 mM CaCl2 were added and incubated for 1 hour at room temperature. After washing with TBS containing 5 mM CaCl2, 100 μL 125I-IgG raised in rabbits (200 000 cpm, 0.04 μg/mL) specific for human IgG diluted in TBS-0.1% BSA containing 5 mM CaCl2 was added to the plates and incubated for 1 hour at room temperature. The bound radioactivity was counted in a gamma counter (LKB Instruments SA, Bromma, Sweden). In all experiments, media from cells expressing recombinant wild-type FX and purified IgG from healthy volunteers were used as controls.
FIX binding to lipospheres
Lipospheres were prepared from glass microspheres coated with phosphatidylserine/phosphatidylcholine (1:1, mol/mol) and characterized as described.27 Lipospheres, with a final lipid concentration of 0.76 μM, were incubated with various concentrations of 125I-FIX in 25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% (vol/vol) Tween 20, 5 mM CaCl2, and 0.1% BSA. After 1 hour of incubation at room temperature to ensure equilibrium had been reached, duplicate aliquots (100 μL) were layered onto 300 μL 10% sucrose in conical polyethylene tubes and centrifuged for 3 minutes at 12 000g. The tips of the tubes containing the pellets were cut and bound, and free radioactivity was counted. Unspecific binding determined on uncoated glass microspheres was subtracted. Dissociation constant (Kd) and the maximum number of binding sites (Bmax) were estimated from equilibrium-binding assays by fitting experimental data into a model describing the interaction of FIX with a single class of binding sites. Binding of125I-FIX either on coated or uncoated glass microspheres was totally inhibited by an excess of unlabeled FIX.
Inhibition of FIX binding to lipospheres
Lipospheres (0.76 μM lipid concentration) were mixed with125I-FIX (40 000 cpm, 1.5 nM) preincubated 1 hour with various concentrations of patients' IgG as competitor in 25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% BSA containing 5 mM CaCl2. After 1 hour the bound and free radioactivities were counted as described earlier. Nonspecific binding determined on uncoated glass microspheres was subtracted. Results were expressed as the percentage of binding measured in the absence of competitor (100%). Controls were performed in the presence of normal IgG.
Inhibition of FX activation
FX activation was assayed as described previously28with minor modifications. FVIII (1 U/mL) was added to phospholipid vesicles (0.1 mM), Ca++ (5 mM), thrombin (5 nM), and FIXa (0.1 nM). FIXa was preincubated 1 hour with various concentrations of patients' IgGs. After 1 minute of incubation, FX (0.1 μM) and hirudin (2 U/mL) were added. FXa formation was stopped by the addition of EDTA (10 mM) and subsequently quantified by the chromogenic substrate S2765 (Chromogenix AB, Mölndal, Sweden). The relationship between substrate hydrolysis and FXa concentration was determined by using an active site titrated FXa reference preparation. During the activation period, less than 5% FX was converted, and FXa formation was linear in time. Results were expressed as the percentage of FIXa activity measured in the absence of competitor (100%). Controls were performed in the presence of normal IgG.
FVIII light chain binding to FIXa
Purified FIXa was immobilized on microtiter plates (0.5 μg/well). After blocking with 3% (wt/vol) BSA in 100 mM NaCl, 25 mM Tris, pH 7.4, 0.1% (vol/vol) Tween 20, 5 mM CaCl2, and 10 mM benzamidine, dilutions of FVIII light chain were added and incubated for 2 hours at 37°C in the same buffer with 1% (wt/vol) BSA. The effect of patients' IgG concentration on FVIII light chain binding to FIXa was assessed by incubating various concentrations of such IgG premixed with FVIII light chain (50 nM) under the same conditions. After washing with 100 mM NaCl, 25 mM Tris, pH 7.4, 0.1% (vol/vol) Tween 20, 5 mM CaCl2, and 10 mM benzamidine, the peroxidase-conjugated anti-FVIII light chain antibody CLB-CAg 117 was added and incubated 10 minutes at room temperature. Wells were washed and peroxidase was detected with the substrate 3-3′-5-5′-tetramethylbenzidine. Nonspecific binding was estimated in uncoated wells and subtracted from the total binding. Inhibition of FVIII light chain binding to FIXa was expressed as the percentage of binding measured in the absence of competitor (100%). Controls were performed in the presence of mouse monoclonal antibody CLB-FIX 11 and CLB-FIX 14, or normal human IgG. Mouse monoclonal antibodies CLB-FIX 11 and CLB-FIX 14 are IgG1.18
Results
Identification of immunoglobulin class and subclass of anti-FIX antibodies
Class and subclass determination of antibodies against FIX was performed on microtiter plates using class- and subclass-specific antibodies. All 8 patients showed IgG antibodies against FIX, whereas none displayed any anti-FIX of the IgA or IgE classes. In one patient, P4, IgM antibodies against FIX were identified (data not shown). IgG subclass-specific antibodies showed that IgG1 and IgG4 antibodies against FIX were present (Table 2). These 2 subclasses were detectable in all patients. In contrast, IgG2 and IgG3 against FIX were not detectable in any of the patients (Table 2).
IgG subclass of patients' anti-FIX antibodies
| Patient no. . | IgG1 . | IgG2 . | IgG3 . | IgG4 . |
|---|---|---|---|---|
| 1 | + | − | − | ++ |
| 2 | ++ | − | ++ | |
| 3 | ++ | − | − | +++ |
| 4 | +++ | − | − | +++ |
| 5 | +++ | − | − | +++ |
| 6 | +++ | − | − | +++ |
| 7 | ++ | − | − | ++ |
| 8 | +++ | − | − | +++ |
| Patient no. . | IgG1 . | IgG2 . | IgG3 . | IgG4 . |
|---|---|---|---|---|
| 1 | + | − | − | ++ |
| 2 | ++ | − | ++ | |
| 3 | ++ | − | − | +++ |
| 4 | +++ | − | − | +++ |
| 5 | +++ | − | − | +++ |
| 6 | +++ | − | − | +++ |
| 7 | ++ | − | − | ++ |
| 8 | +++ | − | − | +++ |
IgG indicates immunoglobulin G; FIX, factor IX.
Localization of the anti-FIX antibody binding sites
To localize epitopes recognized by anti-FIX antibodies, binding assays using FX/FIX chimeras were performed. The chimeras were produced in mammalian cells (MDCK) and are schematically represented in Figure1. The interaction between IgG fractions purified from the serum of the patients and FX/FIX chimeras immunosorbed on microtiter plates coated with polyclonal anti-FX antibody was studied.
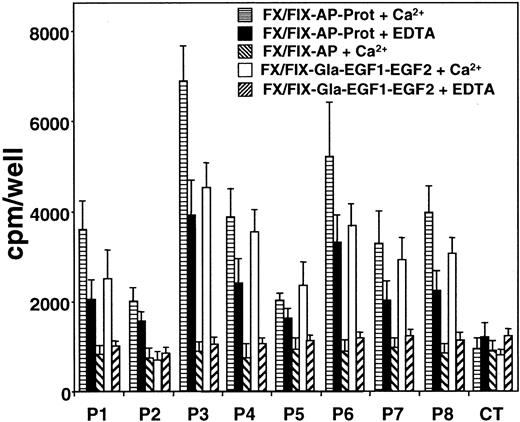
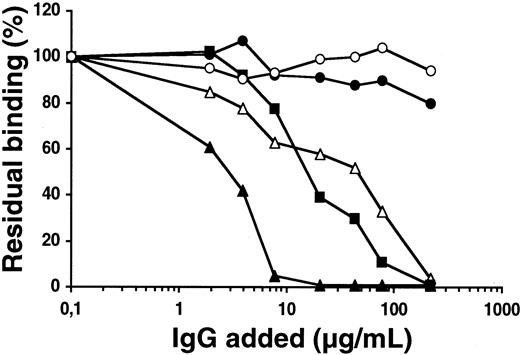
The ability of patient anti-FIX IgGs to detect chimeras FX/FIX-AP-Prot and FX/FIX-AP in the presence of Ca++ or EDTA was investigated. As shown in Figure 2, IgG from all 8 patients recognized the FIX protease domain, whereas the binding of the patients' IgGs to chimera FX/FIX-AP was comparable to the one obtained with control IgG. For the 8 patients, IgG binding to the FIX protease domain was higher in the presence of Ca++than in the presence of EDTA.
Reactivity of patients' anti-FIX IgGs against chimeras FX/FIX-AP-Prot, FX/FIX-AP, and FX/FIX-Gla-EGF1-EGF2.
To distinguish the reactivity of patients' IgGs toward the domains of FIX, medium from cells expressing chimeras FX/FIX-Gla-EGF1-EGF2, FX/FIX-AP-Prot, and FX/FIX-AP was added to microtiter plates coated with polyclonal anti-FX antibody. The response of patients' anti-FIX IgG against the chimeras was investigated in the presence of Ca++ and EDTA as described in “Patients, materials, and methods.” Data represent the mean values from 3 to 6 experiments (± SD).
Reactivity of patients' anti-FIX IgGs against chimeras FX/FIX-AP-Prot, FX/FIX-AP, and FX/FIX-Gla-EGF1-EGF2.
To distinguish the reactivity of patients' IgGs toward the domains of FIX, medium from cells expressing chimeras FX/FIX-Gla-EGF1-EGF2, FX/FIX-AP-Prot, and FX/FIX-AP was added to microtiter plates coated with polyclonal anti-FX antibody. The response of patients' anti-FIX IgG against the chimeras was investigated in the presence of Ca++ and EDTA as described in “Patients, materials, and methods.” Data represent the mean values from 3 to 6 experiments (± SD).
The ability of patients' IgGs to recognize chimera FX/FIX-Gla-EGF1-EGF2 was also investigated in the presence of Ca++ or EDTA. As shown in Figure 2, patients' IgGs bound to the FIX Gla domain linked to the EGF-like domains of the protein only in the presence of Ca++. In contrast, in the presence of EDTA binding of patients' IgGs to FX/FIX-Gla-EGF1-EGF2 was comparable to binding observed with control IgG. One exception was noticed with patient 2's IgGs in which no significant binding was observed in the presence of Ca++ or EDTA.
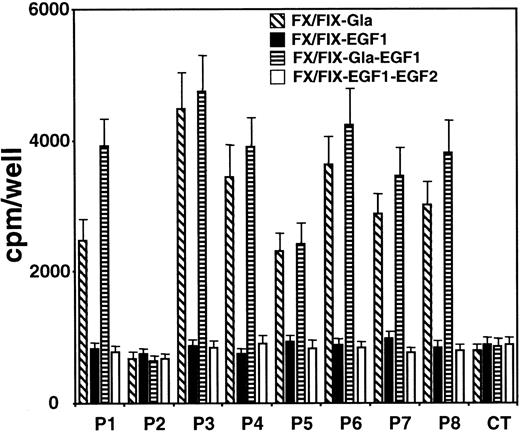
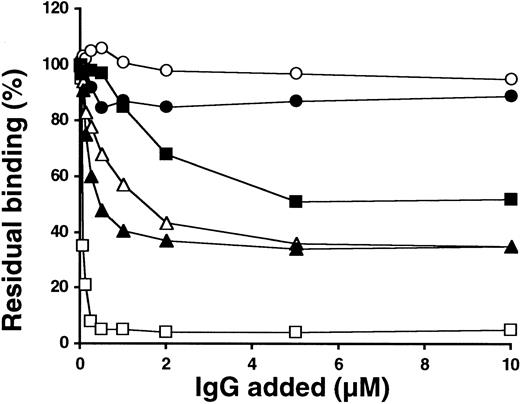
To further localize the recognized epitopes on FIX light chain in the presence of Ca++ by patients' anti-FIX IgGs, FX/FIX chimeras containing several combinations of FIX Gla and EGF-like domains were used. The results obtained (Figure3) showed that patients' IgGs, except from patient 2, recognized chimeras containing FIX Gla domain. In contrast, binding of patients' IgGs to chimeras containing only FIX EGF-like domains was comparable to IgG controls.
Reactivity of patients' anti-FIX IgGs against FX/FIX chimeras containing Gla and/or EGF-like domains of FIX.
To distinguish the reactivity of patients' IgGs toward Gla-containing domain and EGF-like domains of FIX, medium from cells expressing chimeras FX/FIX-Gla, FX/FIX-EGF1, FX/FIX-Gla-EGF1, and FX/FIX-EGF1-EGF2 was added to microtiter plates coated with polyclonal anti-FX antibody. The response of patients' anti-FIX IgG against the chimeras was investigated in the presence of Ca++ as described in “Patients, materials, and methods.” Data represent the mean values from 3 to 6 experiments (± SD).
Reactivity of patients' anti-FIX IgGs against FX/FIX chimeras containing Gla and/or EGF-like domains of FIX.
To distinguish the reactivity of patients' IgGs toward Gla-containing domain and EGF-like domains of FIX, medium from cells expressing chimeras FX/FIX-Gla, FX/FIX-EGF1, FX/FIX-Gla-EGF1, and FX/FIX-EGF1-EGF2 was added to microtiter plates coated with polyclonal anti-FX antibody. The response of patients' anti-FIX IgG against the chimeras was investigated in the presence of Ca++ as described in “Patients, materials, and methods.” Data represent the mean values from 3 to 6 experiments (± SD).
These results led to the conclusion that all patients possess anti-FIX IgG directed against the Gla domain, except patient 2, and against the protease domain. Patient 2 is dissimilar in that only the antibody against the protease domain was detected. It is noteworthy that the level of recognition by patients' IgGs of FIX domains is correlated with their inhibitor titer.
Inhibition of FX activation in the presence of FVIIIa
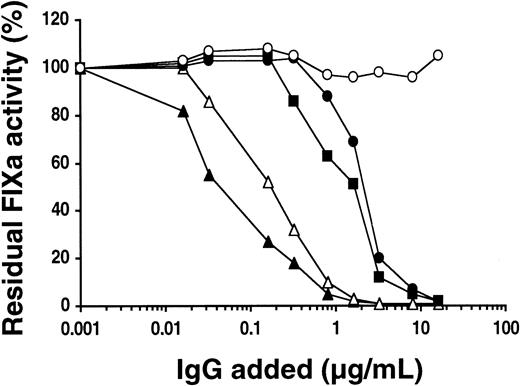
The ability of purified patients' IgGs to inhibit the FVIIIa-dependent activation of FX was first examined. In the blood coagulation cascade, a complex consisting of the serine protease FIXa and the cofactors Ca++, FVIIIa, and a membrane surface, which exposes anionic phospholipids, activates FX. The activation of FX was studied in vitro in the presence of various concentrations of patients' IgGs (see “Patients, materials, and methods”). Under these conditions, inhibition of FIXa by all patients' IgGs was dose dependent and higher than 90% at the maximum concentration of IgG tested as illustrated in Figure 4 for patients 1, 2, 3, and 4. The ability of IgGs from patients 5, 6, 7, and 8 to inhibit the FVIIIa-dependent activation of FX was between that observed for patients 2 and 3 (data not shown). Comparison of the inhibition induced by patients' IgGs revealed that differences exist in their abilities to inhibit FIXa function. FIXa activity was 50% reduced by approximately 0.24, 2.24, 0.06, and 1.44 μg/mL IgG for patients 1, 2, 3, and 4, respectively. Moreover, a significant proportion of purified IgG from the 3 patients with anti-FIX IgG directed against the Gla domain and against the protease domain as well as the patient with only anti-FIX IgG directed against the protease domain inhibits the proteolytic activity of FIXa toward FX induced by FVIIIa in the presence of phospholipids.
Effect of patients' anti-FIX IgGs on FVIIIa-dependent FX activation by FIXa.
The ability of various concentrations of patient1 (▵), patient 2 (●), patient 3 (▴), patient 4 (▪), and healthy control (○) IgGs to inhibit the FVIIIa-dependent activation of FX was investigated in vitro (see “Patients, materials, and methods”). Data represent the mean values from 4 experiments.
Effect of patients' anti-FIX IgGs on FVIIIa-dependent FX activation by FIXa.
The ability of various concentrations of patient1 (▵), patient 2 (●), patient 3 (▴), patient 4 (▪), and healthy control (○) IgGs to inhibit the FVIIIa-dependent activation of FX was investigated in vitro (see “Patients, materials, and methods”). Data represent the mean values from 4 experiments.
Inhibition of FIX binding to lipospheres
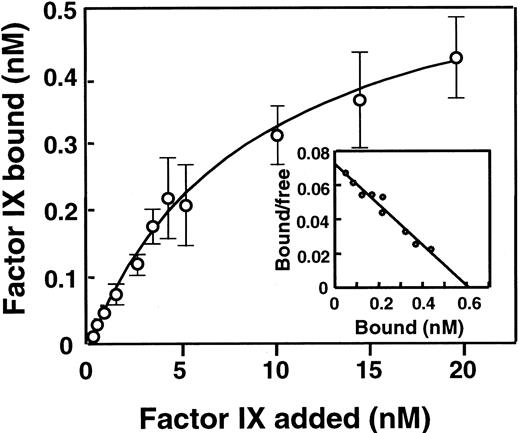
To further investigate the selective capacity of patients' IgGs to inhibit the interaction of FIXa with components of the FX-activating complex, the capacity of the IgG to interfere with FIX phospholipid interactions was studied. Equilibrium binding studies were performed employing glass microspheres coated with phosphatidylcholine and phosphatidylserine in 1/1 molar proportions. In preliminary experiments, 125I-FIX binding to lipospheres was observed to increase until a maximum was reached after less than 15 minutes (results not shown). In all further experiments, 1-hour incubation periods were employed to ensure that equilibrium had been attained. In equilibrium conditions, bound and free radioactivities were counted, and the specific binding of 125I-FIX to lipospheres was concentration dependent (Figure 5). Binding of 125I-FIX on either coated or noncoated glass microspheres was totally inhibited by an excess of nonlabeled FIX (data not shown). The dissociation constant was calculated to be 8.9 ± 0.8 nM, whereas the maximum number of 125I-FIX binding sites involved was found to be 0.6 ± 0.2 mmol/mol phospholipids. Similar results could be derived from the Scatchard analysis of the same data (Figure 5 inset).
Binding of FIX to lipospheres.
The graph represents the specific binding of 125I-FIX to lipospheres. Binding parameters were estimated as described in “Patients, materials, and methods.” The inset shows a Scatchard plot of the same data. Data represent mean values (± SD) from 4 independent experiments.
Binding of FIX to lipospheres.
The graph represents the specific binding of 125I-FIX to lipospheres. Binding parameters were estimated as described in “Patients, materials, and methods.” The inset shows a Scatchard plot of the same data. Data represent mean values (± SD) from 4 independent experiments.
The specific binding of 125I-FIX to lipospheres was inhibited in a dose-dependent manner by IgG from patients 1, 3, and 4 and not by IgG from patient 2 (Figure 6). The ability of IgGs from patients 5, 6, 7, and 8 to inhibit the binding of FIX to lipospheres was similar to the one observed for patients 1, 3, and 4 (data not shown). FIX binding to lipospheres was 50% reduced by approximately 48, 3, and 17 μg/mL IgG for patients 1, 3, and 4, respectively. With regard to patient 2, no inhibition was observed, even at concentrations higher than 500 μg/mL IgG. These results demonstrate that differences exist between patients with and without IgG directed against the FIX Gla domain. Only the former patients have IgG inhibiting the interaction between FIX and anionic phospholipids. The rate of inhibition of IgGs by patients 1, 3, and 4 are related to the patients' inhibitor titer.
Effect of patients' anti-FIX IgGs on FIX binding to phospholipids.
The ability of various concentrations of patient 1 (▵), patient 2 (●), patient 3 (▴), patient 4 (▪), and healthy control (○) IgGs to inhibit 125I-FIX binding to lipospheres was investigated as described in “Patients, materials, and methods.” Data represent the mean values from 4 experiments.
Effect of patients' anti-FIX IgGs on FIX binding to phospholipids.
The ability of various concentrations of patient 1 (▵), patient 2 (●), patient 3 (▴), patient 4 (▪), and healthy control (○) IgGs to inhibit 125I-FIX binding to lipospheres was investigated as described in “Patients, materials, and methods.” Data represent the mean values from 4 experiments.
Inhibition of FVIII light chain binding to FIXa
The possibility that patients' IgG could interfere with the capacity of FIXa to interact with its specific cofactor, FVIIIa, was also addressed. The interaction between FIXa and thrombin-cleaved FVIII light chain using nonequilibrium conditions was first examined, and a saturable and dose-dependent binding of FVIII light chain to FIXa was observed (data not shown). An apparent dissociation constant of 55 ± 8 nM was found. Subsequently, these conditions were used to study the inhibition induced by patients' IgG on the interaction between FIXa and thrombin-cleaved FVIII light chain. Two patients were selected, patient 1 who possessed IgG directed against both the FIX Gla domain and the FIX protease domain and patient 2 who possessed only IgG directed against the FIX protease domain. The mouse monoclonal antibody CLB-FIX 11 was used as positive control. It was found to recognize a Ca++-dependent epitope on FX/FIX chimeras carrying the FIX Gla domain (G.C. et al, unpublished results, October 2000). As shown in Figure 7, the binding of FVIII light chain to FIXa was inhibited in a dose-dependent manner by IgG from patient 1. In contrast, and as observed with FIX-binding inhibition to phospholipids (Figure 5), no inhibition was observed with patient 2's IgGs, as with IgG from a healthy donor even at high concentrations of competitor (Figure 7). As previously reported,18 the antibody CLB-FIX 11 strongly inhibits the interaction of FVIII light chain with FIXa. These results demonstrate that patients with and without IgG inhibiting the FIXa binding to FVIII light chain can be distinguished. The ability of patients' IgG to inhibit FIXa-FVIII light chain interaction is directly linked to the capacity of such IgG to bind to the FIX Gla domain.
Effect of patients' anti-FIX IgGs on FVIII light chain binding to FIX.
The ability of various concentrations of mouse monoclonal anti-FIX antibody CLB-FIX 11 (■), mouse monoclonal anti-FIX antibody CLB-FIX 14 (○), and IgGs from patient 1 (▵), patient 2 (●), patient 3 (▴), patient 4 (▪) to inhibit thrombin-cleaved FVIII light chain binding to immobilized FIXa was investigated (see “Patients, materials, and methods”). The data represent mean values from 3 independent experiments.
Effect of patients' anti-FIX IgGs on FVIII light chain binding to FIX.
The ability of various concentrations of mouse monoclonal anti-FIX antibody CLB-FIX 11 (■), mouse monoclonal anti-FIX antibody CLB-FIX 14 (○), and IgGs from patient 1 (▵), patient 2 (●), patient 3 (▴), patient 4 (▪) to inhibit thrombin-cleaved FVIII light chain binding to immobilized FIXa was investigated (see “Patients, materials, and methods”). The data represent mean values from 3 independent experiments.
Discussion
The development of FIX inhibitors in hemophilia B is a rare but severe complication of FIX substitution therapy. Insight into the epitope specificity and mechanism of action of these inhibitory antibodies has been poorly documented. In this study, the epitopes of IgG purified from 8 patients with hemophilia B with inhibitors were localized within the FIX molecule by using recombinant FX/FIX chimeras carrying FIX domains. The 2 regions identified as containing epitopes recognized by patients' IgGs were the Gla domain and the protease domain, whereas the EGF-like domains and the activation peptide did not.
Patients' IgGs binding to the FIX Gla domain occurred only in the presence of Ca++ and not in the presence of EDTA (Figures2,3). Analysis by nuclear magnetic resonance spectroscopy has revealed different conformations between the structures of the FIX Gla domain in the presence or in the absence of Ca++.29,30Results depicted in Figure 3 demonstrate that epitopes recognized by patients' IgG on the Gla domain are exposed only in the presence of Ca++. This finding is in agreement with a previous report31 in which recognition of antibodies from 12 patients with hemophilia B with inhibitors was evaluated. In that study, the authors showed that a majority of the patients' response is directed against the Gla domain in a Ca++-dependent conformation. As we tested the majority of patient plasmas from France and the Netherlands, the results represent most of the existing inhibitor pattern. It is striking that patient 2 differed from the other patients in that no antibody against the Gla domain could be detected (Figures 2,3). This finding does not necessary mean that B-cell epitopes on the Gla domain have not been recognized. It should be noted that in our study the information is from a single time point in the history of the patient. Its is possible that epitope specificity of anti-FIX antibodies vary over time in one individual as previously described for anti-FVIII antibodies.32 An other explanation is that in patient 2, epitope spreading could not occur as the gene defect presumably gives rise to a truncated protein containing the Gla domain, which might have induced a tolerance for this domain in this patient. The Gla domain is well known to serve a crucial role in the interaction between FIX and phospholipids. Indeed, patients' IgGs directed against the Gla domain are able to interfere with this particular interaction (Figure 5).
Several lines of evidence indicated that the light chain of FIXa interacts with the thrombin-cleaved light chain of FVIII. First, a FIXa fragment containing the Gla-EGF1-EGF2 region33 inhibits the interaction of FIXa with the A1/A3-C1-C2 FVIIIa dimer (where A3-C1-C2 region represents of the FVIII light chain). Second, FVIII light chain binds to the light chain of FIXa in ligand-blotting experiments.18 The finding that the ability of patients' IgGs to inhibit FXIa-FVIII light chain interaction is directly linked to the capacity of such IgG to bind to the FIX Gla domain (Figure 7) suggests that the Gla domain also contributes to the interaction with FVIII light chain. This possibility is supported by several observations. First, a genetic variant in the Gla domain that results in a glycine to arginine mutation at amino acid 12 has a reduced affinity for FVIIIa.34 Second, the mouse monoclonal antibody CLB-FIX 11 only recognizes the Ca++-dependent conformation of the Gla domain (G.C. et al, unpublished results, October 2000) and strongly inhibits the interaction of FIXa with the FVIII light chain18 (Figure 7). However, our results do not exclude that EGF domain residues are part of the patients' IgGs binding site. Several previous studies indicated that FIX EGF-like domains are involved in FVIII light chain interaction.35 36 Thus, patients' IgGs could interfere with the interaction of FIXa light chain and FVIII light chain by steric hindrance or by modulating the conformation of the FIXa light chain.
Opposite results concerning the ability of patients' IgGs to bind to the activation peptide of FIX were shown by 2 previous studies.31,37 In one study the researchers did not observe any binding of 1 of the 12 patients' IgGs tested to purified FIX activation pepide.31 In contrast the other study showed that 2 synthetic peptides corresponding to FIX residues of the activation peptide were recognized by 3 patients' IgGs.37The approach using FX/FIX chimeras demonstrated that none of the 8 patients' IgG tested interacted with FIX activation peptide (Figure2). It cannot be excluded that patients' IgGs against the activation peptide have a very low affinity for FIX and/or very few clones that produce antibodies against this domain, and they were therefore not detected in this study. However, oligopeptides are not always reliable for the mapping of IgGs. Cocrystallography studies of antigen and antibody complexes have provided convincing evidence that IgG epitopes on globular proteins are made of discontinuous peptide sequences.38 Therefore, we consider the approach using FX/FIX chimeras carrying FIX regions as being more appropriate to map the physiologic epitopes of antibodies directed against FIX. This approach is supported by several studies employing FIX chimeras with substitutions derived from FVII for epitope mapping of monoclonal and oligoclonal antibodies.39 40
The strategy using FX/FIX chimeras revealed the presence of patients' IgGs with a significantly lower recognition of the protease domain in the presence of EDTA than in the presence of Ca++ (Figure2). Interestingly, one structural feature of the FIX protease domain is the presence of a high-affinity Ca++-binding site.8 Site-directed mutagenesis of residues involved in this binding site revealed the influence of Ca++ binding on the conformation of the protease domain and on complex formation with FVIIIa.41 42 Thus, one can conclude from the results presented here that the immune response toward the protease domain is multiple. Moreover, some of these clones interfere with FIXa functions like the binding to FVIIIa and the enzymatic activity toward FX. This finding is supported by the observation that IgG from patient 2 totally inhibited the capacity of FIXa to activate FX in the presence of Ca++, FVIIIa, and phospholipids (Figure 4), whereas these IgGs were not directed against the FIX Gla domain (Figure 2) and did not inhibit the binding of FIX to phospholipids (Figure 6).
In conclusion, a significant number of patients with inhibitors were studied. Mapping of the epitopes revealed that FIX-specific IgGs recognize independent sites on the protease and the Gla domains. The functional characterization of patients' IgGs demonstrated the presence of antibodies against the functional part of the FIX molecule. Further studies will be needed to explore the details of the immune response against FIX, including reactivity to different epitopes or the epitopes spreading over time. The restricted epitope specificity found in this study suggests that only a limited number of gene segments is used during assembly of the anti-FIX immunoglobulin repertoire. One approach to characterize this would be by the use of phage display technology to isolate monoclonal antibodies from the immunoglobulin repertoires of patients with hemophilia B with inhibitors as recently reported for patients with hemophilia A.43
Supported in part by a travel grant from NWO-INSERM, by a grant from the Stichting Haemophilia, and by a grant from la Fondation pour la Recherche Médicale.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Olivier D. Christophe, INSERM U143, Bâtiment Claude Bernard, Hôpital de Bicêtre, 84 rue du Général Leclerc, 94276 Le Kremlin-Bicêtre Cedex, France; e-mail: olivierchristophe@usa.net.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal