Since the cloning in 1990 of complementary DNA corresponding to messenger RNA transcribed at the blood group ABO locus, polymorphisms and phenotype-genotype correlations have been reported by several investigators. Exons 6 and 7, constituting 77% of the gene, have been analyzed previously in samples with variant phenotypes but for many subgroups the molecular basis remains unknown. This study analyzed 324 blood samples involved in ABO grouping discrepancies and determined their ABO genotype. Samples from individuals found to have known subgroup alleles (n = 53), acquired ABO phenotypes associated with different medical conditions (n = 65), probable chimerism (n = 3), and common red blood cell phenotypes (n = 109) were evaluated by ABO genotype screening only. Other samples (n = 94) from apparently healthy donors with weak expression of A or B antigens were considered potential subgroup samples without known molecular background. The full coding region (exons 1-7) and 2 proposed regulatory regions of the ABO gene were sequenced in selected A (n = 22) or B (n = 12) subgroup samples. Fifteen novelABO subgroup alleles were identified, 2 of which are the first examples of mutations outside exon 7 associated with weak subgroups. Each allele was characterized by a missense or nonsense mutation for which screening by allele-specific primer polymerase chain reaction was performed. The novel mutations were encountered in 28 of the remaining 60 A and B subgroup samples but not among normal donors. As a result of this study, the number of definable alleles associated with weak ABO subgroups has increased from the 14 previously published to 29.
Introduction
In clinical transfusion medicine, ABO is the most important blood group system. Depending on a person's ABO blood group, preformed IgM anti-A and/or anti-B can be present in serum, constituting a major barrier against ABO-incompatible blood transfusions and organ transplantation. The ABO haptens are terminally located carbohydrate residues on the cell surface and in secretions that are biosynthesised by blood group-specific glycosyltransferases (reviewed by Watkins1) encoded by the ABO locus on the long arm of chromosome 9.2,3 Ninety years after the discovery of the ABO blood groups4 the molecular genetic basis of the system was defined5 and the polymorphisms of the common alleles at this blood group locus established.6However, for many of the variant phenotypes of the ABO system our knowledge of the molecular basis for altered antigen expression is sparse.
In 1993, molecular alterations at the ABO locus in individuals with inherited subgroup phenotypes were reported.7-10 Only one or 2 individuals with each phenotypic variant were investigated by sequencing of exons 6 and 7 in the ABO gene. Investigation of other subgroup alleles followed,11-15 but several examples exist in which no deviation from the consensus sequence was found in exons 6 and 7.7,15,16 In total, 8 A (including cis-AB) and 6 B subgroup [including 2 B(A)] alleles defined by missense mutations in exon 7 have been described to date. However, no investigation of exons 1 to 5 in samples with unusual ABO phenotypes has been published despite mutations located in the transmembrane or stem regions of other glycosyltransferases being shown to affect the resulting carbohydrate structures.17 18
ABO genotyping is a valuable complement to serology for correct determination of donor and patient ABO blood group status.19-24 As an independent tool in the clinical laboratory it can confirm a weakly expressed A or B antigen in inherited subgroups14 and also exclude B allele markers in the acquired B antigen phenotype.25 Although monoclonal antibodies recognizing and agglutinating most weak subgroups exist, the risk for mistyping (eg, Ax donors as blood group O) remains when using commercially available routine reagents. This may have adverse consequences when transfusing such blood to group O recipients.26
ABO allele nomenclature poses significant problems that are still under consideration by the International Society of Blood Transfusion. In this paper alleles are referred to by their serologic activity. Different alleles associated with the same serology have been numbered sequentially.
We evaluated the molecular genetic basis for ABO discrepancies in clinical samples including suspected inherited subgroups and acquired variant phenotypes sent to our laboratory during the past 7 years. The full ABO coding region (exons 1-7) and 2 potential regulatory 5′-untranslated regions (UTRs)27 28 were analyzed by direct DNA sequencing in selected samples for which the reason for the ABO discrepancy was still unexplained afterABO genotype screening and evaluation of medical records. In this way, 15 novel A and B subgroup alleles were identified.
Materials and methods
Blood samples and DNA preparation
Blood was drawn by venipuncture at the referring centers. DNA was prepared in Lund, Sweden from EDTA or acid-citrate-dextrose blood with a simple salting-out method.29 Blood from apparently healthy random donors was used to screen for mutations associated with the novel subgroup alleles.
Bloodgroup serology
ABO serology was performed with commercially available monoclonal, polyclonal, and lectin anti-A, anti-B, anti-AB, anti-A1, and anti-H reagents according to the routine of the referring laboratories. Serologic investigations were performed in Lund, Sweden as previously described14 if the red blood cell (RBC) condition allowed it. Saliva inhibition studies were performed according to modern blood grouping practice30when required for subgroup diagnosis, although in many cases either saliva samples were unavailable or the Lewis phenotype indicated nonsecretor status of the propositus.
ABO genotyping
A duplex polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method19 was used for the initialABO genotype screening. The presence of mutations associated with known alleles was confirmed by methods described earlier (Table1). A method36 linking mutations in exons 6 and 7 by allele-specific primer PCR (PCR-ASP) across intron 6 was used when exclusion of recombinant hybrid alleles was required. Primers in Table 2 (labeled ‡) were used under the PCR conditions described below for primer mixes W2/W3.
Polymorphic nucleotides detectable by restriction endonuclease digestion or sequence-specific primer PCR as used forABO genotyping in this study
| Polymorphism detected by PCR- . | Nucleotide position . | Mutation . | Alleles detectable* . | Allele reference . | Method reference . | |
|---|---|---|---|---|---|---|
| RFLP . | ASP . | |||||
| BstUI | − | 188-189 | GC → AT | O1v | 31 | 31 |
| KpnI | − | 261 | G → − | O1, O1v, O1 hybrids | 6,32 | 19 |
| HpaII | − | 467 | C → T | AsianA1, A2, O3, cis-AB, O1-A2 hybrid | 8,32-35 | 19 |
| NheI | + | 526 | C → G | B, O1-B hybrids | 6,32 | 14,36 |
| MboI | − | 646 | T → A | Ax, Ax hybrids, O1v | 9,15,31 | 14 |
| HpaII | − | 703 | G → A | B, O1-B hybrids | 6,32 | 19 |
| DdeI | − | 771 | C → T | Ax hybrids,O1v | 31 | 15,31 |
| — | + | 796 | C → A | B | 6 | 36 |
| — | + | 802 | G → A | O2 | 10 | 36 |
| — | + | 803 | G → C | B | 6 | 36 |
| — | + | 798-804 | +G | Ael, O3 | 11,35 | 11 |
| SalI | − | 871 | G → A | A3, Bx | 7,13 | 14 |
| — | + | 1059-1061 | −C | A2, O3 | 34,35 | 14 |
| HpaII | − | 1096 | G → A | B, O2 | 19 | 19 |
| Polymorphism detected by PCR- . | Nucleotide position . | Mutation . | Alleles detectable* . | Allele reference . | Method reference . | |
|---|---|---|---|---|---|---|
| RFLP . | ASP . | |||||
| BstUI | − | 188-189 | GC → AT | O1v | 31 | 31 |
| KpnI | − | 261 | G → − | O1, O1v, O1 hybrids | 6,32 | 19 |
| HpaII | − | 467 | C → T | AsianA1, A2, O3, cis-AB, O1-A2 hybrid | 8,32-35 | 19 |
| NheI | + | 526 | C → G | B, O1-B hybrids | 6,32 | 14,36 |
| MboI | − | 646 | T → A | Ax, Ax hybrids, O1v | 9,15,31 | 14 |
| HpaII | − | 703 | G → A | B, O1-B hybrids | 6,32 | 19 |
| DdeI | − | 771 | C → T | Ax hybrids,O1v | 31 | 15,31 |
| — | + | 796 | C → A | B | 6 | 36 |
| — | + | 802 | G → A | O2 | 10 | 36 |
| — | + | 803 | G → C | B | 6 | 36 |
| — | + | 798-804 | +G | Ael, O3 | 11,35 | 11 |
| SalI | − | 871 | G → A | A3, Bx | 7,13 | 14 |
| — | + | 1059-1061 | −C | A2, O3 | 34,35 | 14 |
| HpaII | − | 1096 | G → A | B, O2 | 19 | 19 |
This list of alleles is not complete.
Oligonucleotide primer pairs used for amplification and direct sequencing of ABO gene fragments
| Primer designation . | Primer pair . | Primer sequence (5′ → 3′) . | Primer location . | ABO region amplified . | Fragment size* (bp) . |
|---|---|---|---|---|---|
| Pro-F | 137 | ggaaacaaatcctacccctac | 5′-UTR | Enhancer (CBF-NFY) | 215/344 |
| Pro-R | gtgctgcctgtgcctgttac | 5′-UTR | |||
| mo-12F | 2 | ggcgccgtcccttcctag | 5′-UTR | Promoter, exon 1 | 267 |
| mo-12R | cctgcggtagcggctcct | Intron 1 | |||
| mo-21F | 3 | ggtgagagaaggagggtgag | Intron 1 | Exon 2-3 | 935 |
| mo-31R | ccagcaccccggccagca | Intron 3 | |||
| mo-41F | 431 | taaatcctgctcctagactaaac | Intron 3 | Exon 4 | 148 |
| mo-42R | ggacaattctgtgacatgggag | Intron 4 | |||
| mo-51F | 5 | tgcatcccacgctttccatgc | Intron 4 | Exon 5-6 | 823 |
| mo-46R19 | actcgccactgcctgggtctc | Intron 6 | |||
| mo-57a | 6 | gggtttgttcctatctctttgc | Intron 5 | Exon 6-7 | 2273 |
| mo-71R19 | gggcctaggcttcagttactc | 3′-UTR | |||
| ABO-261F-C | 7a | AGGAAGGATGTCCTCGTGGTG | Exon 6 | Exon 7 (non-O1/O1v) | 2002 |
| mo-71R19 | gggcctaggcttcagttactc | 3′-UTR | |||
| ABO-261F-C | 7b | AGGAAGGATGTCCTCGTGGTG | Exon 6 | Exon 7 (A2-specific) | 1888 |
| ABO-1060R-A2 | CCTGGCAGCCGCTCACGGT | Exon 7 | |||
| ABO-261F-C | 7c | AGGAAGGATGTCCTCGTGGTG | Exon 6 | Exon 7 (B-specific) | 1923 |
| ABO-1096R-B | GAGGGGGACGGGGCTGCT | 3′-UTR | |||
| ABO-297F-B† | — | CCATTGTCTGGGAGGGCACG | Exon 6 | — | — |
| ABO-467F-A2† | — | CTATGTCTTCACCGACCAGCT | Exon 7 | — | — |
| ABO-467R-A2‡ | CGGGGCACCGCGGCCAG | Exon 7 | |||
| ABO-526R-B‡ | GCCAGCGCTTGTAGGCGCC | Exon 7 | |||
| ABO-796R-B‡ | CCCCGAAGAACGCCCCCAT | Exon 7 | |||
| ABO-802R-O2‡ | CCGACCCCCCGAAGAACCT | Exon 7 | |||
| ABO-803R-B‡ | ACCGACCCCCCGAAGAACG | Exon 7 |
| Primer designation . | Primer pair . | Primer sequence (5′ → 3′) . | Primer location . | ABO region amplified . | Fragment size* (bp) . |
|---|---|---|---|---|---|
| Pro-F | 137 | ggaaacaaatcctacccctac | 5′-UTR | Enhancer (CBF-NFY) | 215/344 |
| Pro-R | gtgctgcctgtgcctgttac | 5′-UTR | |||
| mo-12F | 2 | ggcgccgtcccttcctag | 5′-UTR | Promoter, exon 1 | 267 |
| mo-12R | cctgcggtagcggctcct | Intron 1 | |||
| mo-21F | 3 | ggtgagagaaggagggtgag | Intron 1 | Exon 2-3 | 935 |
| mo-31R | ccagcaccccggccagca | Intron 3 | |||
| mo-41F | 431 | taaatcctgctcctagactaaac | Intron 3 | Exon 4 | 148 |
| mo-42R | ggacaattctgtgacatgggag | Intron 4 | |||
| mo-51F | 5 | tgcatcccacgctttccatgc | Intron 4 | Exon 5-6 | 823 |
| mo-46R19 | actcgccactgcctgggtctc | Intron 6 | |||
| mo-57a | 6 | gggtttgttcctatctctttgc | Intron 5 | Exon 6-7 | 2273 |
| mo-71R19 | gggcctaggcttcagttactc | 3′-UTR | |||
| ABO-261F-C | 7a | AGGAAGGATGTCCTCGTGGTG | Exon 6 | Exon 7 (non-O1/O1v) | 2002 |
| mo-71R19 | gggcctaggcttcagttactc | 3′-UTR | |||
| ABO-261F-C | 7b | AGGAAGGATGTCCTCGTGGTG | Exon 6 | Exon 7 (A2-specific) | 1888 |
| ABO-1060R-A2 | CCTGGCAGCCGCTCACGGT | Exon 7 | |||
| ABO-261F-C | 7c | AGGAAGGATGTCCTCGTGGTG | Exon 6 | Exon 7 (B-specific) | 1923 |
| ABO-1096R-B | GAGGGGGACGGGGCTGCT | 3′-UTR | |||
| ABO-297F-B† | — | CCATTGTCTGGGAGGGCACG | Exon 6 | — | — |
| ABO-467F-A2† | — | CTATGTCTTCACCGACCAGCT | Exon 7 | — | — |
| ABO-467R-A2‡ | CGGGGCACCGCGGCCAG | Exon 7 | |||
| ABO-526R-B‡ | GCCAGCGCTTGTAGGCGCC | Exon 7 | |||
| ABO-796R-B‡ | CCCCGAAGAACGCCCCCAT | Exon 7 | |||
| ABO-802R-O2‡ | CCGACCCCCCGAAGAACCT | Exon 7 | |||
| ABO-803R-B‡ | ACCGACCCCCCGAAGAACG | Exon 7 |
Primer sequences corresponding to untranslated sequences are shown in lower case.
The exact number of nucleotides may vary slightly between different alleles.
Used as sequencing primers only.
Each primer was used with ABO-261F-C for coupling of mutations in exons 6 and 7.36 The nucleotide- and allele-specificity is indicated by the name of the primers.
Amplification of the ABO gene for DNA sequencing
Oligonucleotide primers were synthesized by DNA Technology ApS (Aarhus, Denmark). Primers used to amplify DNA fragments for direct sequencing are shown in Table 2. PCR with primer pair 1 was described elsewhere37; PCR with primer pairs 2 to 5 was performed as follows: For one reaction 2 pmol of each primer (Table 2) was mixed with 200 ng genomic DNA, 4 nmol of each dNTP, and 1 U AmpliTaq Gold (Perkin-Elmer/Roche molecular system, Branchburg, NJ) in the buffer supplied. The final volume was 20 μL. Thermocycling was undertaken in GeneAmp PCR system 2400 (Perkin-Elmer/Cetus, Norwalk, CT): Initial denaturation at 96°C for 10 minutes was followed by 10 cycles at 94°C for 20 seconds, 63°C for 30 seconds, 72°C for 1 minute, then 25 cycles at 94°C for 20 seconds, 60°C for 30 seconds, and 72°C for 1 minute. PCR products were excised from 1.5% to 3% agarose gels (Seakem, FMC Bioproducts, Rockland, ME), stained with ethidium bromide (0.56 mg/L gel, Sigma Chemicals, St Louis, MO), and purified using Qiaquick gel extraction kit (Qiagen, Hilden, Germany). Primer pairs 6 and 7a-c (depending on the ABO genotype of the sample, Table2) were used to amplify allele-specific fragments covering exon 6, intron 6, and exon 7 of the ABO gene under the following cycling conditions: Initial denaturation at 96°C for 10 minutes followed by 15 cycles at 94°C for 20 seconds, 65°C for 2.5 minutes, then 25 cycles at 94°C for 20 seconds, 61°C for 30 seconds, 72°C for 2.5 minutes, and a final extension for 5 minutes. PCR products obtained with primer pair 6 from samples heterozygous forO1 or O1v alleles were digested by addition of 2 U of the restriction endonucleaseKpnI (Life Technology, Gaithersburg, MD) in the appropriate buffer and incubated at 37°C for 6 hours. Fragments were separated in 1% agarose and the undigested fragments (lacking the 261delG ofO1 /O1v alleles) subjected to sequencing after purification from gel.
The Big Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA) and an ABI PRISM 310 Genetic Analyzer (Applied Biosystems) were used for direct DNA sequencing according to the manufacturer's instructions. Besides the PCR primers in Table 2, the following internal primers were used as sequencing primers: mo-46,19 mo-75,11 EPB79,3 and fy-476 for sequencing of exons 6 and 7. In addition, allele-specific primers (Table 2) were used as sequencing primers to ensure that the sequencing results obtained for exons 6 and 7 reflected only the relevant allele. Sequence analysis was performed with SeqEd software 1.03 (Applied Biosystems).
Screening for subgroup-specific mutations by PCR-ASP
Three different PCR mixes (W1, W2, W3) containing the primers in Table 3 were used for subgroup mutation screening. Each primer (2 pmol) was mixed with 100 ng genomic DNA, 2 nmol dNTP, and 0.5 U of AmpliTaq Gold in the buffer supplied under the following cycling conditions: initial denaturation at 96°C for 10 minutes, followed by either 30 cycles at 94°C for 20 seconds, 65°C for 30 seconds, and extension at 72°C for 40 seconds (W1), or 32 cycles at 94°C for 30 seconds, 65°C for 30 seconds, and extension at 72°C for 90 seconds with a final extension at 72°C for 2 minutes (W2/W3). PCR products were separated for 30 minutes at 150 V using 2% ethidium bromide-stained agarose gels and visualized as described.19
Oligonucleotide primers used to screen for novelABO gene mutations found in this study
| Primer designation . | Reaction ID . | Primer sequence (5′ → 3′) . | Primer location in ABO gene . |
|---|---|---|---|
| JK-F3 | W1 | CATGCTGCCATAGGATCATTGC | 3-150 |
| JK-R3 | W1 | GAGCCAGGAGGTGGGTTTGC | 3-150 |
| ABO-in3F | W1 | gctggccgttacagggtctg | Intron 3 |
| ABO-203CR | W1 | AGGGAGGCACTGACATTATACG | Exon 4 |
| mo-57a | W1 | gggtttgttcctatctctttgc | Intron 5 |
| ABO-350CR | W1 | CTTGATGGCAAACACAGTTAACG | Exon 6 |
| mo-101s | W2-3 | ccgtccgcctgccttgcag | Intron 6 |
| ABO-407R-Aw | W2 | CCATGAAGTGCTTCTCCGCCA | Exon 7 |
| ABO-965R-Aw | W2 | GCTGCTGGTCCCACAAGTACC | Exon 7 |
| ABO-996R-Aw | W2 | TCCTCAGGACGGCGGGTC | Exon 7 |
| ABO-539R-Bw | W3 | CATGGACACGTCCTGCCAGT | Exon 7 |
| ABO-548R-Bw | W3 | ATGCGGCGCATGGACACGC | Exon 7 |
| ABO-721R-ABw | W3 | CTCGTAGGTGAAGGCCTCCCA | Exon 7 |
| ABO-863R-Bw | W3 | GTTGGCCTGGTCGACCATCC | Exon 7 |
| ABO-873R-Bw | W3 | CTCGATGCCGTTGGCCTGC | Exon 7 |
| ABO-1036R-Bw | W3 | CGGACCGCCTGGTGGTTCC | Exon 7 |
| ABO-1055R-Bw | W3 | GCAGCCGCTCACGGGTTCT | Exon 7 |
| Primer designation . | Reaction ID . | Primer sequence (5′ → 3′) . | Primer location in ABO gene . |
|---|---|---|---|
| JK-F3 | W1 | CATGCTGCCATAGGATCATTGC | 3-150 |
| JK-R3 | W1 | GAGCCAGGAGGTGGGTTTGC | 3-150 |
| ABO-in3F | W1 | gctggccgttacagggtctg | Intron 3 |
| ABO-203CR | W1 | AGGGAGGCACTGACATTATACG | Exon 4 |
| mo-57a | W1 | gggtttgttcctatctctttgc | Intron 5 |
| ABO-350CR | W1 | CTTGATGGCAAACACAGTTAACG | Exon 6 |
| mo-101s | W2-3 | ccgtccgcctgccttgcag | Intron 6 |
| ABO-407R-Aw | W2 | CCATGAAGTGCTTCTCCGCCA | Exon 7 |
| ABO-965R-Aw | W2 | GCTGCTGGTCCCACAAGTACC | Exon 7 |
| ABO-996R-Aw | W2 | TCCTCAGGACGGCGGGTC | Exon 7 |
| ABO-539R-Bw | W3 | CATGGACACGTCCTGCCAGT | Exon 7 |
| ABO-548R-Bw | W3 | ATGCGGCGCATGGACACGC | Exon 7 |
| ABO-721R-ABw | W3 | CTCGTAGGTGAAGGCCTCCCA | Exon 7 |
| ABO-863R-Bw | W3 | GTTGGCCTGGTCGACCATCC | Exon 7 |
| ABO-873R-Bw | W3 | CTCGATGCCGTTGGCCTGC | Exon 7 |
| ABO-1036R-Bw | W3 | CGGACCGCCTGGTGGTTCC | Exon 7 |
| ABO-1055R-Bw | W3 | GCAGCCGCTCACGGGTTCT | Exon 7 |
Polymorphic nucleotides in primer sequences are shown in bold, primer sequences targeting intron sequences in lower case. In addition to the primers in this table, mo-21F and mo-31R (Table 2) were used as the internal control primer pair in primer mixes W2 and W3.
These primer sequences correspond to exons 8 and 9 sequences in the JK blood group gene.38
Results
We investigated 324 clinical samples referred to our laboratory due to ABO grouping discrepancies since 1994. The samples were divided into different categories based on the medical condition of the donor, the serologic discrepancies, and the genomic typing results as described below. In addition, approximately 1500 bp of ABOgene sequence covering exons 1 to 7 and splice sites, the promoter region (from nucleotide [nt] −118 to −1) and a 215- or 344-bp enhancer region (around nt −3800) from each of 36 selected donors was evaluated.
Acquired variant ABO phenotypes
Sixty-five samples were included in this group based on serologic or medical factors in the presence of which acquired phenotype changes have often been described.26 39 Although the simultaneous presence of such factors and an unusual ABO subgroup allele is theoretically possible, no further analysis beyond ABOgenotype screening was done in this category.
Pregnancy.
Variable weak A expression was observed serologically during pregnancy (n = 13). Based on standard agglutination tests using repeat samples drawn 1 to 6 months after delivery, A antigen expression returned to normal levels in 5 women available for repeat testing. However, the other women were unavailable for repeat sampling or became pregnant again before new samples were taken. In 3 of the unavailable women normal blood grouping results had been obtained before pregnancy. All samples showed heterozygosity for the Aallele (Table 4).
ABO genotype screening results obtained in samples with ABO blood grouping discrepancies and known risk factors for noninherited RBC phenotype variants
| Phenotype/condition . | No. . | ABOgenotype . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1A1 . | A1O1 . | A1O1v . | A1A2 . | A2O1 . | A2O1v . | A1B . | A2B . | B O1 . | BO1v . | O1O1v . | O1vO1v . | ||
| Weak A expression in: | |||||||||||||
| Pregnancy | 13 | 5 | 3 | 1 | 4 | ||||||||
| Hematologic disorders | 14 | 1 | 3 | 2 | 2 | 3 | 2 | 1 | |||||
| Miscellaneous diagnoses | 13 | 3 | 3 | 3 | 2 | 2 | |||||||
| Weak B expression in hematologic disorders | 2 | 1 | 1 | ||||||||||
| Polyagglutination | |||||||||||||
| Acquired B syndrome | 19 | 2 | 5 | 6 | 4 | 1 | 1 | ||||||
| Acquired A-like antigen | 1 | 1 | |||||||||||
| Unspecified T-activation | 3 | 1 | 1 | 1 | |||||||||
| Phenotype/condition . | No. . | ABOgenotype . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1A1 . | A1O1 . | A1O1v . | A1A2 . | A2O1 . | A2O1v . | A1B . | A2B . | B O1 . | BO1v . | O1O1v . | O1vO1v . | ||
| Weak A expression in: | |||||||||||||
| Pregnancy | 13 | 5 | 3 | 1 | 4 | ||||||||
| Hematologic disorders | 14 | 1 | 3 | 2 | 2 | 3 | 2 | 1 | |||||
| Miscellaneous diagnoses | 13 | 3 | 3 | 3 | 2 | 2 | |||||||
| Weak B expression in hematologic disorders | 2 | 1 | 1 | ||||||||||
| Polyagglutination | |||||||||||||
| Acquired B syndrome | 19 | 2 | 5 | 6 | 4 | 1 | 1 | ||||||
| Acquired A-like antigen | 1 | 1 | |||||||||||
| Unspecified T-activation | 3 | 1 | 1 | 1 | |||||||||
Hematologic disorders.
Weak but variable A (or in 2 cases B) antigen expression was also noted in nontransfused patients (n = 16) with a variety of hematologic diseases including acute and chronic myelogenous leukemia, multiple myeloma, non-Hodgkin lymphoma, and myelodysplastic syndrome. In these patients transfusion of high-demand, group O blood components was avoided, partly due to the ABO genotyping results confirming their non-O status (Table 4).
A patient with myelodysplastic syndrome showed serologic agglutination patterns compatible with an acquired A antigen. The historical blood grouping was uncomplicated (blood group O) and ABOgenotyping indicated the O1vO1vgenotype. A patient with non-Hodgkin lymphoma had polyagglutinable RBCs, but was shown to have theO1O1v genotype.
Various medical conditions associated with ABO discrepancies.
Nineteen patients with nonhematologic malignancies (mainly adenocarcinomas of the colon, rectum, or uterus but also 3 cases with brain tumors) and gastrointestinal infections showed serologic results compatible with a form of polyagglutination often referred to as acquired B.30,39 Seventeen of the samples hadABO genotypes compatible with the A1 phenotype. However, a patient with advanced gynecologic carcinoma and another with malignant glioma had A2O1 andA2O1v genotypes, respectively. The presence of B allele markers was investigated byABO genotyping, and 4 missense mutations (526C>G, 703G>A, 796C>A, and 803G>C) differentiating the B from theA allele were excluded (Table 1). Erroneous transfusion of AB blood to this type of blood group A patient has been documented40 but could be avoided.
A patient undergoing orthopedic surgery and another with systemic lupus erythematosus had other forms of polyagglutinable red cells but hadBO1v and A1O1 genotypes, respectively.
Thirteen patients expressed weak A antigen, similar to the situation among patients with hematologic disorders above. Their records revealed a wide variety of medical conditions including septicemia, orthopedic, or abdominal surgery and autoimmune disease.
The genotyping results obtained in samples with known risk factors for noninherited erythrocyte phenotype variants are summarized in Table 4.
Inherited ABO phenotypes due to known subgroup alleles
Genotype screening of 53 samples with suspected weak A subgroup phenotypes revealed heterozygosity for previously recognized variant alleles associated with variant A expression, namelyAel,11B(A),9 and different variants ofAx.9,15 Each of these samples showed the expected serologic reactions according to the subgroup allele found. Two of the Ax alleles had exon 1 to 7 sequences identical to earlier reported Axhybrid alleles15 but different crossing-over regions shown by intron 6 sequencing. These 2 novel alleles are dealt with in a later section.
The B(A) phenotype sample had initially been interpreted serologically as AweakB but was genotyped as B(A) O1 , that is, heterozygous for a B allele lacking the B-specific mutation at nt 703.
A sample defined serologically as Ax by a major European reference laboratory was genotyped asA3O2 where theA3 allele (A1 with the 871G>A mutation) is that earlier described in 2 individuals with the A3B phenotype.7 These genotypes were confirmed by sequencing exons 1 to 7 in the B(A) andA3 alleles. In agreement with the originalA3 and B(A) reports consensusA and B sequences were found with the exception of 871G>A and 703A>G, respectively. The number of known subgroup alleles encountered in this study is summarized in Tables 5 and 6.
Previously published A subgroup alleles and the 8 novel A subgroup alleles reported here
| Allele description . | Nucleotide change(s) . | Amino acid change(s)5-150 . | Individuals (families)5-151 . | Geographic origin5-152 . | Reference/GenBank accession no. . |
|---|---|---|---|---|---|
| A3-1 | 871G>A | D291N | 15-153 | France (United States) | A37 |
| A3-2 | 829G>A | V277M | — | (Brazil) | A312 |
| 1060C− | P354 frameshift | ||||
| Ax-1 | 646T>A | F216I | 12 (6) | Various | Ax9,Ax(1)15, A10813 |
| Ax-25-155 | 646T>A | F216I | 6 (2) | Sweden | Ax(2), AF01662515 |
| 681G>A | |||||
| 771C>T | |||||
| 829G>A | V277M | ||||
| Ax-3 | 297A>G | 3 (1) | Sweden | Ax(3), AF01662415 | |
| 646T>A | F216I | ||||
| 681G>A | |||||
| 771C>T | |||||
| 829G>A | V277M | ||||
| ▸Ax-45-155 | 646T>A | F216I | 1 | Poland | AF324006 |
| 681G>A | |||||
| 771C>T | |||||
| 829G>A | V277M | ||||
| ▸Ax-55-155 | 646T>A | F216I | 1 | United States | AF324007 |
| 681G>A | |||||
| 771C>T | |||||
| 829G>A | V277M | ||||
| ▸Ax-6 | 996G>A | W332 stop | 1 | New Zealand | AF324013 |
| Ael-1 | 798-804 (+G) | F269 frameshift | 28 (16) | Various | Ael11, A10913 |
| Ael-2 | 467C>T | P156L | — | (Japan) | A11013 |
| 646T>A | F216I | ||||
| 681G>A | |||||
| ▸Aw-1 | 407C>T | T136M | 2 (2) | United Kingdom | AF324010 |
| 467C>T | P156L | ||||
| 1060C− | P354 frameshift | ||||
| ▸Aw-2 | 350G>C | G117A | 6 (6) | New Zealand, United Kingdom, Australia, United States | AF324009 |
| 467C>T | P156L | ||||
| 1060C− | P354 frameshift | ||||
| ▸Aw-3 | 203G>C | R68T | 28 (> 5) | Scandinavia | AF324008 |
| 467C>T | P156L | ||||
| 1060C− | P354 frameshift | ||||
| ▸Aw-4 | 721C>T | R241W | 2 (2) | Belgium, Germany | AF324011 |
| ▸Aw-5 | 965A>G | E322G | 2 (1) | Finland | AF324012 |
| cis-AB | 467C>T | P156L | — | (mainly Japan) | cis-AB8 41 |
| 803G>C | G268A |
| Allele description . | Nucleotide change(s) . | Amino acid change(s)5-150 . | Individuals (families)5-151 . | Geographic origin5-152 . | Reference/GenBank accession no. . |
|---|---|---|---|---|---|
| A3-1 | 871G>A | D291N | 15-153 | France (United States) | A37 |
| A3-2 | 829G>A | V277M | — | (Brazil) | A312 |
| 1060C− | P354 frameshift | ||||
| Ax-1 | 646T>A | F216I | 12 (6) | Various | Ax9,Ax(1)15, A10813 |
| Ax-25-155 | 646T>A | F216I | 6 (2) | Sweden | Ax(2), AF01662515 |
| 681G>A | |||||
| 771C>T | |||||
| 829G>A | V277M | ||||
| Ax-3 | 297A>G | 3 (1) | Sweden | Ax(3), AF01662415 | |
| 646T>A | F216I | ||||
| 681G>A | |||||
| 771C>T | |||||
| 829G>A | V277M | ||||
| ▸Ax-45-155 | 646T>A | F216I | 1 | Poland | AF324006 |
| 681G>A | |||||
| 771C>T | |||||
| 829G>A | V277M | ||||
| ▸Ax-55-155 | 646T>A | F216I | 1 | United States | AF324007 |
| 681G>A | |||||
| 771C>T | |||||
| 829G>A | V277M | ||||
| ▸Ax-6 | 996G>A | W332 stop | 1 | New Zealand | AF324013 |
| Ael-1 | 798-804 (+G) | F269 frameshift | 28 (16) | Various | Ael11, A10913 |
| Ael-2 | 467C>T | P156L | — | (Japan) | A11013 |
| 646T>A | F216I | ||||
| 681G>A | |||||
| ▸Aw-1 | 407C>T | T136M | 2 (2) | United Kingdom | AF324010 |
| 467C>T | P156L | ||||
| 1060C− | P354 frameshift | ||||
| ▸Aw-2 | 350G>C | G117A | 6 (6) | New Zealand, United Kingdom, Australia, United States | AF324009 |
| 467C>T | P156L | ||||
| 1060C− | P354 frameshift | ||||
| ▸Aw-3 | 203G>C | R68T | 28 (> 5) | Scandinavia | AF324008 |
| 467C>T | P156L | ||||
| 1060C− | P354 frameshift | ||||
| ▸Aw-4 | 721C>T | R241W | 2 (2) | Belgium, Germany | AF324011 |
| ▸Aw-5 | 965A>G | E322G | 2 (1) | Finland | AF324012 |
| cis-AB | 467C>T | P156L | — | (mainly Japan) | cis-AB8 41 |
| 803G>C | G268A |
Nucleotide and amino acid changes are given in comparison to the consensus A1 allele.
▸ indicates novel A subgroup allele reported in this paper.
Charged residues involved in amino acid changes are shown in bold.
The number of samples identified by sequencing or PCR-ASP in this study is given for each allele (the number of apparently unrelated families investigated is given in brackets).
Country from which the samples were referred (countries in parentheses refer to samples reported elsewhere).
The phenotype of this sample was reported to be Ax, not A3.
These alleles differ by their intron 6 polymorphism showing 3 different cross-over regions between A andO1v, namely, nt 189-225 in intron 6 forAx-2, nt 236-445 in intron 6 forAx-4, and nt 298 in exon 6 to nt 41 in intron 6 for Ax-5.
Previously published B/B(A) subgroup alleles and the 7 novel B subgroup alleles reported here
| Allele description . | Nucleotide change . | Amino acid change6-150 . | Individuals (families)6-151 . | Geographic origin6-152 . | Reference/GenBank accession no. . |
|---|---|---|---|---|---|
| B3-1 | 1054C>T | R352W | — | (United States) | B37 |
| Bel-1 | 641T>G | M214R | — | (Japan) | B10513 |
| Bel-2 | 669G>T | E223D | — | (Japan) | B10613 |
| Bw-1 | 871G>A | D291N | — | (Japan) | B10413 |
| ▸Bw-2 | 873C>G | D291E | 1 | France | AF324018 |
| ▸Bw-3 | 721C>T | R241W | 8 (2) | Sweden | AF324014 |
| ▸Bw-4 | 548A>G | D183G | 5 (2) | Sweden | AF324016 |
| ▸Bw-5 | 539G>A | R180H | 1 | United States | AF324015 |
| ▸Bw-6 | 1036A>G | K346E | 3 (1) | Finland | AF324019 |
| ▸Bw-7 | 1055G>A | R352Q | 2 (1) | United States | AF324020 |
| ▸Bw-8 | 863T>G | M288R | 1 | Turkey | AF324017 |
| B(A)-1 | 703A>G | S235G | 1 | Germany | 9 |
| B(A)-2 | 700C>G | P234A | — | (Asia) | 42 |
| Allele description . | Nucleotide change . | Amino acid change6-150 . | Individuals (families)6-151 . | Geographic origin6-152 . | Reference/GenBank accession no. . |
|---|---|---|---|---|---|
| B3-1 | 1054C>T | R352W | — | (United States) | B37 |
| Bel-1 | 641T>G | M214R | — | (Japan) | B10513 |
| Bel-2 | 669G>T | E223D | — | (Japan) | B10613 |
| Bw-1 | 871G>A | D291N | — | (Japan) | B10413 |
| ▸Bw-2 | 873C>G | D291E | 1 | France | AF324018 |
| ▸Bw-3 | 721C>T | R241W | 8 (2) | Sweden | AF324014 |
| ▸Bw-4 | 548A>G | D183G | 5 (2) | Sweden | AF324016 |
| ▸Bw-5 | 539G>A | R180H | 1 | United States | AF324015 |
| ▸Bw-6 | 1036A>G | K346E | 3 (1) | Finland | AF324019 |
| ▸Bw-7 | 1055G>A | R352Q | 2 (1) | United States | AF324020 |
| ▸Bw-8 | 863T>G | M288R | 1 | Turkey | AF324017 |
| B(A)-1 | 703A>G | S235G | 1 | Germany | 9 |
| B(A)-2 | 700C>G | P234A | — | (Asia) | 42 |
Nucleotide and amino acid changes are given in comparison to the consensus B allele.
▸ indicates novel B subgroup allele reported in this paper.
Charged residues involved in amino acid changes are shown in bold.
The number of samples identified by sequencing or PCR-ASP in this study is given for each allele (the number of apparently unrelated families investigated is given in parentheses).
Country from which the samples were referred (countries in parentheses refer to samples reported elsewhere).
CommonABO phenotypes
Relatives of subgroup individuals.
Samples from parents, siblings, or children of the suspected subgroup propositus fell into 2 groups: samples with common ABO phenotypes (n = 81) and those with the same weak A or B subgroup as the propositus. The former group was not studied further followingABO genotyping when the expected inheritance and phenotype/genotype correlation was observed. In 2 cases the serologic consequences of the A subgroup allele were masked by a normalA1 allele (A1Ael orA1Ax).
Unexpected absence or very low titer of anti-A or anti-B despite common RBC phenotype.
Twenty-eight samples lacking (or having very low titers of) anti-A and/or anti-B despite the absence of serologic proof of B and/or A antigen, respectively, were genotyped. Adsorption/elution tests had not usually been performed or else gave negative or inconclusive results. This group included patients undergoing immunosuppressive therapy and also apparently healthy blood donors lacking one of their isoagglutinins (in most cases anti-A). The genotypes found included only well-known ABO alleles compatible with common red cell phenotypes, thus not confirming the suspicion of weak A or B expression.
Suspectedchimerism
Two samples showed signs of more than 2 alleles followingABO genotyping. In both cases the third allele gave weak reactions in PCR-ASP tests and represented an A orB allele compatible with the weak A or B expression also observed serologically. A third suspected chimeric sample was referred due to mixed-field reactions in agglutination tests involving antigenic markers for several blood group systems. Unfortunately, ABOgenotyping was uninformative because only 2 ABO alleles were detected. Re-examination of the clinical records revealed that the first 2 individuals had a twin sibling, thus making chimerism due to intrauterine exchange of hematopoietic precursor cells the most likely explanation for their discrepant ABO typing result. Twin status could not be confirmed for the third patient. Because chimerism was not the main topic of this study, these samples were not examined further.
Variant ABO phenotypes potentially due to unknown subgroup alleles
The remaining samples came from apparently healthy donors without identifiable risk factors for acquired ABO phenotype changes to explain their variant phenotypes. Mutations associated with known subgroup alleles were excluded by ABO genotype screening as described above (Table 1). These samples were potentially A (n = 72) or B (n = 22) subgroups with an unknown molecular basis. Inheritance of the phenotype was shown in 17 and 18 cases, respectively (data not shown).
Serologic classification.
Several of the donors had been blood grouped repeatedly over a number of years in different laboratories and always gave the expected reactions for the subgroup assigned.26 30 Subgroup status was confirmed with samples from relatives, if available. Frequently, however, no assignment was possible, usually due to lack of saliva or the donor's nonsecretor status.
ABO genotyping.
All A and B subgroup samples were heterozygous for A-and B-like alleles, respectively. For the A subgroup samples, A1- or A2 -like alleles were predicted. In the presence of the 721C>T and 1055G>A mutations (see below) anomalous bands appeared using the PCR-RFLP method with simultaneous digestion with KpnI andHpaII endonucleases,19 thus preventing the erroneous prediction of normal A and B alleles in these cases. The anomalous bands were the result of allele-specific mutations removing HpaII sites present in the consensusA1 and B sequences.
Sequencing of novel ABO subgroup alleles.
The full coding region (exons 1-7) and 2 proposed regulatory motifs of the ABO gene were sequenced in genomic DNA from selected A (n = 22) or B (n = 12) subgroup samples representing different serologic patterns and varying geographic origin. Priority was given to samples shown to have inherited ABO discrepancies.
Except for the mutations stated below, all analyzed A and B subgroup alleles had consensusA1 /A2 and Bsequences, respectively, including regulatory regions. Novel sequences have been deposited in GenBank (accession numbers AF324006-AF324020). The 15 novel A and B variant alleles reported here are summarized and compared to previously published subgroup alleles in Table 5 and Table 6, respectively.
A subgroup alleles.
Ten samples classified as A3 had apparentlyA1 alleles. Two fully sequenced samples had exons and regulatory regions identical to the consensusA1 allele. The same was true for 2 Norwegian sisters with the Ael phenotype reported to have consensus exons 6 and 7.16
The majority of Scandinavian Aend, Aweak, and Ax samples lacking the Ax mutation at nt 6469 15 had an A2 allele. Two Aend and 2 Ax samples were fully sequenced and had the same missense mutation, 203G>C, in exon 4 causing R68T (single-letter amino acid codes). Because these samples had been assigned to different serologic categories, this allele was designatedAw-3.
A donor of Polish origin had the AweakB phenotype and an American donor was suspected to have the B(A) phenotype. Both were heterozygous for Ax alleles and sequencing showed 2 novel variants of the Ax hybrid allele including the missense mutations at nt 646 (F216I) and 829 (V277M) (Ax-4 and Ax-5 in Table5). As in the previously described recombinantAx alleles (Ax-2 andAx-3) the 5′ end of common alleles were fused with the 3′ end of O1v alleles. In both cases exon 7 was derived from O1v, but by sequencing of intron 6, 2 novel crossing-over regions were defined (Table 5 and GenBank accession numbers therein). Another Ax sample from New Zealand had a nonsense mutation, 996G>A, predicting loss of 23 amino acids due to a premature stop codon (Ax-6).
Samples from 2 unrelated British donors categorized as Aweak or weak Ax phenotypes were sequenced and had A2 -like alleles with an additional 407C>T (T136M) (Aw-1), while 2 unrelated American donors with Ax? or Aweak phenotype and one weak Ax? sample from New Zealand hadA2 -like alleles with an additional 350G>C (G117A) (Aw-2).
Two samples from Belgium and Germany were only very weakly positive in agglutination and adsorption/elution tests using anti-A, B and had only anti-B in their sera. Both were heterozygous for anA1 -like allele with the additional presence of 721C>T (R241W) (Aw-4) also noted in some Bweak families (see below).
A Finnish family with very weak erythrocyte A expression (differing from the Afinn phenotype according to the Finnish Red Cross Transfusion Service) had an A1 -like allele (Aw-5) with a 965A>G mutation predicting E322G.
B subgroup samples.
The Bweak samples had weak anti-B in plasma and very similar mixed field agglutination patterns with a low proportion of agglutinated red cells. Saliva studies were performed on the Swedish samples but were uninformative, because one family in each genetic variant had low amounts of secreted B substance, and the other 2 families lacked B substance. No further attempts were made to categorize samples into different subgroups, a decision supported by the vague or sometimes contradictory criteria for classification published in current textbooks. However, a Finnish family (Bw-6) had been classified26 as Bv by the Finnish Red Cross Transfusion Service.
The B alleles studied showed remarkable genetic heterogeneity and were geographically dispersed. Seven novel sequences were found despite the serologic similarity. All were Bconsensus sequences with the additional presence of a single missense mutation (Table 6).
Earlier work by Professor Ch. Salmon (oral communication, Dr LePennec, Paris, July 2000) and Professor W. M. Watkins (written report, London, 1983) showed decreased B-transferase activity in samples now shown to have the D291E and D183G substitutions, respectively (Table 6).
Screening for subgroup-specific mutations by PCR-ASP
Primers specific in their 3′ ends for all the novel mutations detected in this study were synthesized (Table 3). Primer mix W1 was designed to detect mutations outside exon 7, whereas W2 and W3 detect mutations in exon 7 of weak A and B subgroup alleles, respectively. Sixty random blood donor samples were screened with 3 different multiplex primer mixes to see if the novel mutations were found in samples from normal ABO phenotypes. None were found except for a donor with the A1A2 genotype, who also had the 203G>C mutation in the A2 allele (according to a family study). His phenotype was A1 as expected. Some of his relatives had the Aend phenotype due to Aw-3 O1 or Aw-3 O1v genotypes.
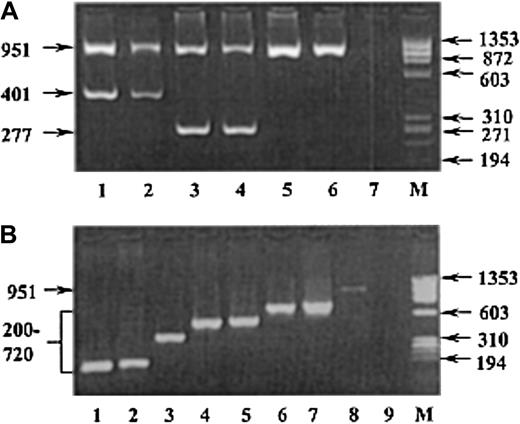
The 60 remaining variant samples (not previously sequenced) were screened by PCR-ASP and 28 were positive for one of the new subgroup markers. The total number of samples found positive for each mutation is given in Tables 5 and 6, as is their geographic distribution. Representative gel electrophoretograms corresponding to primer mixes W1 and W3 are shown in Figure 1. The remaining 32 samples not associated with a definable allele were not further evaluated, in most cases because of unsatisfactory DNA quality or insufficient amounts of available DNA.
Representative gel electrophoretograms showing PCR-ASP screening for subgroup alleles.
PCR-ASP amplification products for A and B subgroup alleles with the W1 (A) and W3 (B) primer mixes, respectively, as separated on 2% agarose gels. The W2 primer mix gel is not shown because it works according to the same principle and looks virtually identical to the W3 gel. Samples from individuals with the following weak A and B subgroup mutations are shown. (A) Lanes 1-2, donors with the Aw-2 (350G>C) allele; lanes 3-4, donors with theAw-3 (203G>C) allele; lanes 5-6,A1O1 andA2O1v random donors; and lane 7, H2O contamination control. (B) Lane 1,Bw-5 (539G>A) allele; lane 2,Bw-4 (548A>G); lane 3,Bw-3 (721C>T); lane 4,Bw-8 (863T>G); lane 5,Bw-2 (873C>G); lane 6,Bw-6 (1036A>G); lane 7,Bw-7 (1055G>A); lane 8,BO1 random donor; and lane 9, H2O contamination control. M is the molecular size marker φX174RF DNAHaeIII (Life Technologies). Arrows indicate the size of DNA fragments in base pairs.
Representative gel electrophoretograms showing PCR-ASP screening for subgroup alleles.
PCR-ASP amplification products for A and B subgroup alleles with the W1 (A) and W3 (B) primer mixes, respectively, as separated on 2% agarose gels. The W2 primer mix gel is not shown because it works according to the same principle and looks virtually identical to the W3 gel. Samples from individuals with the following weak A and B subgroup mutations are shown. (A) Lanes 1-2, donors with the Aw-2 (350G>C) allele; lanes 3-4, donors with theAw-3 (203G>C) allele; lanes 5-6,A1O1 andA2O1v random donors; and lane 7, H2O contamination control. (B) Lane 1,Bw-5 (539G>A) allele; lane 2,Bw-4 (548A>G); lane 3,Bw-3 (721C>T); lane 4,Bw-8 (863T>G); lane 5,Bw-2 (873C>G); lane 6,Bw-6 (1036A>G); lane 7,Bw-7 (1055G>A); lane 8,BO1 random donor; and lane 9, H2O contamination control. M is the molecular size marker φX174RF DNAHaeIII (Life Technologies). Arrows indicate the size of DNA fragments in base pairs.
Discussion
The clinical use of genomic blood grouping in transfusion medicine practice has evolved during the past decade as a result of the cloning of most blood group genes. A recent review43 outlined different situations in the clinical laboratory where blood group genotyping may be valuable and ABO genotyping has been used for many of them, although acquired and inherited variant phenotypes are probably the most important.
In this study we evaluated serologic ABO discrepancies encountered in a consecutive series of clinical samples referred to us forABO genotyping. Two acquired variant phenotypes are of particular interest: weakened A antigen expression (eg, in leukemia and pregnancy) and acquired B antigen. Concern is motivated due to their frequency and the implications in clinical transfusion. The underlying reason for the observed alterations is unknown. Erythrocyte A or B antigen activity decreased in a number of patients while being treated for leukemia or other hematologic disorders. The correlation between leukemia and erythrocyte ABO typing irregularities was first described in 195744 and has been amply confirmed.45 A similar phenomenon has also been reported in patients of blood group B. Why leukemia affects the expression of ABO system antigens is unknown.
An elevated frequency of the Le(a−b−) phenotype has been noted in pregnant women,46 where A1 women were the most likely to lose their Leb antigen on RBCs. Changes during pregnancy and lactation suggest this to be influenced by hormonal factors.39 The decreased A activity in the 13 women is interesting and a 10% decrease in antibody binding to RBCs in pregnant women has been described.47 This is not due to abnormalA alleles because none were found and A expression often reverted to normal. No correlation to A1 (n = 8) or A2 (n = 5) alleles was obvious.
Acquisition of B-mimicking activity on RBCs usually occurs in A1 patients (occasionally A2) with diseases of the digestive tract, in particular colon cancer but also other adenocarcinoma of the female reproductive system and in septic states.39 This phenomenon is found, but rarely, in apparently normal individuals. Interestingly, 3 patients with brain tumors were among the acquired B patients in this study. The connection between bacterial enzyme activity and infection/gastrointestinal disease is more obvious than why erythrocytes would be affected in patients with tumors behind the blood–brain barrier. Two thirds of the acquired B referrals occurred as a result of unexpected RBC agglutination with the monoclonal ES-4 anti-B reagent, which has a high affinity for the deacetylated A antigen.48 Several samples were referred even after the pH adjustment of this reagent, as required by the Food and Drug Administration, was made by the manufacturer, suggesting that the measure taken was inadequate. Subsequent cases (after withdrawal of ES-4) have all occurred as a result of testing with polyclonal human reagents. All samples examined with a serologically suspected acquired variant A or B phenotype were genotyped unambiguously and no unusual or unexpected (eg,Bw) alleles were found.
A striking feature of the genotypes of individuals with weak A and B phenotypes is the diversity of alleles giving the same phenotype, and to some extent the assignment of the same allele to more than one phenotype. A number of practical problems arise as interpretation of incomplete hemagglutination can vary between laboratories and even between individuals in the same laboratory. Furthermore, different clones (monoclonal) or batches (polyclonal) of anti-A, anti-B and anti-A, B reagents show a high grade of interreagent variation. As an example, many of the true A subgroups reported here were initially detected by the referring centers as single weak reactivities against commercially available monoclonal anti-A (eg, A003 or BRIC186) or anti-A, B blend reagents (most often containing the ES-15 clone). Over time, blood grouping reagents may vary and availability changes continually. As a result, the delineation between some of the weak subgroups is diffuse. This is exemplified by theAw-3 allele (Table 5) that was present in numerous samples submitted to us and described as both Aend, Ax, and unspecified Aweak. The relevance of categorical subgroup classification based on serologic phenomena alone is becoming questionable; so use of the more general terms Aweak and Bweak is not unfounded.
In 28 cases submitted due to an anti-A or anti-B deficiency there was no evidence for the presence of a weak A or Ballele. Following genotyping blood units donated by such donors could be issued as regular units labeled either blood group A, B, or O.
Not surprisingly, genotyping of many samples from blood donors and patients throughout the world selected due to irregular serologic findings resulted in detection of several new subgroup alleles. Until recently, ABO genotyping examined exclusively exons 6 and 7 of the gene, representing 77% of the expressed enzyme protein. Very few alleles were actually sequenced in the early reports. Currently, we analyze all 7 exons, intron 6, and promoter and enhancer regions. The 8 new A alleles described here comprised 3 serologically classified as Axand 5 that we prefer to callAweak(Aw) because the serologic results did not allow further classification. Two of theAx alleles (Ax-4 andAx-5) were similar toAx-2 described previously.15 All are hybrid alleles having similar but not identical recombination sites in intron 6 fusing A and O1valleles.
Of the 5 new Aw alleles, 3 were normalA2 alleles with additional missense mutations, 2 of which are the first mutations (203G>C in exon 4 and 350G>C in exon 6) outside exon 7 to be associated with weak subgroup phenotypes. The remaining 2 alleles were A1 but with a missense mutation in exon 7.
As shown in Table 6 each of the 7 new Bw alleles resulted from a single amino acid-changing mutation in the originalB allele. The heterogeneity is striking and it is also interesting to note that the point mutations in allBw alleles described involve charged amino acids. The latter also applies to 4 of the Awalleles reported.
The 721C>T mutation weakens both A and B activity (Aw-4 and Bw-3) through an R241W substitution. A similar effect has also been observed with 871G>A (D291N) that differentiates A3 -1 from the consensus A allele and Bw-1 from a normal B allele. Amino acid substitutions spread along the length of the enzyme protein (Figure 2) have varying effects on enzyme activity resulting in the different ABO subgroup phenotypes. It is worth noting how most of the mutations reported to date cluster in certain regions of the A andB alleles, most predominantly between nt 539 to 548 (2 alleles) and nt 641 to 721 (6 alleles), nt 829 to 873 (5 alleles), and nt 965 to 1060 (8 alleles). Amino acid residues located in these regions may be directly involved in the enzyme's active site or affect protein conformation and could make interesting targets for site-directed mutagenesis.
Schematic representation of single missense amino acid substitutions in A and B glycosyltransferases associated with decreased expression of A and B antigens, respectively.
The 2 horizontal bars represent 20 of the variant A (upper) and B (lower) glycosyltransferases in Tables 5 and 6 with the addition of 4 A2 transferases.49 CisAB andB(A) alleles were not included. Vertical lines separate regions coded for by different exons (numbered 1-7) in theABO gene. The putative transmembrane domain (amino acid 15-39 of the glycosyltransferase) is shown in gray.5 The arrows indicate the stem region susceptible to proteolytic cleavage for generation of soluble enzyme. All amino acid changes published to date are shown as geometric symbols above the bars (except for P156L, which does not affect glycosyltransferase activity). Filled symbols represent substitutions involving a charged amino acid and open symbols involve noncharged residues. Different ABO phenotypes are represented by the symbols as follows: ▪ and ■, A2; ▴, A3/B3; ● and ○, Ax, Aw/Bw; ♦ and ⋄, Ael/Bel. The Ax alleles other than Ax-1 (hybridsAx-2 to Ax-5 andAx-6 with a nonsense mutation at nt 996, Table5) have been excluded from this figure, as has the frame-shiftingAel-1 allele. Three alleles each having 2 mutations resulting in amino acid changes were included: a, this diamond represents Ael-2 that also has the neutral P156L change; b, this triangle represents a V277M substitution in A3 -2 that also has the common A2-related mutation (1060C−); c, one of the 3 squares represents the common A2 -1 allele (a nonsense, not missense, mutation).
Schematic representation of single missense amino acid substitutions in A and B glycosyltransferases associated with decreased expression of A and B antigens, respectively.
The 2 horizontal bars represent 20 of the variant A (upper) and B (lower) glycosyltransferases in Tables 5 and 6 with the addition of 4 A2 transferases.49 CisAB andB(A) alleles were not included. Vertical lines separate regions coded for by different exons (numbered 1-7) in theABO gene. The putative transmembrane domain (amino acid 15-39 of the glycosyltransferase) is shown in gray.5 The arrows indicate the stem region susceptible to proteolytic cleavage for generation of soluble enzyme. All amino acid changes published to date are shown as geometric symbols above the bars (except for P156L, which does not affect glycosyltransferase activity). Filled symbols represent substitutions involving a charged amino acid and open symbols involve noncharged residues. Different ABO phenotypes are represented by the symbols as follows: ▪ and ■, A2; ▴, A3/B3; ● and ○, Ax, Aw/Bw; ♦ and ⋄, Ael/Bel. The Ax alleles other than Ax-1 (hybridsAx-2 to Ax-5 andAx-6 with a nonsense mutation at nt 996, Table5) have been excluded from this figure, as has the frame-shiftingAel-1 allele. Three alleles each having 2 mutations resulting in amino acid changes were included: a, this diamond represents Ael-2 that also has the neutral P156L change; b, this triangle represents a V277M substitution in A3 -2 that also has the common A2-related mutation (1060C−); c, one of the 3 squares represents the common A2 -1 allele (a nonsense, not missense, mutation).
Of all defined ABO alleles to date, only a few [A1 , A2 , B,B(A), O2 and possiblyA3 -1]50-52 have been expressed in synthetic systems to evaluate the causative relationship between the mutations and phenotypes observed. As shown by discrepancies between the initially reported in vitro experiments using BABBplasmid constructs (resulting in pure B antigenicity detected by flow cytometry in HeLa cells50) and the same sequence in genomic DNA [resulting in the B(A) phenotype in vivo9] limited sensitivity or other difficulties may lead to results not necessarily reflecting subgroup phenotypes. Nevertheless, a future aim is to express subgroup alleles to show how they affect the enzymic activities of these subgroup glycosyltransferases.
ABO genotyping is a powerful, independent tool in the reference laboratory for resolution of clinical ABO blood grouping discrepancies. A major use of this technique in transfusion medicine is the unequivocal discrimination between acquired and inherited variant phenotypes. Subgroup alleles can be differentiated from normal alleles temporarily resulting in weak agglutination, for example, during pregnancy or in patients with hematologic disorders. In this way the safety of the ABO grouping is further enhanced so that all donors and patients can be correctly grouped to ensure the safest possible use of the blood supply. The rapidly increasing number of alleles being found enhances the usefulness in paternity testing and other applications where individual-specific genetic markers are of interest.43 In addition, amniotic fluid DNA has been referred to this center for fetal ABO genotyping to optimize the care of pregnant women with a history of severe hemolytic disease of the newborn or neonatal alloimmune thrombocytopenia due to high-titer IgG anti-A in earlier pregnancies.
The frequent occurrence of hybrid ABO alleles, at least in some ethnic groups,32,53 has emphasized the need for better ABO genotyping protocols to achieve safe phenotype prediction. In addition, none of the hitherto published methods for genomic typing has included means to define subgroup alleles.49 Following this study, the number of A and B subgroup alleles has more than doubled, a fact that further accentuates the need for improvement.
We express our gratitude to the following persons for performing serologic investigations on samples referred to our laboratory and obtaining samples from relatives: Ms Imelda Bromilow (Diamed, Switzerland), Ms Rida Chiu (Oakland, CA), Dr Hans-Erik Heier (Oslo, Norway), Dr Stephen Henry (Auckland, New Zealand), Dr France Noizat-Pirenne and Dr PierreYves LePennec (Paris, France), Mr Steven Pierce (Kansas City, MO), Ms Anna Pirkola (Helsinki, Finland), Ms Joyce Poole (Bristol, United Kingdom), Dr Hans Gerhard Ruhl (Dresden, Germany), Dr Karin Schneider and Dr Jan Säfwenberg (Uppsala, Sweden), Dr Hans Sonnenborn (Biotest, Germany), Dr Rudi Steffensen (Aalborg, Denmark), Dr Jill Storry (New York, NY), and Dr Vered Yahalom (Tel Aviv, Israel).
Supported in part by the Claes Högman SAGMAN Stipendium, Georg Danielssons Fond för Blodsjukdomar, Tore Nilsons Stiftelse för Medicinsk Forskning, Universitetssjukhusets donationsfonder, the Medical Faculty at Lund University, Sweden (M.L.O.), and the World Health Organization (N.M.I.).
Part of this work was presented at the Annual Meeting of the American Association of Blood Banks held in San Francisco, CA, November 8, 1999, and published in abstract form.54
M.K.M. is employed by Immucor/Gamma Reference Laboratories, whose products were used in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Martin L. Olsson, Blood Centre, University Hospital, S-221 85 Lund, Sweden; e-mail:martin_l.olsson@transfumed.lu.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal