Abstract
Human acute myelogenous leukemia (AML) is thought to arise from a rare population of malignant stem cells. Cells of this nature, herein referred to as leukemic stem cells (LSCs), have been documented for nearly all AML subtypes and appear to fulfill the criteria for stem cells in that they are self-renewing and give rise to the cells found in many leukemic populations. Because these cells are likely to be critical for the genesis and perpetuation of leukemic disease, the present studies sought to characterize unique molecular properties of the LSC population, with particular emphasis on the transcription factor, nuclear factor-κB (NF-κB). Previous experiments have shown that unstimulated human CD34+ progenitor cells do not express NF-κB. In contrast, primary AML CD34+ cells display readily detectable NF-κB activity as assessed by electrophoretic mobility shift assay and gene expression studies. Furthermore, detailed analyses of enriched AML stem cells (CD34+/CD38−/CD123+) indicate that NF-κB is also active in the LSC population. Given the expression of NF-κB in leukemic, but not normal primitive cells, the hypothesis that inhibition of NF-κB might induce leukemia-specific apoptosis was tested by treating primary cells with the proteasome inhibitor MG-132, a well-known inhibitor of NF-κB. Leukemic CD34+/CD38− cells displayed a rapid induction of cell death in response to MG-132, whereas normal CD34+/CD38− cells showed little if any effect. Taken together, these data indicate that primitive AML cells aberrantly express NF-κB and that the presence of this factor may provide unique opportunities to preferentially ablate LSCs.
Introduction
Recent years have witnessed significant advances in the field of stem cell biology. Numerous studies have appeared describing the biologic relevance of stem cells, as well as their specific cellular and molecular properties.1-6Interestingly, while the focus of recent studies has been predominantly on normal cell types, important progress has also been made in the characterization of malignant stem cells. For example, several recent studies have identified and characterized a leukemic stem cell (LSC) population in patients with acute myelogenous leukemia (AML). LSCs are sufficient to perpetuate human leukemic cell growth in long-term culture assays and in the murine nonobese diabetic/severe combined immunodeficiency model system.7-9 As a consequence of these data, it has been proposed that LSCs are central to the pathogenesis of human myeloid leukemia. However, despite the clear importance of this population, little is currently known about the molecular properties that make LSCs distinct from normal hematopoietic stem cells. In particular, it is critical to understand how malignant stem cells regulate growth and survival. If LSCs use unique mechanisms to control processes such as apoptosis, then it may be possible to target such pathways as a means to specifically destroy leukemic, but not normal hematopoietic cells.
Several recent studies have shown that LSCs can be phenotypically defined as CD34+, CD38−, HLA-DR−, CD90−, CD117−, and CD123+.7-12 This antigenic profile is intriguing in that it overlaps with normal stem cells (CD34+, CD38−, HLA-DR−), but also has leukemia-specific features (CD90−, CD117−, and CD123+). This is important because it permits isolation of highly enriched stem cell populations of leukemic origin, which are not potentially contaminated with normal stem cells. Consequently, it is now possible to pursue detailed molecular analyses of primitive leukemia cells. For example, we have recently shown that primitive AML cells aberrantly express the tumor suppressor genes IRF1 (interferon regulatory factor 1) andDAPK (death-associated protein kinase).13 In seeking to further characterize this observation, we reasoned that transcriptional activators such as nuclear factor-κB (NF-κB) might lie upstream of IRF-1. This appeared plausible due to many previous reports describing active NF-κB in various malignant cell types, including primary lymphoid leukemia specimens.14-19 In addition, one study reported NF-κB activity in some primary AML samples after 16 hours of culture in 10% fetal calf serum.20 Importantly, the active form of NF-κB is known to have antiapoptotic activity and is thereby considered to be a key survival factor for several types of cancer.21,22 Further, inhibition of NF-κB induces apoptosis in several malignant cell types.19,21 This has been accomplished using drugs such as proteasome inhibitors, as well as with dominant-negative alleles of the NF-κB regulatory molecule, IκBα.23 In the present study we demonstrate that primitive human AML cells also express an active form of NF-κB and that treatment of such cells with proteasome inhibitors is sufficient to induce rapid cell death.
Materials and methods
Cell isolation and culture
The AML blood cells, normal bone marrow cells, and umbilical cord blood (CB) cells were isolated and processed as described.12 Briefly, primary AML cells were obtained from the peripheral blood of patients at the Markey Cancer Center, Duke University Medical Center, and the University of Pennsylvania Medical Center. Normal bone marrow was obtained as waste material following pathologic analysis, surgical marrow harvest, or from the National Disease Research Interchange (NDRI). CB was obtained from patients at the University of Kentucky Obstetrics Department or from the NDRI. All tissues were obtained with the approval of the respective institutional review boards and appropriate informed consent. Marrow cells were depleted of erythrocytes by suspending in 150 mM NH4Cl plus 10 mM NaHCO3 for 5 minutes, followed by 2 washes with phosphate-buffered saline (PBS). Blood cells were subjected to Ficoll-Paque (Pharmacia Biotech, Piscataway, NJ) density gradient separation to isolate the mononuclear white blood cell compartment. Resulting leukocytes from marrow or blood were then used for immunoaffinity selection and flow cytometric sorting. For CD34+ cell selection, the Miltenyi immunoaffinity device (VarioMACS) was used according to the manufacturer's instructions (Miltenyi Biotech, Auburn, CA). In some cases, leukocytes were cryopreserved at a concentration of 5 × 107 cells/mL in freezing medium consisting of Iscoves modified Dulbecco medium (IMDM), 40% fetal bovine serum (FBS), and 10% dimethyl sulfoxide (DMSO). For analysis of unstimulated cells, cryopreserved primary samples were thawed and used immediately for isolation of RNA or protein. For apoptosis induction studies, samples were thawed and cultured in serum-free medium24 for 1 hour prior to the addition of carbobenzoxyl-l-leucyl-l-leucyl-l-leucinal (MG-132) or sodium salicylate. MG-132 was obtained from Calbiochem (San Diego, CA) and resuspended in ethanol at 20 mM stock concentration. Sodium salicylate and cytosine arabinoside (Ara-C) were obtained from Sigma (St Louis, MO) and made at a 0.5 M stock concentration in water.
Flow cytometry
The CD34+ cells were obtained from normal or AML specimens using the Miltenyi VarioMACS immunoselection system as per manufacturer's instructions. Purified AML populations were isolated by labeling samples with CD34–fluorescein isothiocyanate (FITC), CD38-allophycocyanin (APC; Becton Dickinson, San Jose, CA), and CD123-phycoerythrin (PE; Pharmingen, San Diego, CA), and sorting using a FACSVantage flow cytometer (Becton Dickinson). A typical purity of at least 95% was obtained. For apoptosis analysis, cells were first stained with antibodies against the surface markers CD34-PE and CD38-APC (Becton Dickinson), and incubated on ice for 15 minutes. Cells were then washed with cold PBS and resuspended in 200 μL annexin binding buffer (10 mM HEPES/NaOH, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2). Annexin V–FITC (Pharmingen) and 0.25 μg/mL 7-aminoactinomycin D (7-AAD; Molecular Probes, Eugene, OR) were added and the tubes were incubated at room temperature in the dark for 15 minutes. Cells were then diluted with 200 μL annexin binding buffer and analyzed immediately by flow cytometry. Each sample was also verified to be CD123+ using the CD123-PE antibody.
Nuclear extracts
Nuclear extracts were prepared as described by Dignam and coworkers25 with some modifications. Briefly, cells were washed with cold PBS followed by 2 additional washes with extract buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). Cells were then lysed in extract buffer containing 0.1% NP-40 and incubated on ice for 5 minutes. Samples were centrifuged at 1400g for 15 minutes and resuspended in 20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, and 20% glycerol. Next, samples were vortexed gently and incubated for 15 minutes on ice. Crude nuclear extracts were centrifuged at 14 000g to remove debris, and the supernatant was aliquoted and stored at −80°C.
Electrophoretic mobility shift assay
The NF-κB consensus and mutant oligonucleotide sequences were obtained from Santa Cruz Biotechnologies (Santa Cruz, CA). The probes were labeled with T4 polynucleotide kinase (Life Technologies, Gaithersburg, MD) and γ-[32P]-ATP (NEN, Boston, MA) and purified over a Sephadex G25 (Pharmacia Biotech) column. Protein nuclear extracts equivalent to 100 000 cells were incubated with 2 μg poly d(I-C) (Roche Molecular Biochemicals, Indianapolis, IN) and 10−14 mol 32P-labeled probe in 10 mM HEPES, 5 mM Tris, 50 mM KCl, 1.2 mM EDTA, and 10% (vol/vol) glycerol (pH 7.8) for 15 minutes at room temperature. A 200-fold molar excess of unlabeled oligonucleotide (NF-κB consensus or mutant) was added for competition assays. For supershift assays, 2 μg of antibodies to NF-κB p65 and p50 (Santa Cruz Biotechnologies) was added to the reaction. Protein/DNA complexes were resolved on 6% native polyacrylamide gels in 0.25 × TBE (25 mM Tris, 22.5 mM boric acid, and 0.25 mM EDTA). Gels were visualized by autoradiography using MS-BioMax film and intensifying screens (Kodak, Rochester, NY).
Reverse transcription–polymerase chain reaction
The RNA samples were prepared with the use of the Miltenyi μMACS messenger RNA (mRNA) isolation kit (Miltenyi Biotech) as per the manufacturer's instructions and reverse transcribed with Superscript II (Life Technologies, Carlsbad, CA). Polymerase chain reactions (PCRs) were performed using a Perkin Elmer (Shelton, CT) 9700 thermal cycler and the following primers: β-actin sense 5′ ATCTGGCACCACACCTTCTACAATGAGCTGCG 3′, β-actin antisense 5′ CGTCATACTCCTGCTTGCTGATCCACATCTGC 3′, c-IAP2 sense 5′ GCTTCTGTTGTGGCCTG 3′, c-IAP2 antisense 5′ CACCTTGGAAACCAC 3′. For each reaction the complementary DNA (cDNA) equivalent of 200 cells was amplified for 35 cycles using cycle parameters of 94°C for 30 seconds, 60°C for 60 seconds, and 72°C for 60 seconds. Interleukin-6 (IL-6) and IL-8 PCR primers were obtained from Maxim Biotech (San Francisco, CA). The cDNA equivalent of 200 cells was amplified for 35 cycles as per the manufacturer's recommended cycle parameters.
Western blot analysis
For the analysis of IκBα shown in Figure 3, cells were cultured in serum-free medium alone or in the presence of 1.0 μM MG-132 for the indicated number of minutes. Specimens were then harvested, prepared, and analyzed exactly as described in Jordan and coworkers.12 Blots were probed with antiphospho-specific IκBα from Cell Signaling Technology (Beverly, MA).
Cell-cycle analysis
Cell-cycle analysis was performed as previously described with some modifications.26 Cells were harvested, washed, and resuspended in PBS plus 0.5% FBS. Cell surface staining was performed using CD34-PE-Cy5 (Coulter, Miami, FL) and CD38-PE (Becton Dickinson). Afterward, cells were fixed in PBS plus 0.5% formaldehyde and incubated on ice for 30 minutes. Next, to permeabilize the cells, an equal volume of PBS plus 0.2% Triton X-100 was added and samples were incubated for 15 minutes on ice. Then, cells were washed and resuspended in PBS plus 0.5% FBS and labeled with Ki-67–FITC (Coulter). Lastly, each sample was washed and resuspended in PBS plus 0.5% FBS plus 10 μM 4′, 6-diamidino-2-phenylindole (DAPI; Molecular Probes) and incubated for 1 hour before analysis by flow cytometry using a FACSVantage instrument configured to emit 355-nm, 488-nm, and 633-nm laser lines.
Results
NF-κB binding is detected in AML but not normal hematopoietic cells
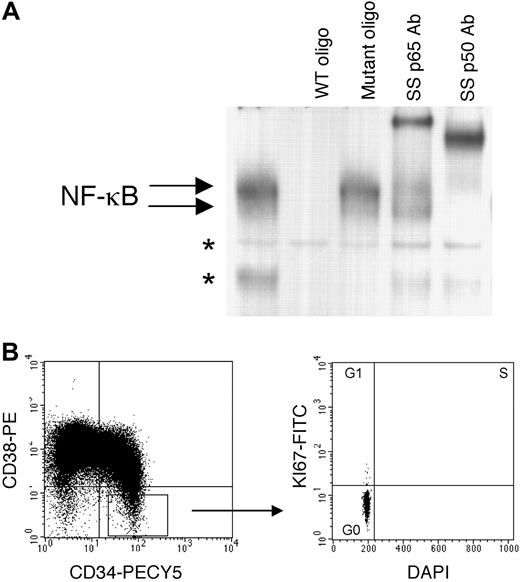
For studies described in this report, AML cells were obtained from the 11 patient specimens listed in Table1. Importantly, the specimens derive from several different subtypes (French-American-British [FAB] classifications M1, M2, M4, and M5), and therefore represent a broad cross-section of commonly detected AML types. Based on previous studies, we hypothesized that NF-κB might be active in primary human AML cells. To address this question, electrophoretic mobility shift assays (EMSAs) were used to measure binding to a32P-labeled NF-κB consensus oligonucleotide in nuclear extracts obtained from primary AML cells, normal bone marrow, or CB cells. Prior to the isolation of nuclear extracts, both the AML and normal cells were preselected for expression of the cell surface antigen CD34. For AML specimens, which were derived from peripheral blood samples, selection of CD34+ cells ensures a virtually pure AML population, with no contamination from normal cells (the presence of normal CD34+ cells in peripheral circulation is extremely rare). Expression of CD34 also selects for relatively immature components of the AML clone. Similarly, selection of CD34+ cells is a common means of enriching for primitive normal hematopoietic cells.27 As shown in Figure1A, significant NF-κB binding was detected in CD34+ AML cell nuclear extracts but not in nuclear extracts obtained from normal CD34+ hematopoietic cells. Each AML specimen showed at least 2 specific species (indicated by arrows). In addition to the 6 independent specimens shown in Figure1A, this observation has been confirmed with an additional 5 samples (specimens 7-11 in Table 1, data not shown). To further analyze the specificity of the binding shown in Figure 1A, all specimens were tested using competition assays and supershift analysis. Figure 1B shows representative data for 3 specimens. In each case the NF-κB binding was effectively competed with a 200-fold molar excess of unlabeled consensus oligonucleotide, but not with a control mutant oligonucleotide. Further, because the NF-κB active complex can consist of homodimers of p50 or heterodimers of p50 and p65,28 the composition of the binding complex was assessed using supershift assays with anti-p50 or anti-p65 antibodies. Both the p50 and p65 subunits were readily detected.
Characteristics of AML specimens
| Case no. . | Age/sex . | FAB/type . | WBC × 109/L . | Cytogenetics . | 34+/38−/G0(%) . |
|---|---|---|---|---|---|
| 1 | 72/M | M2/de novo | > 100 | 46, XY | 93 |
| 2 | 68/M | M5/de novo | 120 | 45, X, −Y | nd |
| 3 | 50/F | M1/relapse | 60 | 46, XX | 96 |
| 4 | 30/M | M5/de novo | 27 | 47, XY, +21, t(17;21) | 97 |
| 5 | 56/F | M4/relapse | 86 | 46, XX | 83 |
| 6 | 61/F | M5/de novo | 380 | 46, XX | 98 |
| 7 | 50/F | M1/de novo | 140 | 46, XX | 94 |
| 8 | 55/F | M4/de novo | > 200 | 46, XX | 98 |
| 9 | 39/M | M2/de novo | 200 | 46, XY | 96 |
| 10 | 53/F | M4/relapse | 280 | 47, XX, +4 | 99 |
| 11 | 63/M | M4/de novo | 190 | 46, XY, del(7) | 93 |
| Case no. . | Age/sex . | FAB/type . | WBC × 109/L . | Cytogenetics . | 34+/38−/G0(%) . |
|---|---|---|---|---|---|
| 1 | 72/M | M2/de novo | > 100 | 46, XY | 93 |
| 2 | 68/M | M5/de novo | 120 | 45, X, −Y | nd |
| 3 | 50/F | M1/relapse | 60 | 46, XX | 96 |
| 4 | 30/M | M5/de novo | 27 | 47, XY, +21, t(17;21) | 97 |
| 5 | 56/F | M4/relapse | 86 | 46, XX | 83 |
| 6 | 61/F | M5/de novo | 380 | 46, XX | 98 |
| 7 | 50/F | M1/de novo | 140 | 46, XX | 94 |
| 8 | 55/F | M4/de novo | > 200 | 46, XX | 98 |
| 9 | 39/M | M2/de novo | 200 | 46, XY | 96 |
| 10 | 53/F | M4/relapse | 280 | 47, XX, +4 | 99 |
| 11 | 63/M | M4/de novo | 190 | 46, XY, del(7) | 93 |
WBC indicates white blood cell; 34+/38−/G0, the percentage of CD34+/CD38−/CD123+ cells residing in the G0 phase of the cell cycle; M, male; F, female; nd, not determined.
EMSA of normal and AML CD34+ cells.
NF-κB binding activity is detected in CD34+ AML cells but not in CD34+ normal hematopoietic cells by electrophoretic mobility shift assays. (A) Two different normal bone marrow specimens (lanes 1 and 2) and one normal CB specimen (lane 3) were analyzed for NF-κB binding activity in comparison to 6 CD34+ AML specimens from patients with different FAB types (specimens 1-6 in Table 1). Nuclear extracts equivalent to 100 000 cells were used for each reaction. (B) Competition assays for 3 of the AML samples from panel A. A 200-fold molar excess of unlabeled oligonucleotide (wild-type or mutant) was used. Supershift assays were done by adding 2 μg per reaction of antibodies against NF-κB subunits p65 or p50. Arrows indicate NF-κB species and asterisks indicate nonspecific bands.
EMSA of normal and AML CD34+ cells.
NF-κB binding activity is detected in CD34+ AML cells but not in CD34+ normal hematopoietic cells by electrophoretic mobility shift assays. (A) Two different normal bone marrow specimens (lanes 1 and 2) and one normal CB specimen (lane 3) were analyzed for NF-κB binding activity in comparison to 6 CD34+ AML specimens from patients with different FAB types (specimens 1-6 in Table 1). Nuclear extracts equivalent to 100 000 cells were used for each reaction. (B) Competition assays for 3 of the AML samples from panel A. A 200-fold molar excess of unlabeled oligonucleotide (wild-type or mutant) was used. Supershift assays were done by adding 2 μg per reaction of antibodies against NF-κB subunits p65 or p50. Arrows indicate NF-κB species and asterisks indicate nonspecific bands.
NF-κB binding activity in primitive AML cells
To directly analyze NF-κB in primitive AML cells, primary specimens were sorted to isolate CD34+, CD38−, and CD123+ cells (> 95% pure). Importantly, these cells were not cultured or stimulated in any way. Therefore, they presumably reflect the status of primitive AML cells in vivo. Figure2A shows representative EMSA analysis for NF-κB in the sorted CD34+/CD38−/CD123+ AML cells. Again, as seen for AML blasts (Figure 1), binding is readily detected, with involvement of both the p50 and p65 subunits. This observation is important for 2 reasons. First, it indicates that NF-κB is relevant to the biology of AML stem cells, a marked departure from the regulation of normal stem cells. Second, it suggests a role for NF-κB that is not commonly observed. In most instances, induction of NF-κB activity is associated with cellular activation, as seen in response to inflammatory cytokines or mitogen stimulation.29 However, we determined that CD34+/CD38−/CD123+ AML cells are almost entirely quiescent. The data shown in Table 1 indicate that, with the exception of sample 5, the average proportion of cells in G0 corresponds to 96% ± 2.3%. Although sample 5 is atypical with only 83% of cells in G0, it should be noted that the remainder of the cells for this specimen were almost completely G1, with only 0.5% showing evidence of active DNA replication (ie, S or G2 phase). The cell-cycle studies were performed using a method of analysis previously developed for characterization of normal stem/progenitor cells.26 This strategy, referred to as surface, intracellular, and DNA (SID) labeling, uses a combination of cell surface markers, nuclear antigens, and DNA dyes to permit detailed analyses of cycle status. Figure 2B shows the cell-cycle profile for a representative specimen using the SID method of labeling. The panel on the left indicates the CD34+/CD38− gate used for cell analysis (CD123 gate not shown). The panel on the right shows Ki-67 versus DAPI labeling of CD34+/CD38− cells. G0is defined as cells that are negative for expression of nuclear antigen Ki-67 and have a normal diploid DNA content (lower left quadrant). Clearly, the vast majority of AML CD34+/CD38−cells fall into this category. These data suggest that NF-κB is active in quiescent and primitive AML cells.
EMSA of primitive AML cells.
Quiescent CD34+38−AML cells demonstrate NF-κB binding activity. (A) Representative example (specimen 1 in Table 1) of an EMSA shows NF-κB binding for AML CD34+/CD38−/CD123+ cells. As shown for Figure 1B, binding is competed by wild-type, but not mutant oligonucleotide, and both p50 and p65 subunits are detected. (B) Representative example of AML cells gated for CD34+/38− (left panel) and labeled by the SID method of cell-cycle analysis (right panel). An identical pattern of NF-κB binding was observed for 3 of 3 CD34+/CD38−/CD123+ AML specimens.
EMSA of primitive AML cells.
Quiescent CD34+38−AML cells demonstrate NF-κB binding activity. (A) Representative example (specimen 1 in Table 1) of an EMSA shows NF-κB binding for AML CD34+/CD38−/CD123+ cells. As shown for Figure 1B, binding is competed by wild-type, but not mutant oligonucleotide, and both p50 and p65 subunits are detected. (B) Representative example of AML cells gated for CD34+/38− (left panel) and labeled by the SID method of cell-cycle analysis (right panel). An identical pattern of NF-κB binding was observed for 3 of 3 CD34+/CD38−/CD123+ AML specimens.
Proteasome inhibitors down-regulate NF-κB activity in primary AML cells
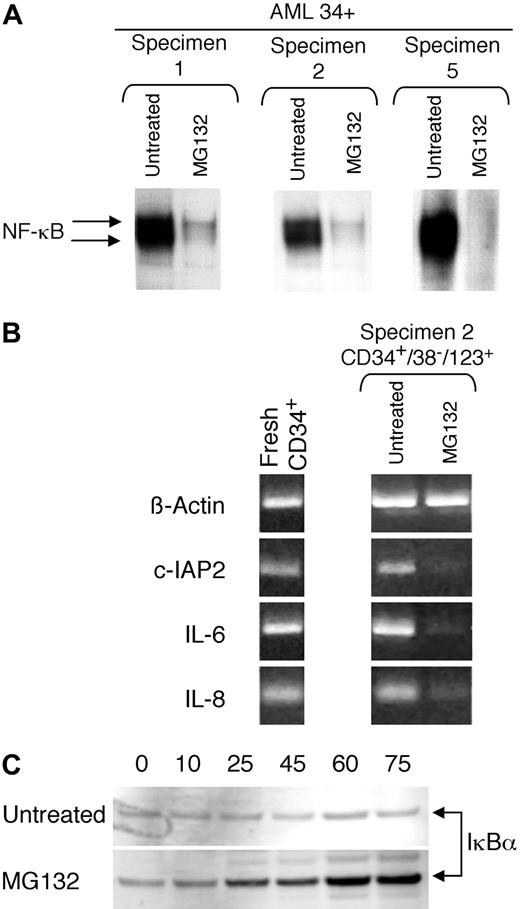
Given the unique presence of NF-κB in primitive AML cells, we sought to further characterize its activity as a transcriptional activator. To this end, one approach is to assess the consequences of NF-κB inhibition. A recently used strategy for NF-κB inhibition involves the use of proteasome inhibitors, which prevent degradation of the regulatory molecule IκBα.30 By maintaining high levels of IκBα, NF-κB remains sequestered in the cytoplasm and is therefore not active. Consequently, primary AML cells were tested with the peptide aldehyde MG-132, a potent proteasome inhibitor31. As shown in Figure3A, a 6-hour treatment with 1 μM MG-132 resulted in strong inhibition of the NF-κB binding activity. This observation was consistent for all of the specimens listed in Table 1(data not shown). To further assess the activity of NF-κB, we also examined downstream events arising from inhibition of NF-κB by determining whether the mRNA levels of NF-κB transcriptional targets were affected. Some known genes that are stimulated by NF-κB include the cellular inhibitor of apoptosis 2 (c-IAP2),22 IL-6 (IL6),32 and IL-8 (IL8).33 Figure 3B shows reverse transcription–PCR (RT-PCR) analysis for each of these 3 genes, as well as for an actin control. The left panel shows that all genes are detected in primary AML CD34+ cells, as would be expected given the activity of NF-κB. The right panel shows an RT-PCR analysis of mRNA isolated from 200 sorted CD34+/CD38−/CD123+ AML cells, plus or minus 6 hours treatment with 1 μM MG-132. The 3 NF-κB–regulated genes are readily detected in the untreated samples, but inhibited in the presence of MG-132, consistent with the down-regulation of NF-κB. As expected, the actin control is unaffected by the presence of MG-132. These data further support the concept that NF-κB is active in primitive AML cells.
Effects of MG-132 treatment on AML cells.
MG-132 inhibits NF-κB activity in AML cells. (A) EMSA of 3 CD34+ AML specimens treated with or without 1 μM MG-132 for 6 hours. (B) RT-PCR analysis of 3 NF-κB transcriptional targets in AML CD34+ (left panel) or CD34+/38− cells (right panel). CD34+/CD38− cells were treated with or without 1 μM MG-132 for 6 hours; β-actin was used as a control. (C) Western blot analysis of phosphorylated IκBα in a control (untreated) versus MG-132–treated specimen. Cells were incubated for the indicated number of minutes (numbers above each lane) and then analyzed as described in “Materials and methods.”
Effects of MG-132 treatment on AML cells.
MG-132 inhibits NF-κB activity in AML cells. (A) EMSA of 3 CD34+ AML specimens treated with or without 1 μM MG-132 for 6 hours. (B) RT-PCR analysis of 3 NF-κB transcriptional targets in AML CD34+ (left panel) or CD34+/38− cells (right panel). CD34+/CD38− cells were treated with or without 1 μM MG-132 for 6 hours; β-actin was used as a control. (C) Western blot analysis of phosphorylated IκBα in a control (untreated) versus MG-132–treated specimen. Cells were incubated for the indicated number of minutes (numbers above each lane) and then analyzed as described in “Materials and methods.”
Because proteasome inhibitors are thought to inhibit NF-κB by preventing the degradation of IκBα, we tested MG-132–treated cells to determine whether this was the case. The signal for IκBα degradation (via a ubiquitin-mediated pathway) is phosphorylation on serines 32 and 36. Thus, treatment with MG-132 should result in the accumulation of phosphorylated IκBα. Representative data in Figure3C show that phosphorylated IκBα rapidly increases in MG-132–treated AML cells, but not in untreated controls. These data indicate that NF-κB is inhibited by increased cytoplasmic levels of the negative regulator IκBα.
MG-132 induces apoptosis in AML but not normal primitive cells
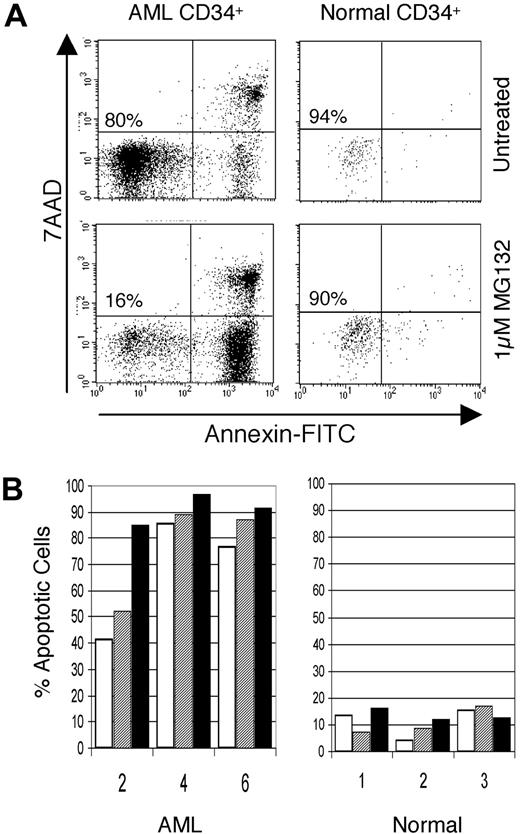
Because NF-κB is known to promote cell survival in many cases, we next determined whether its inhibition by MG-132 could preferentially induce cell death in primary AML cells while sparing normal cells. Figure 4A shows an example of annexin V staining of AML versus normal CD34+ cells in response to a 12-hour treatment with 1 μM MG-132. Cells in the lower left quadrant of each dot plot represent viable cells, with early to mid stages of apoptosis detected in the lower right and upper right quadrants, respectively. Although normal cells show almost no loss of viability in the presence of MG-132, the AML CD34+ cells are strongly induced to undergo apoptosis (reduction from 80% to 16% viable). Figure 4B shows further analysis of this phenomenon. Primary cells were cultured for 12 hours with MG-132 alone (0.5 or 1.0 μM) or in a combination of 1.0 μM MG-132 and 5 mM sodium salicylate. Sodium salicylate is also a known inhibitor of NF-κB,34and previous studies have shown that it can also be toxic to AML cells, albeit with slower kinetics (M.L.G. and C.T.J., unpublished observations, 2000). The figure shows the percentage of apoptotic cells for 3 AML versus normal CD34+ cell specimens, as assessed by staining with annexin V. The data indicate a strong induction of apoptosis in the AML specimens, with almost no effect on the normal cells. Interestingly, even the lower concentration of MG-132 (0.5 μM) was sufficient to achieve high levels of apoptosis for AML specimens 4 and 6. In contrast, AML specimen 2 had a more moderate response to MG-132 alone, but a strong response to the combination of MG-132 and salicylate. Because 5 mM salicylate alone has almost no discernible effect within a 12-hour time frame (data not shown), this observation suggests that in some cases the actions of MG-132 and sodium salicylate may be additive or synergistic.
MG-132 induces apoptosis in AML but not normal CD34+ cells.
Inhibitors of NF-κB induce apoptosis in AML but not in normal CD34+ hematopoietic cells. (A) Representative flow cytometric profile of primary AML cells (left panels) compared to normal cord blood cell (right panels) after a 6-hour treatment with or without 1 μM MG-132. Cells failing to label with annexin or the DNA dye 7-AAD represent viable cells (lower left). As apoptosis begins, cells start to label with annexin (lower right quadrant). As apoptosis proceeds, membrane integrity breaks down, and cells also label with 7-AAD (upper right quadrant). (B) Percent apoptotic cells in primary CD34+ AML cells (left panel, specimens 2, 4, and 6) compared to normal cord blood CD34+ cells (right panel) treated with 0.5 μM MG-132 (open bars), 1 μM MG-132 (hatched bars), or 1 μM MG-132 plus 5 mM sodium salicylate (dark bars).
MG-132 induces apoptosis in AML but not normal CD34+ cells.
Inhibitors of NF-κB induce apoptosis in AML but not in normal CD34+ hematopoietic cells. (A) Representative flow cytometric profile of primary AML cells (left panels) compared to normal cord blood cell (right panels) after a 6-hour treatment with or without 1 μM MG-132. Cells failing to label with annexin or the DNA dye 7-AAD represent viable cells (lower left). As apoptosis begins, cells start to label with annexin (lower right quadrant). As apoptosis proceeds, membrane integrity breaks down, and cells also label with 7-AAD (upper right quadrant). (B) Percent apoptotic cells in primary CD34+ AML cells (left panel, specimens 2, 4, and 6) compared to normal cord blood CD34+ cells (right panel) treated with 0.5 μM MG-132 (open bars), 1 μM MG-132 (hatched bars), or 1 μM MG-132 plus 5 mM sodium salicylate (dark bars).
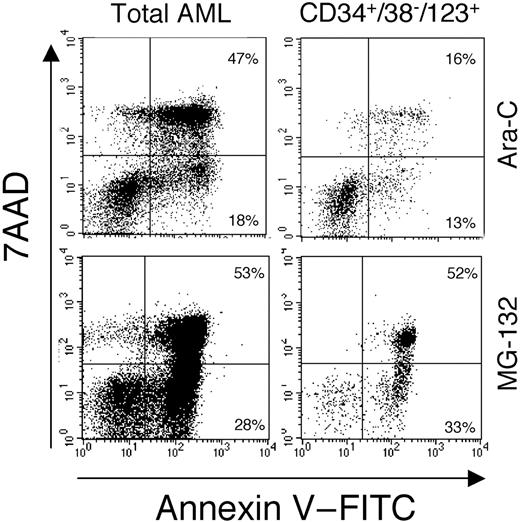
Finally, we compared the effect of MG-132 to the standard chemotherapeutic drug, Ara-C. Like most chemotherapeutic drugs, Ara-C preferentially kills actively cycling cells. As described above, most of the LSC population is in the G0 phase of cell cycle and, therefore, is not expected to be effectively targeted by Ara-C. Figure5 shows annexin V labeling of total versus CD34+/CD38−/CD123+ AML cells in response to 100 μg/mL Ara-C or 1 μM MG-132. Treatment with Ara-C shows a potent induction of apoptosis for total AML cells, but a much less effective targeting of primitive CD34+/CD38− cells (compare top left and right panels). In contrast, treatment with MG-132 shows a strong induction of apoptosis for both the total cell sample, as well as the CD34+/CD38− compartment (87% in early to mid stages of apoptosis). These data further indicate that MG-132 effectively and preferentially targets primitive, quiescent AML cells.
MG-132 induces apoptosis in primitve AML cells.
MG-132 induces potent apoptosis in primitive AML cells (specimen 7 in Table 1). Annexin V/7-AAD profiles of total AML (left panels) and CD34+/CD38− cells (right panels) treated with 100 μg/mL Ara-C (top panels) or 1 μM MG-132 (bottom panels).
MG-132 induces apoptosis in primitve AML cells.
MG-132 induces potent apoptosis in primitive AML cells (specimen 7 in Table 1). Annexin V/7-AAD profiles of total AML (left panels) and CD34+/CD38− cells (right panels) treated with 100 μg/mL Ara-C (top panels) or 1 μM MG-132 (bottom panels).
Discussion
Acute myelogenous leukemia arises from a stem cell precursor with a phenotype that was originally described as CD34+, CD38−, HLA-DR−.7-9 Because this phenotype is identical to that of normal hematopoietic stem cells, it has previously been difficult to clearly distinguish the malignant stem cell population. However, we and others have recently described surface antigens, CD90, CD117, and CD123, that are differentially expressed between normal and leukemic populations, thereby allowing isolation of primitive AML cells without normal cell contamination.10-12 With this capability in hand, we sought to identify differences between normal and malignant stem cell populations at the molecular level. Of course, AML is known to be a heterogeneous disease, with many different histologically defined subtypes (FAB subtypes M0 to M7). Although the blast cell populations from the various subtypes clearly have differing cellular and molecular properties, we suggest that at the stem/progenitor cell level many commonalities are present. For example, in addition to a similar phenotype, most AML specimens show a quiescent cell-cycle status in the CD34+/CD38− population (Jordan and coworkers, manuscript in preparation). This observation is consistent with previous studies that have suggested leukemic stem cells are likely to be quiescent.35,36 Further, as shown in this study and others,13 the expression of certain genes appears to be conserved among many different types of AML. Given these similarities, we propose that those features held in common among varying subtypes may in fact represent the most important characteristics of AML stem cells and are therefore potentially key elements of the leukemogenic process.
For most cell types, NF-κB is found as an inactive form in the cytoplasm. On stimulation with a large variety of different factors, the regulatory molecule IκBα is degraded and NF-κB is free to translocate to the nucleus and form an active transcriptional complex.30 In this regard, NF-κB plays an important role in inflammation and stress responses, and is a major mediator of cell survival.28 Indeed, it can be activated by numerous different kinds of stimuli, and regulates the transcription of over 150 genes.37 Importantly, NF-κB has been reported to be active in a number of different malignancies.14-20However, most of these studies were conducted with tumor cell lines and may not necessarily reflect the biology of primary tumors in vivo. The experiments described in this article, and recent studies in acute lymphoblastic leukemia (ALL) are among the few that actually examine NF-κB in primary tumor tissue.18 Interestingly, like our studies in AML, the analysis of primary ALL specimens by Kordes and colleagues observed NF-κB activity in 39 of 42 samples without any subtype restriction. Further, they also observed complexes composed of both p50 homodimers and p50-p65 heterodimers. Given the central role of NF-κB as a transcriptional regulator, expression of this factor in both AML and ALL cells represents a striking biologic distinction between leukemic and normal tissue.
One recent study detected NF-κB in normal CD34+ cells; however, its activity was only evident on stimulation with phorbol ester (PMA).38 Our data, in agreement with Pyatt and colleagues, shows no NF-kB activity in naı̈ve (ie, unstimulated) CD34+ cells from normal tissue. The fact that NF-κB is highly activated in leukemic cells suggests that an intrinsic aspect of AML biology resides in constitutive activation of various pathways. Indeed, this observation is consistent with previous studies showing activation of mitogen-activated protein kinase and signal transducer and activator of transcription 3 in primary AML specimens.39,40 However, it is noteworthy that we also observe NF-κB activity in the noncycling subpopulation of primitive CD34+/CD38− AML cells. Although some studies have indicated a role for NF-κB in quiescent cells,41 it is most commonly associated with growth and inflammatory responses.29 Thus, activity of NF-κB in primitive AML cells appears to represent a unique and previously undocumented role for this factor in the biology of stem cells.
An important question for future studies will be to determine why NF-κB is activated in leukemic cells. One possibility is that autocrine production of cytokines may play a role in stimulating NF-κB. This idea is supported by studies showing spontaneous expression of IL-1β and IL-6 in AML blast cells, as well as elevated levels of IL-6 in the serum of most patients with AML.20,42 Alternatively, dysregulation of NF-κB regulatory components may also be responsible, as has recently been reported for Hodgkin lymphoma and melanoma cells.43 44Further analysis of this issue is an important priority for determining the underlying cause of leukemia.
Regardless of the reason for NF-κB activation, given its known activity as a survival factor for many normal and malignant cell types, we hypothesized that the presence of NF-κB could represent a unique opportunity for inducing apoptosis in the LSC compartment. Indeed, treatment of AML cells with the proteasome inhibitor MG-132 produced a rapid down-regulation of NF-κB and a concomitant induction of apoptosis. Importantly, this apoptosis was also readily evident in CD34+/CD38− populations, thereby suggesting that quiescent AML cells are not spared. In contrast, normal CD34+/CD38− cells showed no significant degree of apoptosis in response to MG-132. It should be noted that normal CD34+ cells are eventually impaired by treatment with MG-132 (culture at higher concentrations or for > 24 hours)45; however, in the time frame used for the current studies, this was not observed. Moreover, the mechanism of cell death is apparently independent of NF-κB inhibition.
Although definitive proof of LSC apoptosis will await detailed analysis of MG-132–treated cells in appropriate animal model systems, we propose that the data in this report strongly suggest that LSCs are preferentially sensitive to inhibition of NF-κB. Currently, we are attempting to use a mutant (dominant-negative) allele of IκBα as a means to directly assess the role of NF-κB in malignant cells and to determine whether inhibition of its activity is sufficient to mediate apoptosis induction. If NF-κB is a central regulator of LSC survival, it clearly represents an exciting potential target for future therapeutic studies.
We gratefully acknowledge the generous support of the McDowell Cancer Foundation and the Donatina Colachicco Cancer Research Fund. We also thank Drs Gary Van Zant, Charlotte Kaetzel, Hartmut Geiger, and Stephen J. Szilvassy for helpful discussions and critical evaluation of the manuscript. Further, we acknowledge the NDRI for help in procuring normal bone marrow and CB specimens.
Supported by grants to C.T.J. from the Leukemia and Lymphoma Society (translational grant 6057-99), and the American Cancer Society (RPG-99-206-01-LBC).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Craig T. Jordan, Markey Cancer Center, 800 Rose St, Rm CC407, Lexington, KY 40536-0093; e-mail: cjordan@uky.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal