Abstract
Dendritic cells (DCs) are important targets for human immunodeficiency virus (HIV) because of their roles during transmission and also maintenance of immune competence. Furthermore, DCs are a key cell in the development of HIV vaccines. In both these settings the mechanism of binding of the HIV envelope protein gp120 to DCs is of importance. Recently a single C-type lectin receptor (CLR), DC-SIGN, has been reported to be the predominant receptor on monocyte-derived DCs (MDDCs) rather than CD4. In this study a novel biotinylated gp120 assay was used to determine whether CLR or CD4 were predominant receptors on MDDCs and ex vivo blood DCs. CLR bound more than 80% of gp120 on MDDCs, with residual binding attributable to CD4, reconfirming that CLRs were the major receptors for gp120 on MDDCs. However, in contrast to recent reports, gp120 binding to at least 3 CLRs was observed: DC-SIGN, mannose receptor, and unidentified trypsin resistant CLR(s). In marked contrast, freshly isolated and cultured CD11c+ve and CD11c−ve blood DCs only bound gp120 via CD4. In view of these marked differences between MDDCs and blood DCs, HIV capture by DCs and transfer mechanisms to T cells as well as potential antigenic processing pathways will need to be determined for each DC phenotype.
Introduction
Dendritic cells (DCs) play a major role in human immunodeficiency virus (HIV) pathogenesis. Peripheral or surveillance mucosal DCs are one of the first cell types infected and are distributed in the vaginal, ectocervical, and anal mucosa,1,2 allowing contact with HIV during mucosal exposure. Thus, after vaginal inoculation with simian immunodeficiency virus in macaques, DCs are the predominant cell type infected.3 Furthermore, the ability of DCs to cluster with and stimulate T cells may also play a key role in establishing infection. DCs from skin, mucosa, and blood of humans and macaques can participate in highly productive HIV and simian immunodeficiency virus infection in DC–T-cell cocultures and illustrates the importance of this natural DC–T-cell synergy.4-7
Key aspects of HIV binding to DC via gp120 are ill-defined, particularly to the different types of DCs. CD11c+ve and CD11c−ve blood DCs, Langerhans cells (LCs), and in vitro–derived monocyte-derived DCs (MDDCs) all express CD4 and CCR5 and can be productively infected in vitro.8-12 However, HIV also bound several DC populations independently of CD4.8,13,14 The heavy glycosylation of gp120 with mannose and fucose saccharides suggested HIV bound to cells also via lectin receptors. Binding of gp120 to a novel C-type lectin receptor (CLR), originally identified from a placental complementary DNA (cDNA) library15 on the basis of HIV gp120 binding and named clone 11, on MDDCs was recently reported.14,16 The adhesion properties of this CLR were also defined and the receptor subsequently renamed DC-SIGN (dendritic cell specific ICAM-3 grabbing nonintegrin). Although MDDCs express a diverse and abundant array of CLRs in addition to DC-SIGN,16-24 and given substantial overlap in saccharide recognition by such CLRs, they may also serve as receptors for gp120 on MDDCs. The roles of CD4 and CLRs on most other in vivo DC types are unknown.
This study aimed to define the contributions of CD4 and CLRs in binding gp120, to address and identify the capacity of other CLRs including DC-SIGN during monocyte differentiation to mature MDDCs and, more importantly, to compare such populations with ex vivo blood DCs. Understanding the mechanisms of gp120 binding to different DC populations would help define the early events of HIV transmission via DCs in blood or mucosal tissue and improve intervention strategies. Definition of the mechanisms of HIV/gp120 binding and processing by DCs will also assist future HIV vaccine strategies and immunotherapy.
Materials and methods
MDDC generation and culture
Monocytes were isolated from 500 mL of blood (Parramatta Blood Bank, Australia) by countercurrent elutriation as previously described.25,26 Monocytes were further depleted of contaminating cells by using a monocyte-enrichment cocktail (StemCell Technologies, Vancouver, BC, Canada). Monocyte fractions were at least 97% CD11c+ve, at least 90% CD14+ve, and 0.1% or less CD3+ve. DCs were converted as previously described27 28 using 500 U/mL interleukin-4 and 400 U/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) (Schering-Plough, Kenilworth, NJ). At day 6 cells were at least 95% CD1a+ve, CD11c+ve with no detectable CD14, CD3, or CD83 populations. MDDCs were matured by culture for 48 hours with 10 ng/mL tumor necrosis factor α (TNF-α) (R&D Systems, Minneapolis, MN).
Isolation and culture of blood DCs
Blood DCs were isolated from 500 mL of blood (Mater Hospital, Brisbane, Australia) using Ficoll-Paque (Amersham Pharmacia-Biotech, Uppsala, Sweden). Residual erythrocytes were removed by Vitalyze as per the manufacturer's instructions (BioErgonomics, St Paul, MN). Peripheral blood mononuclear cells (PBMCs) were labeled with a mixture of anti-CD3 (OKT3), CD14 (CMRF31), CD11b (OKM1), CD16 (HUNK-2), and CD19 (FMC63) monoclonal antibodies (mAbs). After incubation with Biomag goat antimouse immunoglobulin–coated magnetic beads (Polysciences, Warrington, PA), labeled cells were removed by first preclearing with a MPC-1 magnet (Dynal, Oslo, Norway) and then passing through a Miltenyi cell separation column using a Variomacs magnet (Miltenyi Biotech, Gladbach, Germany). Depleted PBMCs were labeled with fluorescein isothiocyanate (FITC)–goat antimouse (Becton Dickinson, San Jose, CA) and negative cells separated by sorting on a FACSVantage (Becton Dickinson). For cultured blood DCs, DCs were incubated overnight at a concentration of 1 × 106 cells per milliliter in RPMI 1640 supplemented with 10% fetal calf serum and 10 ng/mL interleukin-3 (Gibco, Grand Island, NY) and 200 U/mL GM-CSF (Novartis, Basel, Switzerland).
HIV gp120 binding and inhibition studies
Purified HIV gp120 from the BaL isolate (courtesy of Ray Sweet, SmithKline Beecham, King of Prussia, PA) was biotinylated with EZ-Link NHS-LC-Biotin as per the manufacturer (Pierce, Rockford, IL). Biotinylation of gp120 did not affect the ability of the molecule to bind to CD4 and was confirmed in an sCD4 capture enzyme-linked immunosorbent assay with detection via streptavidin horseradish peroxidase (data not shown). In addition, nonbiotinylated gp120 material from the isolates BaL and 92MW959, using detection with purified and biotinylated human polyclonal antibodies from HIV-seropositive patients (Cellular Products, Buffalo, NY), produced equivalent results to biotinylated gp120 from respective isolates. In particular, the saturating concentrations of gp120 and the relative binding of gp120 by CD4 and CLR on MDDCs were the same by both methods. However, biotinylated gp120 binding assay was routinely used because it reduced one additional antibody staining step, reduced the variability of antibody binding, and allowed for flexibility when working with blood DCs, which are labeled with multiple antibodies for detection of multiple DC subsets.
For binding and inhibition studies, cells were preincubated for 40 minutes in binding media (RPMI 1640 without sodium bicarbonate [Gibco] with 1% bovine serum albumin and 10 mM HEPES [Calbiochem, San Diego, CA] pH 7.4) as above at 4°C with stated concentrations of inhibitors, followed by incubation with b-gp120 (2-fold the predetermined concentration for cellular saturation). Levels of inhibitors, with the exception of mAbs, were initially determined using a broad range of concentrations to assess the maximal level of gp120 blocking. In the cases of mAb, concentrations were routinely 5-fold that of cellular saturation. Cells were then washed twice, and measurement of bound b-gp120 was carried out by incubation of 1 × 106 cells (2 × 105 cells/200 μL) with 5 μg/mL streptavidin Oregon Green 488 (Molecular Probes, Eugene, OR) or avidin FITC (Becton Dickinson) and detected by flow cytometry.
Flow cytometric analysis
For surface staining, cells were treated as previously described.29 In gp120 binding studies, cells were preincubated with b-gp120 at various concentrations for 40 minutes at 4°C in binding media. Antibodies used were CD14-phycoerythrin (PE), immunoglobulin G1 (IgG1)–PE, IgG1-FITC, CD3-FITC, IgG1, goat antimouse FITC (all from Becton Dickinson), CD83, CD86, CD1a-FITC, MR (clones 19 and 3.29), and HLA-DR–PE/P5 (all from PharMingen, San Diego, CA, except anti-MR 3.29, which is from Immunotech, Marseille, France). The CD4 mAbs used were Leu3a (Becton Dickinson), OKT4 (American Type Culture Collection, Manassas, VA), and Q4120 (a generous gift from Quentin Sattentau). The mAbs to DC-SIGN (AZN-D1 and AZN-D2) and associated experiments were a part of the 7th Leukocyte Differentiation DC Antigen Workshop (kindly donated by Yvette van Kooyk). Detection of b-gp120 and biotinylated polyclonal sera to HIV (Cellular Products) was via strepdavidin Oregon Green 488 or avidin FITC.
DC-SIGN reverse transcriptase–polymerase chain reaction
Cells were prepared as above apart from monocytes that were positively selected over a magnetic-activated cell separation column according to the manufacturer (Miltenyi Biotech). The CD11c+ve and CD11c−ve blood DCs selected Vantage fluorescent cell sorting. Total RNA was prepared from 10 000 cells using TRIzol (Gibco) as per the manufacturer. The cDNA was synthesized from DNaseI-treated RNA with oligo-dT primers and Superscript II (Gibco). From 40 μL of RNase H–treated cDNA, 1 μL was polymerase chain reaction (PCR)–amplified with Taq polymerase (Qiagen, Germany) using either the GAPDH primers, 5′-ATGGGGAAGGTGAAGGTCGGA-3′ and 5′-AGGGGCCATCCACAGTCTTCTG-3′, to ensure equivalent amounts of cDNA in each cell type or using the first-round DC-SIGN primers, 5′AGAGTGGGGTGACATGAGTG-3′ and 5′-GAAGTTCTGCTACGCAGGAG-3′, which yielded a fragment approximately 1.2 kilobases in size. A seminested round of PCR was performed for DC-SIGN using the former 5′ primer and 5′-AGCTCCTGGTAGATCTCCTGC-3′. Electrophoresed products were transferred from a 1% agarose gel to Hybond N+ and probed with digoxigenin-labeled internal oligonucleotide 5′-CCAGAGAAATCTAAGCTGCAGG-3′ as per the manufacturer (Roche Biochemicals, Basel, Switzerland).
HIV gp120 internalization and tracking
To examine gp120 internalization, cells were labeled with b-gp120 as described above, washed, and subsequently incubated at 37°C. For short-term incubations (< 2 hours) cells were incubated in a 37°C water, and for longer incubations (> 2 hours) cells were replated and cultured at 37°C in a 5% CO2 incubator. Aliquots were removed at the times outlined in “Results” and terminated by incubation in 0.25% (wt/vol) paraformaldehyde in phosphate-buffered saline at 4°C for 30 minutes. For internal staining, cells were permeabilized with 0.2% (vol/vol) Tween 20, 1% (vol/vol) fetal calf serum in phosphate-buffered saline for 15 minutes at 37°C. Detection of external or internal gp120 was via streptavidin Oregon Green 488 as described above.
Results
HIV gp120 binding to CLR and/or CD4 on immature MDDCs
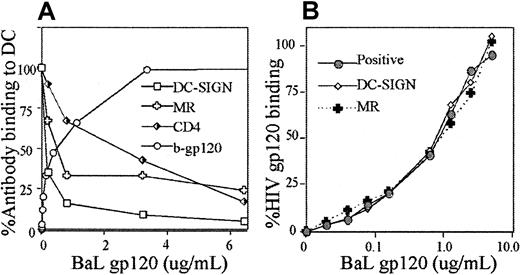
Because of its potent inhibition of CLRs15 and lack of interference with gp120-CD4 binding, mannan was chosen as an inhibitory ligand to determine the proportion of gp120 bound to CLRs in MDDCs.15 In MDDCs, mannan inhibited gp120 by up to 84% (Figure 1A). Higher levels of mannan were also used (up to 25 mg/mL), but further gp120 blocking was not observed (data not shown). Nonbiotinylated Chinese hamster ovary cell–expressed gp120 (detected via anti-HIV polyclonal antibodies) from the primary R5 isolate MW959 was also inhibited with mannan by up to 80% (data not shown). The other CLR inhibitor, α-methyl-mannopyranoside, and the calcium chelator, ethyleneglycotetraacetic acid (EGTA), inhibited gp120 binding by 82% and 77%, respectively (Figure2). The residual gp120 binding was initially attributed to CD4. Therefore, the gp120-blocking CD4 mAbs Leu3a and Q4120, with the nonblocking mAb OKT4 as a negative control, were used to determine CD4 binding. However, neither Leu3a nor Q4120 could block gp120 binding at concentrations up to 25 μg/mL (Figure1B). In view of this CLR-gp120 binding predominance, incubation with CD4 mAbs after prior blocking of CLR binding was examined. To achieve this, MDDCs were preincubated with 5 mg/mL mannan and then with increasing amounts of the Leu3a. In the absence of CLR binding, anti-Leu3a was successful at inhibiting the residual 10% to 20% gp120 binding to less than 1% of gp120 binding (Figure 1B).
Inhibition of gp120 binding on MDDC.
(A) Inhibition of gp120 binding to MDDCs by mannan; 1 × 106 cells/mL were incubated with mannan ranging from 50 μg to 5 mg/mL for 30 minutes at 4°C. The b-gp120 was added at 3-fold excess (9 μg/mL) and incubated for 30 minutes at 4°C. The b-gp120 was detected via streptavidin Oregon Green 488 and fluorescence measured by flow cytometry as described. (B,C) Inhibition of gp120 binding to MDDCs by CD4 mAbs (Leu3a, Q4120, OKT4). In panel B, cells were preincubated with mAbs to CD4 (ranging from 0.05 μg/mL to 25 μg/mL) for 30 minutes at 4°C. In panel C, cells were also incubated with 5 mg/mL mannan in addition to the CD4 mAb Leu3a. The b-gp120 was incubated and detected as in panel A. Percent DC-gp120 binding was calculated as follows: [(sample fluorescence intensity − mean negative control fluorescence intensity)/mean positive control fluorescence intensity] × 100. Positive control cells were treated with b-gp120 in the absence of inhibitors. Negative controls consisted of cells with identical inhibitors but no b-gp120.
Inhibition of gp120 binding on MDDC.
(A) Inhibition of gp120 binding to MDDCs by mannan; 1 × 106 cells/mL were incubated with mannan ranging from 50 μg to 5 mg/mL for 30 minutes at 4°C. The b-gp120 was added at 3-fold excess (9 μg/mL) and incubated for 30 minutes at 4°C. The b-gp120 was detected via streptavidin Oregon Green 488 and fluorescence measured by flow cytometry as described. (B,C) Inhibition of gp120 binding to MDDCs by CD4 mAbs (Leu3a, Q4120, OKT4). In panel B, cells were preincubated with mAbs to CD4 (ranging from 0.05 μg/mL to 25 μg/mL) for 30 minutes at 4°C. In panel C, cells were also incubated with 5 mg/mL mannan in addition to the CD4 mAb Leu3a. The b-gp120 was incubated and detected as in panel A. Percent DC-gp120 binding was calculated as follows: [(sample fluorescence intensity − mean negative control fluorescence intensity)/mean positive control fluorescence intensity] × 100. Positive control cells were treated with b-gp120 in the absence of inhibitors. Negative controls consisted of cells with identical inhibitors but no b-gp120.
Inhibition of gp120 binding to MDDCs with a range of ligands.
The mAbs to CD4 2-Leu3a and 3-Q4120), MR (4-clones 19 and 5-3.29), and DC-SIGN (AZN-5-D1 and 6-AZN-D2) were preincubated 5-fold above predetermined saturating concentrations (5 μg/mL). Inhibitors mannan (8-mannan), α-methyl-mannopyranoside (9-M/P), and EGTA (10-EGTA) were incubated in excess at 5 mg/mL, 125 mM, and 5mM, respectively. Dual Leu3a and mannan inhibition (11-2 and 8) included Leu3a and mannan at levels used above for treatments 2 and 8. Positive and negative controls (treatments 1 and 12) consisted of MDDCs incubated with or without gp120, respectively. The b-gp120 was added and detected as in the legend to Figure 1A.
Inhibition of gp120 binding to MDDCs with a range of ligands.
The mAbs to CD4 2-Leu3a and 3-Q4120), MR (4-clones 19 and 5-3.29), and DC-SIGN (AZN-5-D1 and 6-AZN-D2) were preincubated 5-fold above predetermined saturating concentrations (5 μg/mL). Inhibitors mannan (8-mannan), α-methyl-mannopyranoside (9-M/P), and EGTA (10-EGTA) were incubated in excess at 5 mg/mL, 125 mM, and 5mM, respectively. Dual Leu3a and mannan inhibition (11-2 and 8) included Leu3a and mannan at levels used above for treatments 2 and 8. Positive and negative controls (treatments 1 and 12) consisted of MDDCs incubated with or without gp120, respectively. The b-gp120 was added and detected as in the legend to Figure 1A.
Inhibition of mAb binding to specific CLRs by gp120
Candidate CLRs on MDDCs and other DCs for gp120 binding were DC-SIGN and MR.14,30 Therefore, mAb DC-SIGN (AZN-D2)14,16 and MR (clone 19)31 were used because they have been shown previously to block ligand binding. Preincubation of MDDCs with gp120 inhibited DC-SIGN (AZN-D2), MR (clone 19), and CD4 (Leu3a) mAbs in a dose-dependent manner (Figure 3A). As gp120 approached cellular saturation, binding of the mAbs to all 3 receptors approached zero. The gp120 concentrations that inhibited mAb binding by 50% (Ki) mAb were, for DC-SIGN (AZN-D2), 1 nM; MR (clone 19), 4 nM; and CD4 (Leu3a), 14 nM. The approximate dissociation constant (Kd) for BaL gp120 from the gp120 saturation curve is 6 nM for 1 × 106/mL MDDCs.
Interaction between gp120 and mAbs to CD4, DC-SIGN, and MR.
(A) Inhibition of mAbs with increasing concentrations of gp120. MDDCs were incubated with increasing concentrations of b-gp120 under conditions outlined in Figure 1A. The availability of CD4 and CLR epitopes (those not blocked by gp120 binding) was detected by mAbs to CD4 (Leu3a), DC-SIGN (AZN-D2), and MR (clone 19) all at 1 μg/mL. For comparison, binding of b-gp120 alone at increasing concentrations is shown. Detection and incubation of bound b-gp120 was performed as outlined in Figure 1A. Percent binding of mAbs to DCs was calculated as per Figure 1 with positive and negative controls defined as follows. Positive controls were cells incubated with mAbs in the absence of gp120. Negative controls were cells incubated with the appropriate mAb isotype control (IgG1 for all 3 mAbs listed above). (B) Inhibition of gp120 binding to CLRs by mannan, DC-SIGN (AZN-D2), and MR (clone 19) mAbs at various concentrations of gp120. MDDCs were preincubated with mAbs as in Figure 2. After washing, b-gp120 was incubated with cells at concentrations ranging from 20 ng/mL to 5 μg/mL and detected as outlined in Figure 1A.
Interaction between gp120 and mAbs to CD4, DC-SIGN, and MR.
(A) Inhibition of mAbs with increasing concentrations of gp120. MDDCs were incubated with increasing concentrations of b-gp120 under conditions outlined in Figure 1A. The availability of CD4 and CLR epitopes (those not blocked by gp120 binding) was detected by mAbs to CD4 (Leu3a), DC-SIGN (AZN-D2), and MR (clone 19) all at 1 μg/mL. For comparison, binding of b-gp120 alone at increasing concentrations is shown. Detection and incubation of bound b-gp120 was performed as outlined in Figure 1A. Percent binding of mAbs to DCs was calculated as per Figure 1 with positive and negative controls defined as follows. Positive controls were cells incubated with mAbs in the absence of gp120. Negative controls were cells incubated with the appropriate mAb isotype control (IgG1 for all 3 mAbs listed above). (B) Inhibition of gp120 binding to CLRs by mannan, DC-SIGN (AZN-D2), and MR (clone 19) mAbs at various concentrations of gp120. MDDCs were preincubated with mAbs as in Figure 2. After washing, b-gp120 was incubated with cells at concentrations ranging from 20 ng/mL to 5 μg/mL and detected as outlined in Figure 1A.
The role of individual CLRs in binding gp120
In reciprocal experiments, the effects of prior incubation with MR (clones 19 and 3.29) and DC-SIGN (AZN-D1 and AZN-D2) blocking mAbs on gp120 binding14,16,31 32 were examined to determine relative importance of DC-SIGN and MR in gp120 binding. However, anti-MR (clones 19 and 3.29) and anti–DC-SIGN (AZN-D1 and AZN-D2) mAbs could not inhibit gp120 binding (at levels up to 5 μg/mL). The antibody bound was confirmed in each assay by goat antimouse PE, and it was confirmed that gp120 and the blocking antibodies were each bound to saturating levels on the entire MDDC population (data not shown). However, in the same assay mannan successfully reduced gp120 binding below 20% and combined mannan and Leu3a to below 1%. Because gp120 was used in excess in the above experiments, further inhibitory studies with 5-fold saturating concentrations of mAb (5 μg/mL) were carried out over a range of gp120 concentrations (20 ng/mL to 5 μg/mL) to observe the effects of MR and DC-SIGN mAbs (Figure 3B). However, no significant inhibition of gp120 binding by MR and/or DC-SIGN antibodies was observed at any concentration.
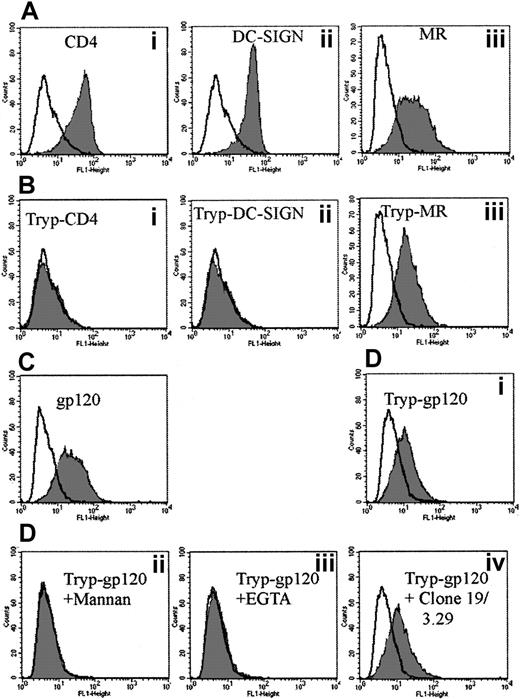
HIV gp120 binding to trypsin-insensitive CLRs
To address the possibility that MDDCs express several CLRs capable of binding gp120, cells were trypsinized to denude them of both the CD4-gp120 binding site and the carbohydrate recognition domains (CRDs) of either DC-SIGN and/or the MR. As expected, the CD4 Leu3a epitope was cleaved. The CRD for DC-SIGN was also trypsin-sensitive, whereas the MR clone 19 epitope was not (Figure 4Biii). When trypsinized MDDCs were exposed to gp120, they retained the ability to bind gp120 at a reduced level (Figure 4Di). If trypsinized cells were preexposed to mannan or EGTA, they lost their ability to bind to gp120, indicating binding was carbohydrate- and calcium-dependent, characteristic of a trypsin-resistant CLR but clearly not DC-SIGN (Figure 4Dii, iii, respectively). To address whether this CLR might be MR, the anti-MR mAb clones 19 and 3.29 were used to block the trypsin-insensitive gp120 binding. However, MR mAb clones 19 and 3.29 could not significantly reduce trypsin-insensitive gp120 binding (Figure 4Div). To ensure that the mAbs can block gp120 binding, parallel studies were carried out with a transfected cell line expressing macrophage mannose receptor (MMR).33 The MMR mAbs could inhibit gp120 binding to 50% regardless of whether these cells were trypsinized (data not shown); ie, the mAbs were partial inhibitors of gp120 binding to MMR on the transfected cell line but had no effect on MDDCs regardless of whether they were trypsinized.
Effect of trypsin on CD4, MR, and DC-SIGN mAbs and b-gp120 binding to MDDCs.
(A) CD4 (Leu3a) (Ai), DC-SIGN (AZN-D2) (Aii), and MR (clone 19) (Aiii) staining before trypsinization. A total of 2 μg/mL of mAb to CD4, DC-SIGN, and MR was added as outlined in “Materials and methods.” The mAb binding was detected via goat antimouse FITC (1 μg/mL) (Becton Dickinson) and fluorescence measured as in Figure 1. Gray histograms represent antibody staining with open overlaid histogram staining by matching isotype controls. (B) CD4 (Leu3a) (Bi), DC-SIGN (AZN-D2) (Bii), and MR (clone 19) (Biii) staining after trypsinization. Cells were treated with 0.25% trypsin at 37°C for 5 minutes and subsequently washed in normal media before the addition of mAbs to CD4, DC-SIGN, and MR as in panel A. (C) The b-gp120 binding before trypsinization. (D) The b-gp120 binding to MDDCs after trypsinization: effect of inhibitors. Trypsinized cells were mock-treated (Di) or treated with excess mannan (5 mg/mL) (Dii), EGTA (5 mM) (Diii), or anti-MR (clones 19 and 3.29) (5 μg/mL) (Div) for 30 minutes at 4°C. The b-gp120 was added and detected as in Figure 1A. Gray histograms represent gp120 staining and open overlays matched negative controls (treatment without addition of b-gp120).
Effect of trypsin on CD4, MR, and DC-SIGN mAbs and b-gp120 binding to MDDCs.
(A) CD4 (Leu3a) (Ai), DC-SIGN (AZN-D2) (Aii), and MR (clone 19) (Aiii) staining before trypsinization. A total of 2 μg/mL of mAb to CD4, DC-SIGN, and MR was added as outlined in “Materials and methods.” The mAb binding was detected via goat antimouse FITC (1 μg/mL) (Becton Dickinson) and fluorescence measured as in Figure 1. Gray histograms represent antibody staining with open overlaid histogram staining by matching isotype controls. (B) CD4 (Leu3a) (Bi), DC-SIGN (AZN-D2) (Bii), and MR (clone 19) (Biii) staining after trypsinization. Cells were treated with 0.25% trypsin at 37°C for 5 minutes and subsequently washed in normal media before the addition of mAbs to CD4, DC-SIGN, and MR as in panel A. (C) The b-gp120 binding before trypsinization. (D) The b-gp120 binding to MDDCs after trypsinization: effect of inhibitors. Trypsinized cells were mock-treated (Di) or treated with excess mannan (5 mg/mL) (Dii), EGTA (5 mM) (Diii), or anti-MR (clones 19 and 3.29) (5 μg/mL) (Div) for 30 minutes at 4°C. The b-gp120 was added and detected as in Figure 1A. Gray histograms represent gp120 staining and open overlays matched negative controls (treatment without addition of b-gp120).
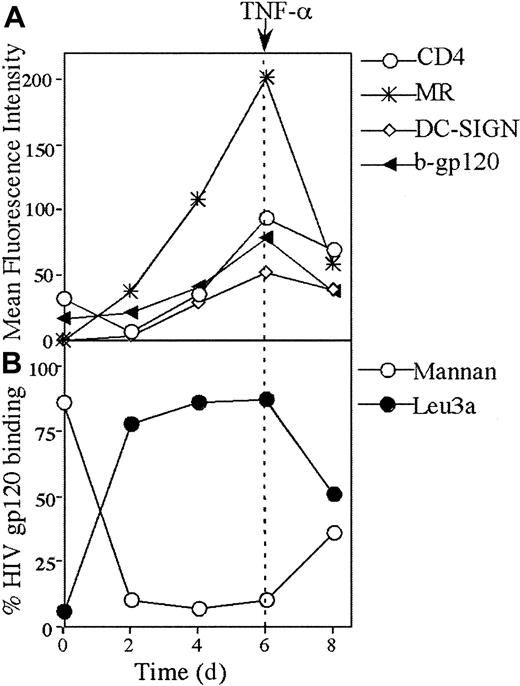
HIV gp120 binding during differentiation of monocytes to MDDCs
The switch from gp120 binding to CD4 on monocytes to CLRs on MDDCs was examined during in vitro differentiation over 6 days. By day 2, CLR binding was predominant (Figure 5B) and correlated with a rise in MR expression and CD4 down-regulation (Figure5A). Over day 2 to day 6 of differentiation, there was a continuous increase in binding of gp120 to CLR with a corresponding decrease in CD4 binding. Over the same period, there was a continuous increase in DC-SIGN, CD4, and MR expression. The peak expression of all 3 receptors at day 6 coincided with the peak in gp120 binding (Figure 5A). Mature MDDCs were generated by stimulation with TNF-α for 2 days. After maturation, MR, DC-SIGN, and CD4 were all down-regulated, but this was more marked with MR (Figure 5A). In mature MDDCs, the pattern of gp120 binding to CLRs and CD4 converged, with intermediate levels of binding to both (Figures 5B and 7B).
Kinetics of CD4, DC-SIGN, and MR expression and gp120 binding during differentiation of monocytes to immature and mature MDDCs.
(A) CD4 and CLR expression and b-gp120 binding. Monocytes were stimulated to immature MDDCs as described in “Materials and methods.” Mature MDDCs were generated from day 6 to 8 by addition of 10 ng/mL TNF-α and expressed both maturation markers CD83 and CD86 by day 8 (> 70% +ve for both markers). The b-gp120, MAb to CD4 (leu3a), DC-SIGN (AZN-D2), and MR (clone 19) were added, incubated, and detected at days 0, 2, 4, 6, and 8 as outlined in Figures 1A and 4. The mean relative intensity of the isotype or negative control was subtracted from the mean fluorescent intensity for 10 000 cells. (B) HIV gp120 binding to CD4 and CLRs. At days 0, 2, 4, 6, and 8, cells were preincubated with either saturating levels of Leu3a (10 μg/mL) or mannan (5 mg/mL) and subsequently exposed to saturating levels of b-gp120 as outlined in Figures 1 and 2.
Kinetics of CD4, DC-SIGN, and MR expression and gp120 binding during differentiation of monocytes to immature and mature MDDCs.
(A) CD4 and CLR expression and b-gp120 binding. Monocytes were stimulated to immature MDDCs as described in “Materials and methods.” Mature MDDCs were generated from day 6 to 8 by addition of 10 ng/mL TNF-α and expressed both maturation markers CD83 and CD86 by day 8 (> 70% +ve for both markers). The b-gp120, MAb to CD4 (leu3a), DC-SIGN (AZN-D2), and MR (clone 19) were added, incubated, and detected at days 0, 2, 4, 6, and 8 as outlined in Figures 1A and 4. The mean relative intensity of the isotype or negative control was subtracted from the mean fluorescent intensity for 10 000 cells. (B) HIV gp120 binding to CD4 and CLRs. At days 0, 2, 4, 6, and 8, cells were preincubated with either saturating levels of Leu3a (10 μg/mL) or mannan (5 mg/mL) and subsequently exposed to saturating levels of b-gp120 as outlined in Figures 1 and 2.
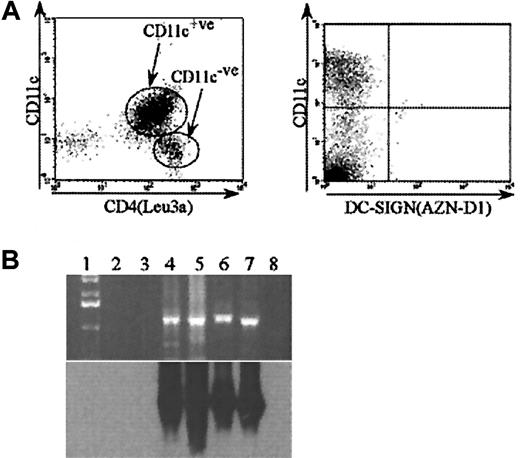
HIV gp120 binding on ex vivo blood DCs
Because MDDCs are derived in vitro, it was important to determine the gp120 binding receptors on ex vivo blood DCs. Blood DCs were separated, incubated with gp120, and triple-stained for b-gp120, CD11c, and HLA-DR, which allowed identification of 2 blood DC populations based on the presence or absence of CD11c expression. The CD11c−ve population expressed much higher levels of CD4 (Figure 6A) and bound greater amounts of gp120 than the CD11c+ve population (Figure7A). CD4 was down-regulated on both blood DC subsets after overnight culture (data not shown) and was reflected by the reduced capacity to bind gp120 (Figure 7A). The importance of CLRs and CD4 for gp120 binding was determined by blocking experiments with mannan and anti-CD4 (Leu3a) mAbs (Figure 7B). The pattern of binding was similar on both blood DC subsets—both fresh and after overnight culture—with a predominance of gp120 binding to CD4 rather than CLRs. The lack of CLR binding was supported by the lack of MR (data not shown) and DC-SIGN surface expression (Figure 6A). Seminested reverse transcriptase (RT)–PCR for DC-SIGN confirmed lack of messenger RNA transcripts in both blood DC subsets. However, transcripts were seen in PBMC and CD14+ve monocyte populations (Figure 6B).
CD4 and DC-SIGN expression on blood DC.
(A) CD4 and DC-SIGN surface expression on blood DC subsets. Blood DCs were freshly isolated as outlined in “Materials and methods.” The mAb to CD4 (Leu3a) or DC-SIGN (AZN-D1) was added and then incubated and detected as outlined in Figure 4. Cultured DCs were DC-incubated overnight in the presence of interleukin-3/GM-CSF. Blood DC subsets were further distinguished by CD11c staining. (B) DC-SIGN expression on blood DCs by RT-PCR. Top panel: ethidium bromide–stained gel (top) of PCR products for DC-SIGN from cDNA. Lane 1: 1-kilobase ladder; lane 2: CD11c+ve blood DCs; lane 3: CD11c−ve blood DCs; lane 4: monocytes; lane 5: MDDCs; lane 6: MDDCs cultured with lipopolysaccharide; Lane 7: PBMCs; lane 8: H2O. Bottom panel: the autoradiograph of the Southern blot probed with a digoxigenin oligonucleotide specific for DC-SIGN.
CD4 and DC-SIGN expression on blood DC.
(A) CD4 and DC-SIGN surface expression on blood DC subsets. Blood DCs were freshly isolated as outlined in “Materials and methods.” The mAb to CD4 (Leu3a) or DC-SIGN (AZN-D1) was added and then incubated and detected as outlined in Figure 4. Cultured DCs were DC-incubated overnight in the presence of interleukin-3/GM-CSF. Blood DC subsets were further distinguished by CD11c staining. (B) DC-SIGN expression on blood DCs by RT-PCR. Top panel: ethidium bromide–stained gel (top) of PCR products for DC-SIGN from cDNA. Lane 1: 1-kilobase ladder; lane 2: CD11c+ve blood DCs; lane 3: CD11c−ve blood DCs; lane 4: monocytes; lane 5: MDDCs; lane 6: MDDCs cultured with lipopolysaccharide; Lane 7: PBMCs; lane 8: H2O. Bottom panel: the autoradiograph of the Southern blot probed with a digoxigenin oligonucleotide specific for DC-SIGN.
Binding of gp120 on several DC subsets.
(A) Relative gp120 binding levels within blood DC subsets. Blood DCs were freshly isolated as outlined in “Materials and methods.” The b-gp120 (Leu3a) was added and then incubated and detected as outlined in Figure 1A. Cultured blood DCs were incubated as outlined in Figure 6. Blood DC subsets were further analyzed by CD11c staining after b-gp120 staining. (B) Inhibition of gp120 binding by anti-CD4 (Leu3a) and mannan to MDDCs and blood DCs. Inhibitors were preincubated with blood DCs as follows: (1) No inhibitors, (2) CD4 (Leu3a) (10 μg/mL), (3) mannan (5 mg/mL), and (4) dual mannan/Leu3a were preincubated with blood DCs. The b-gp120 was added as outlined in Figure 1A and DC subsets analyzed for gp120 binding after CD11c staining.
Binding of gp120 on several DC subsets.
(A) Relative gp120 binding levels within blood DC subsets. Blood DCs were freshly isolated as outlined in “Materials and methods.” The b-gp120 (Leu3a) was added and then incubated and detected as outlined in Figure 1A. Cultured blood DCs were incubated as outlined in Figure 6. Blood DC subsets were further analyzed by CD11c staining after b-gp120 staining. (B) Inhibition of gp120 binding by anti-CD4 (Leu3a) and mannan to MDDCs and blood DCs. Inhibitors were preincubated with blood DCs as follows: (1) No inhibitors, (2) CD4 (Leu3a) (10 μg/mL), (3) mannan (5 mg/mL), and (4) dual mannan/Leu3a were preincubated with blood DCs. The b-gp120 was added as outlined in Figure 1A and DC subsets analyzed for gp120 binding after CD11c staining.
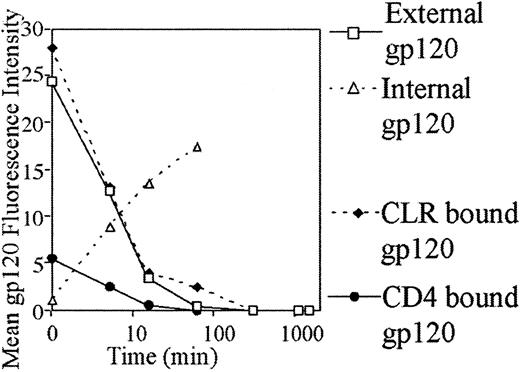
HIV gp120 internalization
Internalization was rapid, with less than 50% of surface gp120 present after 5 minutes. After 1 hour no external gp120 could be observed on MDDCs (Figure 8). MDDCs were reexamined for surface gp120 over 2, 6, 18, and 24 hours. There was no reappearance of external gp120 over the period of 1 to 24 hours. The kinetics of gp120 internalization mediated by CD4 and CLRs was also investigated. First, the CLR pathway was blocked by mannan, and gp120 bound to CD4 was examined for internalization. Conversely, the role of CLRs in internalization was also examined by blocking CD4 with Leu3a and gp120 subsequently tracked. Both CD4 and CLR pathways exhibited rapid internalization with no external gp120 evident after 60 minutes. The CD4-mediated internalization pathway showed a single rapid phase, but CLR internalization was biphasic. The first phase rapidly internalized most of the gp120 within the first 15 minutes, and the second phase internalized the residual gp120 over the 15- to 60-minute period. Rapid external loss of gp120 correlated with rapid appearance of internalized gp120 as observed in permeabilized MDDCs (Figure 8).
Internalization of gp120 by MDDCs.
Cells were preincubated with anti-CD4 (Leu3a) to detect CLR-bound gp120 and with mannan to detect CD4-bound gp120 or binding media for total external or internal gp120 for 30 minutes at 4°C. Cells were then incubated with 5 μg/mL b-gp120 for 30 minutes at 4°C, washed twice in binding media, and incubated in culture media for the indicated times and stained to detect extracellular or intracellular gp120 as outlined in “Materials and methods.”
Internalization of gp120 by MDDCs.
Cells were preincubated with anti-CD4 (Leu3a) to detect CLR-bound gp120 and with mannan to detect CD4-bound gp120 or binding media for total external or internal gp120 for 30 minutes at 4°C. Cells were then incubated with 5 μg/mL b-gp120 for 30 minutes at 4°C, washed twice in binding media, and incubated in culture media for the indicated times and stained to detect extracellular or intracellular gp120 as outlined in “Materials and methods.”
Discussion
MDDCs were used in the current studies as a model for immature tissue DCs such as skin LCs and mucosal DCs. They are a convenient model for in vitro studies but also may have relevance in vivo: Monocytes are observed to develop into MDDCs at sites of inflammation as a second recruitment of antigen-presenting cells.34Nethertheless, they show marked phenotypic difference to other blood and tissue DCs.35 Therefore, we defined the receptors for binding gp120 on MDDCs in vitro and then compared them with ex vivo blood DCs.
In MDDCs, 2 groups of receptors capable of binding gp120 were defined. MDDCs bound gp120 predominantly via CLRs: the mannose saccharides, mannan and mannopyranoside, and also calcium depletion were capable of markedly inhibiting gp120 binding. Monocytes only bound gp120 via CD4 and did not express MR or DC-SIGN. Conversion to the predominant CLR binding pattern seen in MDDCs occurred on monocytes after 2 days of culture in interleukin-4/GM-CSF and peaked at day 6. During MDDC differentiation, the kinetics of DC-SIGN, MR, and CD4 expression and gp120 binding via CLRs were discordant, which supports a more complex gp120 binding pattern than previously described. TNF-α–induced MDDC maturation increased CD4-gp120 binding at the expense of CLR binding and also significantly reduced MR but only slightly decreased CD4 and DC-SIGN expression.
Both CD11c+ve and CD11c−ve blood DCs lacked both DC-SIGN and MR expression, and gp120 bound exclusively by CD4. Culture of both blood DC subsets down-regulated CD4 expression and gp120 binding but did not induce MR, DC-SIGN expression, or gp120 binding via CLRs.
The 2 CLRs, DC-SIGN and the MR, have been previously observed to bind gp120,14,15,30 and both are expressed on MDDCs. HIV gp120 bound to the surface of MDDCs and inhibited anti-CD4, anti-MR, and anti-DC-SIGN mAb binding, supporting gp120 binding to the above 3 receptors. DC-SIGN mAb was most readily inhibited at low gp120 concentrations, consistent with high affinity for gp120.15However, neither CD4, DC-SIGN, nor MR mAb inhibited gp120 binding to MDDCs. Trypsin treatment of MDDCs completely cleaved both the CD4 (Leu3a) and DC-SIGN (AZN-D2) mAb epitopes but only partially inhibited gp120 binding to MDDCs. Both MR mAb clones 19 and 3.29 still bound to trypsinized MDDCs, probably to CRDs 4 or 5, which are protease-insensitive.33 Residual gp120 binding, in trypsinized MDDCs, was blocked by mannan and EGTA but not by either MR mAbs. These results suggest gp120 could bind to other CLRs and/or other CRDs of MR (not recognized by the mAbs). However, the latter seems unlikely because both mAbs block binding of mannose ligands to MR31 32 and, more specifically, partially block gp120 in a trypsinized MMR cell line (data not shown). If several CLRs, including DC-SIGN and MR, can bind gp120, blocking one CLR with mAbs may not significantly reduce gp120 binding. This notion is further supported by the inability of either CD4 or CLR mAbs alone to inhibit binding. In addition, the binding of gp120 to CD4 differed in the presence or absence (mannan block) of CLRs. This might reflect the much higher binding affinity of the CLRs (MMR and DC-SIGN,Kd < 4 nM) compared with the CD4 affinity for BaL gp120 (Kd = 30 nM).
Experiments on gp120 internalization independently confirmed that gp120 bound predominantly via CLRs. The rapid internalization of gp120 in COS-7–DC-SIGN transfectants observed by Curtis et al15and in HeLa transfectants (A. J. Watson, written communication, August 2000), together with internalization of the MR,36 supports our observation of a rapid CLR-mediated phase of gp120 internalization. The biphasic nature of this CLR-based internalization could reflect multiple CLRs capable of binding and internalizing gp120. Electron microscopic studies by Blauvelt et al,8 Dezutter-Dambuyant and Schmitt,37 and Hladik et al7 showed internalization of virions into vacuoles and is consistent with current observations of gp120 internalization. In electron microscopy studies by Dezutter-Dambuyant and Schmitt,37 HIV gp120 internalization was correlated with whole virions, because both were observed in clathrin-coated pits of epidermal LCs. Similarly stable HeLa clone 11 (DC-SIGN) transfectants also internalized HIV into vacuoles, suggesting that CLR binding results in endocytosis (A. J. Watson, written communication, August 2000). In our recent work, mannan was also shown to markedly inhibit accumulation of full-length HIV proviral DNA transcripts within MDDCs, showing a close correlation between gp120 internalization and HIV infection (unpublished observations, 2001). In the current study there was no reappearance of gp120 on the surface of MDDCs, suggesting there was degradation after internalization.
There are many reports of the ability of gp120 to bind to various cell types independently of CD4. Macrophages,30 trypsinized LCs,13,38 MDDCs,16 and cells within the placenta15 are examples. Only the studies of Curtis et al15 and Larkin et al30 identified the specific receptors as CLRs. Geijtenbeek et al14,16recently reported that placental CLR clone 11 (DC-SIGN) previously described by Curtis et al15 was expressed on MDDCs. While the observations described here support CLRs as predominant receptors for gp120 binding to MDDCs, CLR binding of gp120 was not restricted to one receptor as reported previously14 but instead to multiple CLRs, including DC-SIGN and MR. A further CLR related to DC-SIGN, named DC-SIGNR, has recently been indentified on MDDCs,21 and the potential expression and binding by numerous other CLRs on MDDCs17-20 further supports our current hypothesis that multiple CLRs can bind gp120.
Although CLRs bound most gp120 in MDDCs, CD4 is the predominant receptor in blood DCs. This observation expands previously described phenotypic differences between MDDCs and blood DCs.35Thus, the fate of internalized gp120 or of HIV is highly likely to be determined by initial binding to CLRs (MDDCs) or CD4 and then the appropriate chemokine receptors (blood DCs). Transfer of HIV from blood DCs to T cells as shown by Cameron et al4 must involve initial binding by CD4. In contrast, Blauvelt et al8observed that in vitro–derived DCs have the capacity to capture and transfer HIV independently of the CD4/chemokine receptor infection pathway. The current work and recent work by Geijtenbeek et al14 suggest that this previously unknown capture pathway is by CLRs. However, both MDDCs and blood DCs capture and transfer HIV to CD4 T lymphocytes effectively in coculture assays. In light of the current observations, it is obvious that blood DCs could not capture and transfer HIV via both pathways. Further viral binding mechanisms independent of CD4 and CLR may also be present. For instance, HIV can acquire T cell–specific molecules during budding,39,40and DCs may be able to bind virions via the same mechanism they use in clustering to T cells. Another DC in vivo, the follicular DC, predominantly binds HIV virions via the adhesion molecules CD54 (ICAM-1) and CD11a (LFA-1).41 Macropinocytosis must also be considered as another mechanism of gp120/viral uptake by DCs.
In view of the discordant findings for gp120 binding between MDDCs and blood DCs, future work must focus on which CLRs are expressed in vivo on LCs and mucosal DCs and whether CLRs or CD4 are the major receptors for gp120 in these cells. LCs do not express DC-SIGN16 and expression of the MR is controversial,24,42 but they do express a mannose-fucose binding receptor(s)42 and can bind gp120 independently of CD4.13 Therefore, other CLRs and/or CD4/CCR5 could be even more important than DC-SIGN in studies of DC-mediated HIV mucosal transmission. The study of the relevant receptors in appropriate surveillance DCs is essential to understanding both mucosal HIV transmission and systemic or mucosal gp120 antigenic processing pathways. These results are relevant to the design of effective antivirals: Care must be taken to ensure that all routes of HIV-DC binding are blocked, because DCs may bind and transfer HIV to responding CD4 T cells via several of their cell surface receptors.
We thank Ray Sweet for his generous gift of BaL gp120, Yvette van Kooyk for the supply of both antibodies AZN-D1 and AZN-D2 to DC-SIGN, Belinda Herring for her help and assistance, Parramatta Red Cross Blood Bank for its continued support, and Nancy Haigwood and Andrew Watson for helpful discussions on the placental clone 11.
Supported by grants from the Westmead Millennium Foundation, Australian National Center in HIV Virology Research, and the Mater Trust.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anthony L. Cunningham, Westmead Millennium Institute, PO Box 412, Darcy Road, Westmead, NSW 2145, Australia; e-mail: tony_cunningham@wmi.usyd.edu.au.

![Fig. 1. Inhibition of gp120 binding on MDDC. / (A) Inhibition of gp120 binding to MDDCs by mannan; 1 × 106 cells/mL were incubated with mannan ranging from 50 μg to 5 mg/mL for 30 minutes at 4°C. The b-gp120 was added at 3-fold excess (9 μg/mL) and incubated for 30 minutes at 4°C. The b-gp120 was detected via streptavidin Oregon Green 488 and fluorescence measured by flow cytometry as described. (B,C) Inhibition of gp120 binding to MDDCs by CD4 mAbs (Leu3a, Q4120, OKT4). In panel B, cells were preincubated with mAbs to CD4 (ranging from 0.05 μg/mL to 25 μg/mL) for 30 minutes at 4°C. In panel C, cells were also incubated with 5 mg/mL mannan in addition to the CD4 mAb Leu3a. The b-gp120 was incubated and detected as in panel A. Percent DC-gp120 binding was calculated as follows: [(sample fluorescence intensity − mean negative control fluorescence intensity)/mean positive control fluorescence intensity] × 100. Positive control cells were treated with b-gp120 in the absence of inhibitors. Negative controls consisted of cells with identical inhibitors but no b-gp120.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/8/10.1182_blood.v98.8.2482/6/m_h82011629001.jpeg?Expires=1765912859&Signature=e4Tx~juP0lSwiPbxJe3kE4V3BLcbtQteCBpuiRoO3bRhZx9F8fBM50kH2YD69QGYVcXxuYmqcKdbJZQBv1xGW5rHUATsRj2dDuWxzNcPB226nkZNY6il6QpQBQuuVJ0in2uwvBziSwpTdR62v2gJPt3OzIJ7z-8q42onLhcHSkiPujUoJoGVXhQWFI-vLX-mnbU8~HLjJNkpwJCgCRigAh6uiLjC8fq2~cNoBu2c2YpE5RJPCG6pw33qxfQTVzN4yL-yn7QzH0U6twd7pTU4sFiPoCNFRiihbGwT4v7bPci-zKIX6WejEkW5qDad6YBS5nDtd~d0xL16HZ1tPs2VJg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal