Abstract
This study cloned and sequenced the complementary DNA (cDNA) encoding of a putative malarial iron responsive element-binding protein (PfIRPa) and confirmed its identity to the previously identified iron-regulatory protein (IRP)–like cDNA from Plasmodium falciparum. Sequence alignment showed that the plasmodial sequence has 47% identity with human IRP1. Hemoglobin-free lysates obtained from erythrocyte-stage P falciparum contain a protein that binds a consensus mammalian iron-responsive element (IRE), indicating that a protein(s) with iron-regulatory activity was present in the lysates. IRE-binding activity was found to be iron regulated in the electrophoretic mobility shift assays. Western blot analysis showed a 2-fold increase in the level of PfIRPa in the desferrioxamine-treated cultures versus control or iron-supplemented cells. Malarial IRP was detected by anti-PfIRPa antibody in the IRE-protein complex fromP falciparum lysates. Immunofluorescence studies confirmed the presence of PfIRPa in the infected red blood cells. These findings demonstrate that erythrocyte P falciparum contains an iron-regulated IRP that binds a mammalian consensus IRE sequence, raising the possibility that the malaria parasite expresses transcripts that contain IREs and are iron-dependently regulated.
Introduction
Iron is required by virtually all organisms and is used in a wide variety of metabolic functions, including the redox reactions of photosynthesis and respiration, the synthesis of deoxynucleotides, and the transport of oxygen.1,2 In spite of its essential nature, iron can also generate damaging free radicals via the Fenton reaction.3 Consequently, organisms have evolved complex mechanisms for regulating iron transport, storage, and utilization.1,2 4
Vertebrates control the intracellular iron concentration mainly through the synergistic regulation of iron storage and import.5,6The messenger RNA (mRNA) encoding ferritin, the major iron-storage protein of the vertebrate cell, contains an iron-responsive element (IRE), an RNA stem-loop structure, at its 5′ end.7 When intracellular iron levels fall, cytosolic iron-regulatory proteins (IRP1 and IRP2) bind to the IRE and block translation of the ferritin mRNA,5,6 leading to a decrease in the amount of iron that is sequestered within ferritin. The mRNA encoding the transferrin receptor, the major receptor on most mammalian cells for import of iron, contains a recognition site for an endonuclease and 5 copies of an IRE within its 3′-untranslated region.8 When the intracellular iron concentration falls, the IRPs bind to the IREs on the transferrin receptor mRNA, masking the endonuclease site and protecting the mRNA from degradation. This protection leads to an increase in receptor number and an elevation in the intracellular iron concentration. IREs are also present in the mRNAs encoding erythroid-5-aminolevulinate synthase,9 mitochondrial aconitase,10,11 succinate dehydrogenase subunit b ofDrosophila melanogaster,12 the recently cloned intestinal iron exporter, ferroportin1/IREG1/MTP1,13-15and one splice form of divalent metal transporter, DMT1.16
The asexual proliferating forms of plasmodia reside within human erythrocytes and use the erythrocytic contents, mainly its iron-containing hemoglobin, for their metabolic needs.17The parasites metabolize host cell globin and detoxify heme together with iron into a dark pigment, hemozoin, which is identical in its chemical structure to β-hematin.18 Although it has been proposed that some small portion of heme iron may be liberated from hemoglobin and mobilized by the parasites,17 direct evidence for mobilization of iron from heme is not available.
It was demonstrated that labile iron pools are present within normal and malaria-infected erythrocytes,19 and such iron pools are also a potential source of iron for the parasite. Yet, it is not clear how the malaria parasite transports iron and maintains iron homeostasis within the highly compartmentalized parasitized erythrocyte.
IRP homologues have been demonstrated for ancient lineages of vertebrates such as the lamprey,20 certain invertebrates (Drosophila melanogaster and Caenorhabditis elegans),21 and bacteria.22 Also, it was recently shown that a soluble protein from the cultured trypanosomatid,Leishmania tarentolae, specifically interacts with a mammalian IRE.23 An IRP-like complementary DNA (cDNA) fromP falciparum was identified and deposited into the GenBank,27 but the authors were unable to demonstrate its binding to targeted IRE-containing mRNA sequences or its iron regulation pattern in this parasite. Here, we present evidence that an IRP-like protein from P falciparum binds to a mammalian IRE.
Materials and methods
Maintenance of P falciparum growth in culture
P falciparum (strain 3D7) was grown in culture flasks containing RPMI-1640 supplemented with 25 mM HEPES, 23 mM sodium bicarbonate, 10 mM glucose, 10% (vol/vol−1) heat-inactivated human plasma (0+ or A+), and washed human erythrocytes (A+) at 2% to 2.5% hematocrit. The growth medium was replaced daily, and the cultures were gassed with a mixture of 90% N2, 5% CO2, and 5% O2.24 Synchronization to the ring stage was achieved by the lysis of cells containing mature parasites with iso-osmotic sorbitol or alanine.25 The morphologic characteristics of the parasites and the degree of parasitemia were assessed by microscopic inspection of thin blood smears stained by Giemsa.
Treatment of parasites with desferrioxamine and iron and preparation of cell extracts
Ring-synchronized erythrocyte cultures of P falciparum were adjusted to 10% to 15% parasitemia, 1.5% hematocrit in 750 cm2 Falcon flasks, and grown under the following conditions: (1) control cells were supplemented with 500 μM fructose; (2) desferrioxamine (DFO) was added to a final concentration of 100 μM from 30 mM stock solution and 500 μM fructose was added; or (3) 25 μM of FeCl3 and 500 μM fructose were added.17 After 18 hours of incubation, the cultures were washed once with phosphate-buffered saline (PBS), the supernatants were discarded, and the erythrocytic pellets were resuspended in 10 mL PBS. Saponin was added to a final concentration of 0.1% on ice, and after 5 minutes of incubation the suspensions were centrifuged for 10 minutes at 3000 rpm. The trophozoite-containing pellets were washed twice with large volumes of PBS. Trophozoite pellets were kept on ice and used promptly in band-shift assays.
Preparation of hemoglobin-depleted red blood cells
Hemoglobin-depleted red blood cells used as a control in gel-retardation experiments were obtained by hemolyzing red blood cells in a hypotonic buffer containing 5 mM Na-phosphate, pH 8.0 at a ratio of 1:40. The membranes of red cells were resealed for 15 minutes at 37°C by the addition of KCl to a final concentration of 150 mM, and the cells were washed 4 times with PBS by centrifugation.
Synthesis of IRE RNAs
Mammalian consensus IRE RNA was transcribed from a synthetic deoxyoligonucleotide template using T7 RNA polymerase.26The specific activity of the radiolabeled transcripts was approximately 7.4 × 106 MBq/mmol (2 × 108μCi/mmol). Full-length transcripts were purified on an 8% polyacrylamide gel, eluted, ethanol precipitated, solubilized in diethyl pyrocarbonate–treated water, and kept frozen before use. A 53-nucleotide template DNA was used to encode the 36-nucleotide RNA and was purchased from Life Technologies (Gaithersburg, MD). The template for the consensus IRE was 5′-GGAGTTCCGT CCAAGCACTG TTGAAGCAGG AACTCCTATAGTGAGT CGT ATTA-3′ (the T7 RNA polymerase promoter region is italicized). A typical 1-ml transcription reaction contained 40 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 5 mM DTT, 1 μM spermidine, 0.01% Triton X-100, 2 mM each NTP, 0.2 μM DNA template, and 1.5 units/μL T7 RNA polymerase (Life Technologies) and was incubated at 37°C for 3 hours. Transcribed RNA was purified on 8% (7 M urea) polyacrylamide gels, extracted from the gel, and desalted by dialysis against a solution of 10 mM potassium phosphate (pH 7.5) and 10 mM KCl, using Spectra/Por CE 500 molecular weight cut-off DispoDialyzers (Spectrum Medical Industries, Houston, TX). The RNA was quantitated by UV absorbance spectroscopy and was folded prior to use by heating to 95°C for 5 minutes, followed by renaturation on ice for 20 minutes.
Synthesis of radiolabeled consensus IRE
Internally 32P-radiolabeled IRE was transcribed from the same DNA template described above. RNA was transcribed in a 25-μL transcription reaction containing 40 mM Tris-HCl (pH 7.5), 25 mM MgCl2, 5 mM DTT, 1 μM spermidine, 0.01% Triton X-100, 4 mM GTP, 4 mM ATP, 4 mM CTP, 0.8 mM UTP, 1.7 μM (4.6 MBq [125 μCi]) α-[32P]UTP (Amersham Pharmacia Biotech, Piscataway, NJ), 0.2 μM DNA template, and 2 U/μL T7 RNA polymerase. RNA was purified as described above except that following isolation from the gel the RNA was ethanol precipitated and resuspended in diethyl pyrocarbonate-treated H2O. The RNA was quantitated and folded as described above.
Gel-retardation assay
Gel-retardation assay reaction mixtures were prepared by mixing 4 to 10 μg lysate in 10 μL lysis buffer with 10 μL32P-IRE mix (7.4 × 106 MBq/mmol [2 × 108 μCi/mmol], 1 pmol/reaction). Lysates were obtained by solubilizing approximately 100 μL of the trophozoite-containing saponin pellets in 100 to 200 μL of the lysis buffer. The lysis buffer contained 0.1 mM DTT, 40 mM KCL, 1% Triton X-100, 25 mM Tris-HCl, pH 8.0, 10 μg/mL leupeptin and aprotinin, and 10 mM aminoethylbenzene sulfonyl fluoride. 32P-IRE mix contained (per each 20-μL sample) 32P-IRE (1 pmol/reaction), 2 μL 50% glycerol, 0.2 μL RNAase inhibitor (Merck, Whitehouse Station, NJ), 0.3 μL yeast tRNA, 0.02 μL 100 mM DTT. The amounts of lysate protein were thoroughly normalized before loading gels. The binding reactions (20 μL) were incubated for 10 minutes on ice and were resolved on 8% nondenaturing polyacrylamide gels. The running buffer contained 45 mM Tris-base, 4.5 mM boric acid, and 0.5 mM disodium EDTA, pH 8.5. The gels were run for about 4 hours at 140 V, dried onto 3M chromatography Whatman paper and analyzed either after exposure to x-ray film or by PhosphorImager scanning (Molecular Dynamics, Sunnyvale, CA). In the competition assay, the increasing amounts of nonradioactive, “cold” IRE were premixed with32P-IRE (at 25:1; 50:1, and 75:1, mol/mol ratio) before the addition to plasmodial extracts, and the gel-retardation assay was performed as described above.
Antibodies used in the study and Western blot analysis
To produce antibodies against the putative malarial iron responsive element-binding protein (PfIRPa), we used a published sequence,27 after confirmation of its authenticity (see below). Three antibodies directed against residues 609-626 (antibody 3950), 699-717 (antibody 3951), and 881-898 (antibody 3952) of the PfIRPa sequence were produced in rabbits (Bio-Synthesis, Lewisville, TX). Protein (10 μg) from cell lysates was separated by sodium dodecyl sulfate (SDS) gel electrophoresis, transferred to nitrocellulose, and probed using these 3 antibodies at 1:500 dilution. A horseradish peroxidase–labeled secondary antibody was then added at a dilution of 1:5000. The signal was detected using an enhanced chemiluminescence kit. The P falciparum anti-BiP (GRP) antibody was obtained from the Malaria Resource and Reference Center at the American Tissue Culture Collection (ATCC, Manassas, VA). It was also used at 1:500 dilution.
Molecular cloning of PfIRPa
The PfIRPa cDNA was obtained by polymerase chain reaction (PCR) using genomic P falciparum DNA (gDNA) as a template. P falciparum gDNA was obtained as described.28 The primers were 5′-CTCGCGTTCGAAACGATGCATTTCCGAAGGTGTATACGAAAATATTG-3′ and 5′-CGACTGGATCCTTATTTGTTAGCTTCATTAACTAATG-3′.
The cDNA was gel purified and cut withHincII-BamHI restriction enzymes into large and small fragments. The large fragment of PfIRPa was first ligated into a pUC19 plasmid, and the vector DNA was expanded in DH5α-Escherichia coli competent cells. Then the DNA was isolated and moved into the pCA10.3 yeast expression vector (C.R.A. and T.A.R., unpublished, July 1998) as aBstBI-BamHI fragment. Finally, the full-length sequence of PfIRPa was assembled by ligating a small 5′-endBstBI-BstBI fragment produced by PCR on P falciparum gDNA into the pCA10.3 vector. The sequence was verified for correct orientation by restriction analysis and by sequencing of both of the strands. A fluorescent dideoxynucleotide terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) was used for automated sequencing.
Immunoprecipitation of PfIRPa and determination of aconitase activity
We used a modified procedure described in Kaptain et al.29 Hemoglobin-free extracts of the parasites were obtained as described above, and the pellets (75-100 μL) were lysed by passing them 12 times through a 26-gauge needle with a lysis buffer containing 10 mM Tris-HCl, pH7.4, 100 μM Na-citrate, 1% Triton X-100, 0.5% Nonidet P-40, 150 mM NaCl, and protease inhibitors. Lysates (250 μL) were tumbled with 60 μL rabbit antisera, containing antibodies 3950, 3951, and 3952 for 1.5 hours at 4°C. Protein G-agarose beads were washed 3 times with the lysis buffer, and 30 μL beads were added to the Eppendorf tubes with the lysates antisera and tumbled for 1 hour at 4°C. The beads were washed 5 times with the washing buffer containing 100 mM Tris-HCl, 100 μM Na citrate, and 0.1% Triton X-100 and were treated with an aconitase-regeneration buffer containing 100 mM Tris-HCl, 100 μM Na-citrate, 0.1% Triton X-100, 2 μM ferrous ammonium sulfate, and 50 mM DTT to activate potential aconitase activity. The aconitase detection mix consisted of 150 mM Tris-HCl, pH 7.5, 8.6 mMcis-aconitate, 60 mM MgCl2, 0.04 IU of isocitrate dehydrogenase per 10 mL mix, 125 μM NADP, 240 μM MTT (3-[4.5-dimethylthiazol-2,5-diphenyl tetrazolium bromide) (Sigma, St Louis, MO), and 80 μM phenazine methosulfate (Sigma). At each time point, the beads were pelleted, and A560 of the supernatant was measured. The A560 value of a tumbled sample containing a prebleed control serum was used as a background control.
Indirect immunofluorescence analysis
Immunostaining with α-PfIRPa 3950 was combined with the mitochondrial staining with a dye, MitoTracker RedCMXRos (Molecular Probes, Eugene, OR). The suspensions of infected erythrocytes (10%-12% parasitemia) were first loaded with 50 nmoles MitoTracker for 30 minutes at 37°C. The suspensions were washed once with RPMI-1640 and were fixed with 4% paraformaldehyde, 0.05% glutaraldehyde for 10 minutes at room temperature. Then the fixed cells (at about 107 cells/mL) were allowed to adhere and to dry to poly-L-lysine–coated microscopic slides (Electron Microscopy Sciences, Ft Washington, PA). The slides were washed 2 times by submerging for 15 minutes in PBS-Tween 20 (0.01%) and once in PBS. Nonspecific epitopes of cells were quenched with 5% bovine serum albumin (BSA) in PBS for 1 hour, followed by 3 washes in PBS-Tween and PBS as described above. Afterward, the slides were probed with a primary antibody (rabbit polyclonal 3950, 1:100) in the blocking solution for 1 hour, washed 2 times with PBS-Tween, once with PBS, 15 minutes each, and probed with a secondary antibody (goat antirabbit, alexa488, 1:500). The slides were washed as above, dried briefly, and mounted in the presence of VectaShield liquid (Vector Laboratories, Burlingame, CA) before taking fluorescence images. Fluorescence microscopy and digital image collection were performed on the Nikon Eclipse E600 fluorescent microscope and a digital photocamera DKC-ST5 (Image Systems, North Potomac, MD). The images were taken at the excitation wavelengths of 488-nm for alexa488, and at 680-nm for MitoTracker.
Sequence analysis
Alignment of protein sequences and their comparisons were performed using the program ClustalW (Baylor College of Medicine, Houston, TX).
Results
Verification of PfIRPa sequence and its comparison with human IRPs
We cloned the cDNA produced by PCR on P falciparum gDNA into DH5α-competent E coli cells by means of the yeast expression vector, pCA10.3. Figure1A shows the alignment of the putative plasmodial sequence we cloned with the published sequences for the human IRP1 and IRP2. The plasmodial sequence shows 47% identity with human IRP1 and 40% identity with human IRP2, whereas it shows only 22% identity with the porcine mitochondrial aconitase (not shown). The differences among the 3 IRPs are shown in Figure 1B. PfIRPa differs from human IRP1-aconitase only in 2 of the 23 identified active site residues.30 Phenylalanine 88, necessary for hydrogen bond support of the active site residues in human IRP1-aconitase, is replaced with leucine in P falciparum IRP (Leu101). Also, Arg699 in human IRP1 is replaced with Lys711 in PfIRPa. Importantly, Arg536, Arg541, and Arg780 in human IRP1 that are indispensable for high-affinity IRE binding,31 are present in PfIRPa as Arg550, Arg555, and Arg792.
Alignment of the putative plasmodial sequence with the sequences for human IRPs.
(A) Sequence comparison of PfIRPa with human IRP1 and IRP2. On the basis of the comparison with the mitochondrial porcine aconitase,30 the critical residues for substrate recognition, catalysis, cluster ligation and interaction, and hydrogen bond support are shown. The alignment was performed using the program ClustalW (Baylor College of Medicine, Houston, TX). (B) Schematic comparison of PfIRPa with human IRP1 and IRP2. Active site residues that are shared among the IRPs are shown within each domain.32 Active site residues that differ are shown in bold type. The alignment and sequence conservation is high among the 3 proteins.
Alignment of the putative plasmodial sequence with the sequences for human IRPs.
(A) Sequence comparison of PfIRPa with human IRP1 and IRP2. On the basis of the comparison with the mitochondrial porcine aconitase,30 the critical residues for substrate recognition, catalysis, cluster ligation and interaction, and hydrogen bond support are shown. The alignment was performed using the program ClustalW (Baylor College of Medicine, Houston, TX). (B) Schematic comparison of PfIRPa with human IRP1 and IRP2. Active site residues that are shared among the IRPs are shown within each domain.32 Active site residues that differ are shown in bold type. The alignment and sequence conservation is high among the 3 proteins.
Experimental demonstration of PfIRPa in the gel-retardation assay
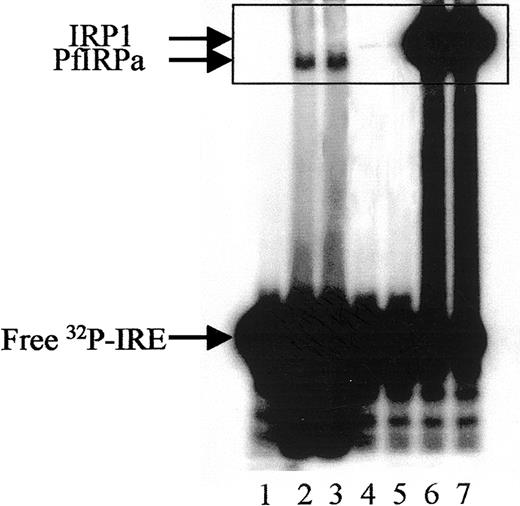
Hemoglobin-free P falciparum lysates retarded the32P-labeled mammalian consensus IRE (Figure2, lanes 2,3), whereas free probe migrated toward the bottom of the gel (lane 1). The position of the IRE-protein complex from P falciparum lysates differs from that generated by IRP1 of mammalian lysates (lanes 6,7), or of lysates from hemoglobin-depleted, uninfected erythrocytes probably harboring some IRP1 from their reticulocyte stage (lanes 4,5). The hemoglobin depletion of uninfected red blood cells was necessary to reduce the interference of hemoglobin with the performance of the gel-retardation assay.
Gel-retardation assay of PfIRPa-mammalian IRE complexes.
Hemoglobin-free lysates from P falciparum–parasitized erythrocytes were incubated with radiolabeled mammalian consensus32P-IRE and were resolved on 8% nondenaturing polyacrylamide gel. Lane 1, consensus IRE alone (1.0 pmol/lane); 2 and 3, 5 μg P falciparum lysate protein plus IRE; 4 and 5, 5 μg lysate protein from hemoglobin-depleted uninfected erythrocytes, 6 and 7, 5 μg lysate protein from a human EBV cell line expressing IRP1. Visible traces of IRP1 on lanes 4 and 5 are probably from the reticulocyte precursors of the erythrocytes as judged on similar mobility with the bands on lanes 6 and 7. The boxed area shows the region of the gel that will be shown on the following figures.
Gel-retardation assay of PfIRPa-mammalian IRE complexes.
Hemoglobin-free lysates from P falciparum–parasitized erythrocytes were incubated with radiolabeled mammalian consensus32P-IRE and were resolved on 8% nondenaturing polyacrylamide gel. Lane 1, consensus IRE alone (1.0 pmol/lane); 2 and 3, 5 μg P falciparum lysate protein plus IRE; 4 and 5, 5 μg lysate protein from hemoglobin-depleted uninfected erythrocytes, 6 and 7, 5 μg lysate protein from a human EBV cell line expressing IRP1. Visible traces of IRP1 on lanes 4 and 5 are probably from the reticulocyte precursors of the erythrocytes as judged on similar mobility with the bands on lanes 6 and 7. The boxed area shows the region of the gel that will be shown on the following figures.
Relative specificity of PfIRPa binding to a mammalian consensus IRE
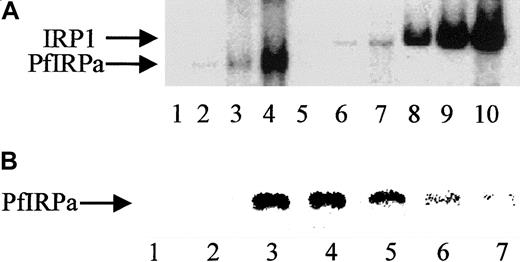
We assume that the specificity of PfIRPa binding to a mammalian consensus IRE is relative, because only a naturally occurring functional plasmodial IRE-containing sequence(s), which is unknown so far, can be considered absolutely specific. Hemoglobin-free P falciparum lysates were preincubated on ice for 10 minutes with the increasing amounts of 32P-IRE (0.125 pmol/lane; 0.5 pmol/lane; 2 pmol/lane). Figure 3A shows the boxed area of the typical gel-retardation assay shown in Figure 2. Free probe has always been present in excess and, without lysates that could contain binding and retarding the probe's proteins, is located close to the bottom of the gel (not shown, lane 1). P falciparum lysates retarded the probe in a concentration-dependent fashion (lanes 2,3,4), similar to the binding of the probe by lysates from uninfected hemoglobin-depleted erythrocytes (lanes 5,6,7) and by an EMB cell line–expressing IRP1 (lanes 8,9,10). Again, the position of the band shifted by PfIRPa-containing lysates was different from the position of the band shifted by erythrocytes and mammalian cell lysates. The relative specificity of PfIRPa binding to a mammalian consensus IRE was also confirmed in the competition assay (Figure 3B). Free IRE runs to the bottom of the gel, as shown in Figure 2 (Figure 3, lanes 1,2). A protein from plasmodial lysate binds a mammalian IRE (lanes 3,4). The position of the labeled IRE retarded on the gel was determined by the size of the bound to it protein. This position in the gel-shift assay coincided with the position of PfIRPa as was detected by transferring the gel-shift on nitrocellulose and probing with antibody 3950 (not shown). Lanes 5, 6, and 7 show that the amounts of the retarded 32P-IRE decreased in the presence of the excess of “cold” IRE (25:1 mol/mol in lane 5; 50:1 mol/mol in lane 6; and 75:1 mol/mol in lane 7). In addition, nonspecific comparable amounts of RNA (tRNA at 75:1) did not reduce the signal of the complex, indicating that the signal reduction is specifically attributable to IRE sequences.
Gel-retardation assay.
(A) Human IRP1 and PfIRPa bind a mammalian IRE in a concentration-dependent manner. Gel-retardation assays of IRP1- and PfIRPa-mammalian IRE complexes are shown. The conditions are as described in the legend to Figure 2. Increasing amounts of the32P-IRE were incubated with 6 μg protein from P falciparum and the human EBV cell line expressing IRP1. Lane 1, IRE alone; 2-4, PfIRPa-IRE; 5-7, uninfected RBC lysate-IRE; 8-10, IRP1-IRE. Lanes 2, 5, and 8, 0.125 pmol 32P-IRE per lane; lanes 3, 6, and 9, 0.5 pmol 32P-IRE per lane; lanes 4, 7, and 10, 2 pmol 32P-IRE per lane. (B) Cold IRE competes with32P-IRE for the binding to PfIRPa. Lanes 1 and 2, free32P-IRE; 3 and 4, 10 μg lysate protein + 1 pmol32P-IRE; 5, 10 μg lysate protein + (1 pmol32P-IRE + 25 pmol cold IRE); 6, 10 μg lysate protein + (1 pmol 32P-IRE + 50 pmol cold IRE); 7, 10 μg lysate protein +(1 pmol 32P-IRE + 75 pmol cold IRE).
Gel-retardation assay.
(A) Human IRP1 and PfIRPa bind a mammalian IRE in a concentration-dependent manner. Gel-retardation assays of IRP1- and PfIRPa-mammalian IRE complexes are shown. The conditions are as described in the legend to Figure 2. Increasing amounts of the32P-IRE were incubated with 6 μg protein from P falciparum and the human EBV cell line expressing IRP1. Lane 1, IRE alone; 2-4, PfIRPa-IRE; 5-7, uninfected RBC lysate-IRE; 8-10, IRP1-IRE. Lanes 2, 5, and 8, 0.125 pmol 32P-IRE per lane; lanes 3, 6, and 9, 0.5 pmol 32P-IRE per lane; lanes 4, 7, and 10, 2 pmol 32P-IRE per lane. (B) Cold IRE competes with32P-IRE for the binding to PfIRPa. Lanes 1 and 2, free32P-IRE; 3 and 4, 10 μg lysate protein + 1 pmol32P-IRE; 5, 10 μg lysate protein + (1 pmol32P-IRE + 25 pmol cold IRE); 6, 10 μg lysate protein + (1 pmol 32P-IRE + 50 pmol cold IRE); 7, 10 μg lysate protein +(1 pmol 32P-IRE + 75 pmol cold IRE).
PfIRPa is iron regulated as demonstrated in a band-shift assay
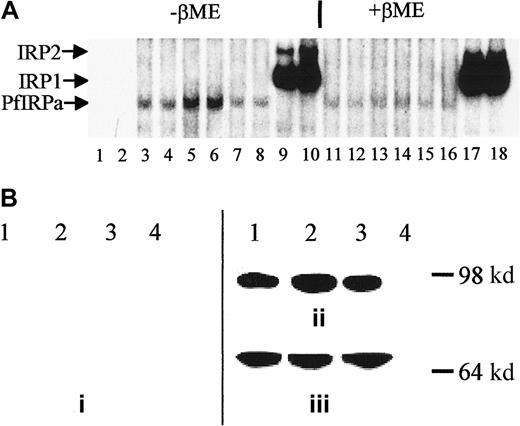
P falciparum cultures were treated for 18 hours with either 100 μM DFO plus fructose or 25 μM FeCl3-fructose, and hemoglobin-free lysates were preincubated with 32P-labeled IRE and electrophoresed (Figure 4A). Free probe migrated toward the bottom of the gel (not shown, lanes 1,2), whereas all of the hemoglobin-free lysates contained IRE-binding activity (lanes 3-8). The maximal intensity of the retained probe is seen with DFO (lanes 5,6) and the minimal intensity with iron-supplemented cultures (lanes 7,8). The amount of PfIRPa (estimated from the amount of radioactivity in the band shift) in lysates from parasites grown in the presence of DFO (iron-limited conditions) was 2 times greater than in lysates from parasites grown under basal conditions (P < .01), whereas the amount of PfIRPa in the lysates from parasites exposed to iron-fructose tended to be 20% less (P = .15, n = 4) than the amount in lysates from parasites grown under basal conditions. The positions of the probe retained by the EBV-cell line lysates correspond with the typical positions for the mammalian IRP1 and IRP2 (lanes 9,10). The addition of β-mercaptoethanol to gel-retardation mixtures of cell lysate-32P-IRE, before the assay, resulted in the previously demonstrated increase of the IRE-binding activity of IRP1,42 no change in the activity of IRP2,32 and reduction in the IRE-binding activity of PfIRPa (Figure 4A, lanes 11-16 versus lanes 3-8).
Expression of PfIRPa is iron dependent.
(A) Gel-retardation assays of human IRP1- and PfIRPa-mammalian IRE complexes are shown. Lanes 1 and 2, 32P-IRE alone; 3, 4, 11, and 12, 2-6 μg protein isolated from fructose-supplementedP falciparum cultures; 5, 6, 13, and 14, 6 μg protein isolated from cultures treated with 100 μM DFO for 18 hours; 7, 8, 15, and 16, 6 μg protein isolated from cultures supplemented with 25 μM FeCl3; 9, 10, 17, and 18, 6 μg lysate protein from a human EBV cell line expressing IRP1 and IRP2. Lanes 3 to 10, no β-mercaptoethanol; lanes 11 to 18, 1% β-mercaptoethanol was added to lysate-IRE incubation mixtures before loading the gel. The lysate proteins were incubated with 1.0 pmol/lane of 32P-IRE and resolved on an 8% polyacrylamide gel. (B) Western blot analysis of expression of P falciparum IRP. Protein (10 μg) isolated from P falciparum cultures was exposed to fructose for 18 hours (lane 1), from cultures treated with 100 μM DFO plus fructose for 18 hours (lanes 2), from cultures supplemented with 25 μM iron as FeCl3 plus fructose (lane 3), or isolated from a human EBV cell line expressing IRP1 (lane 4) were resolved on 8% SDS-polyacrylamide gel. The position of the molecular marker of 98 000 dalton is shown. The proteins were transferred to nitrocellulose and probed with antisera from nonimmune rabbits (prebleed, panel i), or with antibody 3950 (panel ii). Afterward, the nitrocellulose shown in panel B was stripped of antibody 3950 and was probed with an antibody against the plasmodial protein BiP (GRP) (panel iii). The visualization of the proteins was achieved after probing with a secondary, peroxidase-linked antibody in an enhanced chemiluminescence assay.
Expression of PfIRPa is iron dependent.
(A) Gel-retardation assays of human IRP1- and PfIRPa-mammalian IRE complexes are shown. Lanes 1 and 2, 32P-IRE alone; 3, 4, 11, and 12, 2-6 μg protein isolated from fructose-supplementedP falciparum cultures; 5, 6, 13, and 14, 6 μg protein isolated from cultures treated with 100 μM DFO for 18 hours; 7, 8, 15, and 16, 6 μg protein isolated from cultures supplemented with 25 μM FeCl3; 9, 10, 17, and 18, 6 μg lysate protein from a human EBV cell line expressing IRP1 and IRP2. Lanes 3 to 10, no β-mercaptoethanol; lanes 11 to 18, 1% β-mercaptoethanol was added to lysate-IRE incubation mixtures before loading the gel. The lysate proteins were incubated with 1.0 pmol/lane of 32P-IRE and resolved on an 8% polyacrylamide gel. (B) Western blot analysis of expression of P falciparum IRP. Protein (10 μg) isolated from P falciparum cultures was exposed to fructose for 18 hours (lane 1), from cultures treated with 100 μM DFO plus fructose for 18 hours (lanes 2), from cultures supplemented with 25 μM iron as FeCl3 plus fructose (lane 3), or isolated from a human EBV cell line expressing IRP1 (lane 4) were resolved on 8% SDS-polyacrylamide gel. The position of the molecular marker of 98 000 dalton is shown. The proteins were transferred to nitrocellulose and probed with antisera from nonimmune rabbits (prebleed, panel i), or with antibody 3950 (panel ii). Afterward, the nitrocellulose shown in panel B was stripped of antibody 3950 and was probed with an antibody against the plasmodial protein BiP (GRP) (panel iii). The visualization of the proteins was achieved after probing with a secondary, peroxidase-linked antibody in an enhanced chemiluminescence assay.
PfIRPa is iron regulated as demonstrated by Western blot analysis
Proteins from hemoglobin-free lysates from untreated P falciparum cultures exposed to 100 μM DFO, FeCl3-fructose complexes, or fructose alone were electrophoresed on a polyacrylamide gel, transferred to nitrocellulose and probed with 3 polyclonal rabbit antibodies directed against the PfIRPa sequence. Sera from nonimmune rabbits (prebleed sera) served as a negative control. Of the 3 antibodies, antibody 3950 gave better results than the other 2. Figure 4B shows that the control (prebleed) antisera did not recognize any proteins on the gel (panel i), whereas antibody 3950 obtained from rabbits immunized with a peptide specific to the PfIRPa sequence recognized a protein with an approximate size of 100 000 dalton (panel ii, lanes 1-3). This binding is specific for PfIRPa because the antibody 3950 did not recognize a mammalian IRP1 (lane 4). The amount of the protein revealed by the antibody in the DFO-treated sample is 2.4 times greater (P < .02, n = 6) than in the lysates from untreated cultures (lane 1), despite that the equal amounts of lysate protein were initially loaded on the gel. PfIRPa was also shown to conjugate with IRE after the gel-retardation assay of IRE-P falciparum lysate complexes was transferred to nitrocellulose and probed with antibody 3950 (not shown). To confirm that equal amounts of the lysate protein were loaded on the gel, the nitrocellulose membrane was stripped from the α-PfIRPa antibody 3950 shown in Figure 4B (panel ii), and was reprobed with antibody against the plasmodial protein, BiP (GRP) (Figure 4B, panel iii), that is presumed to not be regulated by iron. This plasmodial stress protein belongs to the heat shock protein 70 family and probably plays a role in facilitating the assembly of multimeric protein complexes inside the lumen of endoplasmic reticulum.41 Figure 4B (panel iii) shows that the equal amounts of lysate protein were initially loaded on the gel that was transferred and probed as shown in Figure 4B (panel ii).
Visualization of PfIRPa via indirect immunofluorescence probing
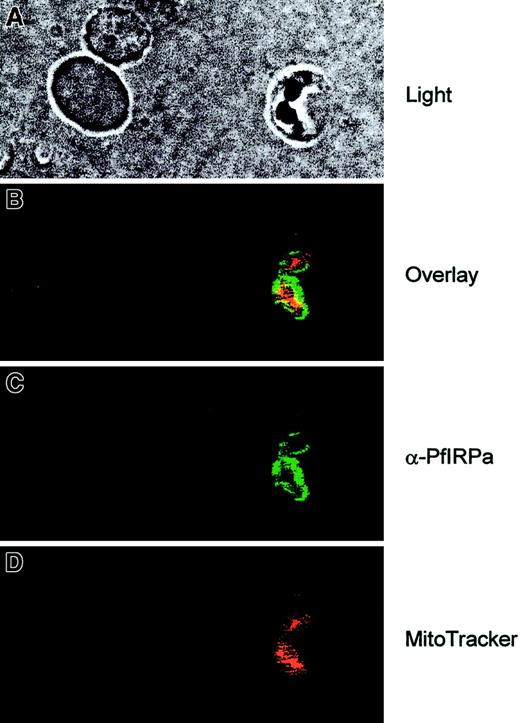
To see the localization of PfIRPa in the parasitized red blood cells, the cell suspensions were first loaded with MitoTracker Red- CMXRos (Molecular Probes), the fluorescent dye that is specifically accumulated in mitochondria and minimally leaks following fixation of cells.43 Then, the slides were fixed and probed with the primary antibody (rabbit polyclonal 3950) and with the secondary antibody (goat antirabbit, alexa488). The images were taken at the excitation wavelengths of 488 nm for alexa488 and 680 nm for MitoTracker, and they were processed and analyzed with the Adobe Photoshop software. Figure 5 (light) shows the double trophozoite-infected erythrocyte (on the right) and 2 uninfected erythrocytes (on the left). It is clearly seen that PfIRPa (green staining) is localized to the cytosolic compartment of the parasite. This protein is most likely not present in the parasitic mitochondrion (red staining) because only marginal yellow staining (resulting from the overlap between the green and the red colors) can be observed.
Visualization of PfIRPa via indirect immunofluorescence probing.
The suspension of infected erythrocytes was first loaded with 50 nmoles MitoTracker for 30 minutes. The suspension was washed and fixed with 4% paraformaldehyde, 0.05% glutaraldehyde. The cells were allowed to adhere and to dry to poly-L-lysine–coated microscopic slides. The slides were washed in PBS-Tween (0.01%) and PBS. Nonspecific epitopes of cells were quenched with 5% BSA in PBS, washed, and probed with a primary antibody (rabbit polyclonal 3950) followed by a secondary antibody (goat antirabbit, alexa488). The slides were washed and mounted in the presence of VectaShield liquid, before taking fluorescence images. The images were taken at the excitation wavelengths of 488 nm for alexa488 and at the 680 nm for MitoTracker.
Visualization of PfIRPa via indirect immunofluorescence probing.
The suspension of infected erythrocytes was first loaded with 50 nmoles MitoTracker for 30 minutes. The suspension was washed and fixed with 4% paraformaldehyde, 0.05% glutaraldehyde. The cells were allowed to adhere and to dry to poly-L-lysine–coated microscopic slides. The slides were washed in PBS-Tween (0.01%) and PBS. Nonspecific epitopes of cells were quenched with 5% BSA in PBS, washed, and probed with a primary antibody (rabbit polyclonal 3950) followed by a secondary antibody (goat antirabbit, alexa488). The slides were washed and mounted in the presence of VectaShield liquid, before taking fluorescence images. The images were taken at the excitation wavelengths of 488 nm for alexa488 and at the 680 nm for MitoTracker.
Discussion
Here we present evidence that hemoglobin-free lysates obtained from erythrocyte-stage P falciparum bind to a consensus mammalian IRE in an iron-dependent fashion. Comparison of the plasmodial IRP with the published sequences for IRP1 and IRP232 shows that the plasmodial sequence has 47% identity with IRP1 and 40% identity with IRP2, whereas it shows only 22% identity with the porcine mitochondrial aconitase, on which the model for the mammalian IRPs was created (Figure 1A).30,33Interestingly, the plasmodial sequence has 49% identity with a number of bacterial aconitases (M. Galperin, personal communication, December 2000). We attempted to assess the aconitase activity ofP falciparum lysates by immunoprecipitating PfIRPa from lysates with antibody 3950 prebound to the protein G-agarose beads, but we were unable to demonstrate aconitase activity. PfIRPa preserves in its structure the majority of the amino acids critical for substrate recognition, catalysis, cluster ligation and interaction, and hydrogen bond support of active site residues for aconitase.30PfIRPa differs from human IRP1-aconitase only in 2 of the 23 identified active site residues (Figure 1B, see “Results”). Previous mutational analysis of porcine mitochondrial aconitase has demonstrated that the Arg699Lys mutation in the mammalian enzyme causes a 250-fold to 30 000-fold decrease in the aconitase activity, relative to the wild-type enzyme.34 PfIRPa, despite conservation of most aconitase active-site residues, carries an analogous substitution (Arg711Lys) (Figure 1A), which could substantially impair aconitase function. Contrary to this finding, in Trypanosoma brucei,aconitase functions with a homologous mutation Arg580Leu.35 Importantly, this same substitution in PfIRPa can be considered as not critical for IRE binding function because the analogous Arg699Gln mutation introduced into human IRP1 did not change the IRE-binding affinity of IRP1.31
It has long been assumed that the asexual-stage malaria parasites rely mainly on glycolysis and use host red blood cell glucose as a major energy source36 rather than on mitochondrial respiration. However, an erythrocytic-stage parasite does have a mitochondrion, and recent experiments in digitonin-permeabilized P bergheitrophozoites suggest that phosphorylation of adenosine 5′-diphosphate occurs on the addition of citrate,37 indicating that the mitochondria are metabolically active. This finding would require that aconitase activity must be present in the parasite's mitochondrion, although this has not been directly documented. Another protozoan parasite, Trypanosoma brucei, expresses an aconitase-related IRP1 in both the cytoplasm and mitochondrion.35
The PfIRPa sequence does not have a mitochondrial recognition targeting sequence (as was concluded by the PSORT program) and it is therefore unlikely that mitochondrial aconitase activity of P falciparum is attributable to PfIRPa. Our immunofluorescence data further support the conclusion that PfIRPa is a cytosolic malarial protein, and it is not present in the mitochondrion of the parasite, at least at the trophozoite stage (Figure 5).
Our results show (Figure 4A) that IRE-binding activity of PfIRPa was diminished in the presence of β-mercaptoethanol, which is known to destabilize the iron-sulfur complex of the aconitase conformation and to enhance IRP1 activity in mammalian systems.42 It is possible that PfIRPa operates like IRP2, which does not contain an iron-sulfur cluster and, therefore, is not activated by reducing agents.23
We found that binding of PfIRPa is concentration dependent (Figure 3A) and is relatively specific because cold, nonradioactive IRE successfully competed with 32P-IRE essentially eliminating its binding to PfIRPa (Figure 3B), whereas competition with equal amounts of nonspecific RNA did not affect binding. However, we have not conducted extensive mutagenesis on the consensus IRE to determine exact sequence requirements.
The critical residues in the structure of IRPs that are known to participate in the RNA binding in human IRP1 and IRP231are present in PfIRPa: Arg536, Arg541,and Arg780 in human IRP1 have their counterparts in PfIRPa as Arg550, Arg555, and Arg792. We demonstrated the binding of PfIRPa to RNA IRE experimentally (Figures2-4A). Antibody raised in rabbits against amino acids 609-626 specifically recognized PfIRPa and confirmed that its molecular mass lies within the range of the predicted mass for this protein of 103 379 dalton (Figure 4B). Most importantly, this same antibody also recognized PfIRPa comigrating with IRE after transfer of gel-retardation assay complexes to nitrocellulose, although in the supershift experiment, when antibody 3950 was added to plasmodial lysate before the addition of 32P-IRE, it did not deplete the IRE-binding activity (not shown). Immunofluorescence probing revealed that PfIRPa is located within the cytosol of the parasite in the infected erythrocyte.
Our results are consistent with the view that the regulation ofP falciparum binding to IRE is iron specific because iron deprivation of the parasite cultures by DFO resulted in the enhancement of binding, whereas the supplementation of iron reduced the IRE-binding activity (Figure 4A) and correlated with a comparable decrease in total PfIRPa bands on Western blots. The iron-rich, potentially pro-oxidant environment of the host erythrocytes38 may dictate the need for the malaria parasite to strictly control the expression of putative iron transport, iron utilization, or iron storage genes. The same type of regulation might be necessary for parasite survival in different iron environments present in the different hosts.
Our demonstration of P falciparum IRP-binding activity differs from the results of Breton et al,39 perhaps because PfIRPa is prone to degradation in P falciparumlysates. This feature also prevented us from determining the precise Kds for the PfIRPa-IRE complexes, which we plan to determine with a recombinant protein. We found that the IRE-binding activity of PfIRPa could be easily overlooked if stringent precautions are not taken to remove the host cell constituents containing large amounts of IRP1 successfully competing with PfIRPa for binding of the mammalian consensus IRE (not shown). Our results indicate that significant changes in the amounts of PfIRPa in response to iron deprivation were found on the level of protein. This type of regulation is reminiscent of the behavior of the mammalian IRP2, rather than the IRP1.32 We can consider further evidence in favor of this type of regulation because of observed lack of PfIRPa-associated aconitase activity, the cytosolic and not the mitochondrial localization of the protein within the parasite (Figure 5), the reduction of IRE-binding activity of PfIRPa in the presence of β-mercaptoethanol (Figure 4A), and because the PfIRPa levels appear to be iron regulated (Figure 4).
We continue analyzing the plasmodial genome regularly as new sequences are being deposited into the GenBank to identify IRE-containing targets for PfIRPa, but there are no other obvious candidates at this point.
Further exploration of IRP-IRE regulation of iron metabolism in P falciparum has significance from different standpoints. First, it will enhance our understanding of the differences in iron regulation between malaria parasites and their mammalian hosts. Second, overexpression of PfIRPa can be used to identify plasmodial transcripts that contain IREs.15 40 Third, this understanding might facilitate the design of novel therapeutic strategies based on insights into P falciparum iron metabolism.
We thank David Keister and Olga Muratova for donating the 3D7 strain of Plasmodium falciparum and for offering technical recommendations. We thank Dr Louis Miller for the fruitful discussion of the results and helpful suggestions. We thank Dr William P. Winter for critically reading the manuscript and for valuable comments. We thank the Malaria Resource and Reference Center at ATCC for the provision of the P falciparum anti-BIP (GRP) antibody.
Supported in part by the Howard University Research Scientist Award 5-UH1-HL03 679-02 from the National Heart, Lung and Blood Institute; and by the RO1 AI 44857-02 grant from the National Institute of Allergic and Infectious Diseases, National Institutes of Health, Bethesda, MD.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mark Loyevsky, Howard University, Center for Sickle Cell Disease, 2121 Georgia Ave, NW, Washington, DC 20059; e-mail: mloyevsky@howard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal