Abstract
The occurrence of clonal T cells in multiple myeloma (MM), as defined by the presence of rearrangements in the T-cell receptor (TCR)–β chains detected on Southern blotting, is associated with an improved prognosis. Recently, with the use of specific anti–TCR-variable-β (anti–TCRVβ) antibodies, the presence in MM patients of expanded populations of T cells expressing particular Vβ regions was reported. The majority of these T-cell expansions have the phenotype of cytotoxic T cells (CD8+CD57+ and perforin positive). Since Vβ expansions can result from either a true clonal population or a polyclonal response, the clonality of CD8+TCRVβ+ T cells was tested by TCRVβ complementarity-determining region 3 length analysis and DNA sequencing of the variable region of the TCR. In this report, the CD57+ and CD57− subpopulations within expanded TCRVβ+CD8+ cell populations are compared, and it is demonstrated that the CD57+ subpopulations are generally monoclonal or biclonal, whereas the corresponding CD57− cells are frequently polyclonal. The oligoclonality of CD57+ expanded CD8+ T cells but not their CD57− counterparts was also observed in age-matched controls, in which the T-cell expansions were mainly CD8−. The CD8+CD57+ clonal T cells had a low rate of turnover and expressed relatively lower levels of the apoptotic marker CD95 than their CD57− counterparts. Taken together, these findings demonstrate that MM is associated with CD57+CD8+ T-cell clones, raising the possibility that the expansion and accumulation of activated clonal CD8+ T cells in MM may be the result of persistent stimulation by tumor-associated antigens, combined with a reduced cellular death rate secondary to reduced expression of the apoptosis-related molecule CD95.
Introduction
Expanded populations of T cells, as measured by an increase in the number of cells positively stained by monoclonal antibodies to variable (V) regions of human T-cell receptor (TCR) subfamilies, have been detected in the peripheral blood of patients with multiple myeloma (MM) and smouldering myeloma.1-5These populations consist mainly of CD8+ cells and persist for long periods, suggesting that they are the result of chronic antigenic stimulation.5,6 In contrast, transient antigenic stimulation is associated with temporary CD8+ T-cell expansions.7 Although persistent T-cell expansions in MM may involve up to 25% of total T cells,5 such expansions may not necessarily be clonal. On the basis of Southern blot analysis of the TCRβ, which detects monoclonal expansions representing at least 4% to 5% of the total Ficoll-separated peripheral blood cells,8 our laboratory previously reported on the prognostic significance of true T-cell clonal populations in MM.9 We found that MM patients exhibiting expanded T-cell clones had a better prognosis than those without detectable clonal bands on Southern blot analysis.9
Flow cytometric analysis offers the means to isolate and characterize such clones in terms of specificity and function. Previous flow analysis indicated that the expanded T-cell clones, as judged by specific positivity with anti–TCRVβ antibodies, expressed the phenotype of cytotoxic T cells and were predominantly CD8+, CD57+, CD28−, and perforin positive.5 However, it was not clear from this flow analysis that the expanded populations of cytotoxic T cells corresponded to the clonal populations that related to survival advantage detected by previous Southern blot experiment.9The CD57 antigen is normally present on a minority (16%) of CD8+ T cells,10 but the percentage of CD8+CD57+CD28− cells has been shown to be increased in a number of clinical conditions, including human immunodeficiency virus (HIV)/acquired immunodeficiency syndrome,11,12 cytomegalovirus (CMV) infection,13,14 common variable immunodeficiency,15 and post–bone marrow transplantation,16 in addition to MM.17
The most sensitive way to demonstrate clonality is by analysis of the length of the complementarity-determining region 3 (CDR3) of theTCRV genes, followed by sequencing.18 19 In the present study, we have used this technique to demonstrate the genuine monoclonal or oligoclonal nature of the expanded populations of CD57+CD8+TCRVβ+ cells in patients with MM. Moreover, only a small proportion of CD57+ T cells were undergoing active proliferation, and they had significantly lower surface expression of CD95 (Fas) than did their CD57− counterparts, in both expanded and nonexpanded TCRVβ subfamilies. Taken together, these findings demonstrated that MM, a B-cell clonal disease, is associated with CD8+ T-cell clones, raising the possibility that the expansion and accumulation of activated clonal CD8+ T cells in MM may be the result of persistent stimulation by tumor-associated antigens, combined with a reduced death rate secondary to the reduced expression of the apoptosis-related molecule CD95.
Patients, materials, and methods
Patients
After informed consent, peripheral heparinized blood samples were collected from 16 patients with well-documented myeloma presenting to our clinic for routine assessment (Table1). Patients were treated according to the Australian Leukaemia Study Group myeloma trial protocol, which involved multiagent therapy (prednisone, cyclophosphamide, doxorubicin, and bis-chlornitrosourea) followed by stem cell transplantation if the patient was younger than age 65 years. Seven of 16 patients had had chemotherapy within the previous 6 months before testing (Table 1). Four patients had autologous stem cell transplantation. However, none of these transplantations occurred within 2 years prior to this study (Table 1). The study was approved by the Central Sydney Area Health Service Human Ethics Committee. Normal controls used in the CDR3 length analysis experiments and CD95 studies were 27 blood donors (mean age = 66 years) at the Red Cross Blood Transfusion Service, Sydney. The normal control samples used in the proliferation studies were from healthy laboratory staff.
Profiles of myeloma patients in this study
| Patient* . | Age (y) . | Sex . | Disease duration after diagnosis (mo) . | Disease stage† . | M-component type‡ . | Chemotherapy treatment within 6 mo of study entry . | Duration since last SCT (mo)1-153 . |
|---|---|---|---|---|---|---|---|
| A | 70 | M | 109 | IA | IgG-κ | No | — |
| B | 53 | F | 61 | IIA | IgG-κ | No | 27 |
| C | 82 | F | 47 | IIIB | IgG-κ | Yes | — |
| D | 82 | F | 71 | IIIA | IgG-λ | Yes | — |
| E | 37 | M | 67 | IIA | IgG-κ | No | 59 |
| F | 74 | M | 81 | IIIB | IgG-κ | Yes | — |
| G | 71 | F | 46 | IIB | IgG-κ | No | — |
| H | 60 | M | 17 | IIB | IgG-λ | Yes1-155 | — |
| I | 44 | F | 99 | IIIA | κ-LC | No | 46 |
| J | 86 | M | 37 | IA | IgG-λ | No | — |
| K | 74 | M | 145 | IA | IgG-λ | No | — |
| L | 43 | M | 4 | IIA | IgG-λ | Yes | — |
| M | 55 | F | 2 | IIA | IgG-κ | Yes | — |
| N | 56 | F | 35 | IIA | IgG-κ | No | — |
| O | 72 | M | 27 | IIIA | IgG-κ | Yes1-155 | 60 |
| P | 61 | M | 61 | IIA | IgA-κ | No | — |
| Patient* . | Age (y) . | Sex . | Disease duration after diagnosis (mo) . | Disease stage† . | M-component type‡ . | Chemotherapy treatment within 6 mo of study entry . | Duration since last SCT (mo)1-153 . |
|---|---|---|---|---|---|---|---|
| A | 70 | M | 109 | IA | IgG-κ | No | — |
| B | 53 | F | 61 | IIA | IgG-κ | No | 27 |
| C | 82 | F | 47 | IIIB | IgG-κ | Yes | — |
| D | 82 | F | 71 | IIIA | IgG-λ | Yes | — |
| E | 37 | M | 67 | IIA | IgG-κ | No | 59 |
| F | 74 | M | 81 | IIIB | IgG-κ | Yes | — |
| G | 71 | F | 46 | IIB | IgG-κ | No | — |
| H | 60 | M | 17 | IIB | IgG-λ | Yes1-155 | — |
| I | 44 | F | 99 | IIIA | κ-LC | No | 46 |
| J | 86 | M | 37 | IA | IgG-λ | No | — |
| K | 74 | M | 145 | IA | IgG-λ | No | — |
| L | 43 | M | 4 | IIA | IgG-λ | Yes | — |
| M | 55 | F | 2 | IIA | IgG-κ | Yes | — |
| N | 56 | F | 35 | IIA | IgG-κ | No | — |
| O | 72 | M | 27 | IIIA | IgG-κ | Yes1-155 | 60 |
| P | 61 | M | 61 | IIA | IgA-κ | No | — |
M indicates myeloma; SCT, stem-cell transplantation; Ig, immunoglobulin; LC, light chain.
CDR3 length analysis and sequencing were performed on specimens of patients A through F. Propidium iodide–labeling analysis was conducted on specimens of all multiple myeloma patients except G through I. CD95 analysis was performed on specimens of all patients except J through M.
Myeloma staging is based on the Durie and Salmon staging system with 2 additional parameters: plasma cell labeling index and β-2 microglobulin.
κ and λ stand for kappa and lambda LCs, respectively.
Autologous SCT was performed on 4 patients before their entry into this study.
Only these 2 out of 7 chemotherapy-treated patients showed decreased white cell counts and lymphocyte counts at time of entry to this study.
Monoclonal antibodies
The following directly conjugated mouse monoclonal antibodies were used: anti-CD3, anti-CD4, anti-CD8, and anti-CD57, all fluorescein isothiocyanate (FITC) labeled (Becton Dickinson, San Jose, CA); phycoerythrin (PE)–conjugated anti-TCRVβ 1, 2, 3, 5.1, 5.2, 5.3, 7, 8, 9, 11, 12, 13.1, 13.6, 14, 16, 17, 18, 20, 21.3, 22, and 23 (all antibodies are mouse immunoglobulin [Ig] except anti-TCRVβ1 which is a rat IgG1) (Immunotech, Marseille, France); anti-CD3 (Becton Dickinson); and peridinin chlorophyll protein (PerCP)–conjugated anti-CD4 and anti-CD8 (Becton Dickinson). Unconjugated monoclonal antibodies used in the proliferation analysis were anti-TCRVβ antibodies as listed above (Immunotech). Biotinylated sheep anti–mouse immunoglobulin and biotinylated rabbit anti–rat immunoglobulin antibodies were used as secondary reagents for the nonconjugated anti-TCRVβ antibodies. The allophycocyanin (APC)–conjugated anti-CD4 (Pharmingen, San Diego, CA) was used. The following biotinylated mouse monoclonal antibodies were used: anti-CD8, anti-CD95, and biotinylated mouse IgG1 isotype control (Pharmingen). Streptavidin conjugated with Alexa Fluor-488 (Molecular Probes, Eugene, OR) was used in the proliferation studies to increase the fluorescent signal in the FITC channel to allow the TCRVβ subsets to be clearly identified. Streptavidin conjugated with Alexa Fluor-594 (Molecular Probes) was used to demonstrate the biotinylated primary antibodies in the dual-laser system.
Cell surface staining and flow sorting
Ficoll-separated cells were stained with biotinylated anti-CD8 and incubated on ice for 20 minutes. After 2 washes with phosphate-buffered saline (PBS), a cocktail of FITC-conjugated anti-CD57, PE-conjugated anti-TCRVβ, and streptavidin conjugated to Alexa Fluor-594 was added, and the cells were incubated for a further 20 minutes on ice. Stained cells were washed once with PBS and remained on ice until sorting by means of a FACStar Plus Cytometer (Becton Dickinson). CD57+TCRVβ+CD8+ and CD57−TCRVβ+CD8+cells were sorted into 2 separate tubes containing RPMI-1640 containing 10% heat-inactivated fetal calf serum (ICN, Costa Mesa, CA), 1 mM L-glutamine, 100 IU/mL penicillin, and 160 U/mL (160 μg/mL) gentamicin. The tubes were kept cool during the sort.
CDR3 length analysis
Six patients (A through F) with expanded TCRVβsubsets were chosen for CDR3 length analysis. They were selected because their T-cell expansions could be labeled at high fluorescent intensity with PE-conjugated anti-TCRVβ antibodies and could therefore be sorted accurately. The CDR3 length analysis has also been carried out on 6 age-matched controls. The QuickPrep Micro mRNA Purification Kit (Amersham Pharmacia Biotech, Piscataway, NJ) with oligo-deoxythymidine (dT)–cellulose was used to obtain messenger RNA (mRNA) from approximately 103 to 104 sorted cells according to the manufacturer's protocol. Complementary DNA (cDNA) was subsequently prepared by means of the T-primed First-Strand Kit (Amersham), which uses Moloney murine leukemia virus reverse transcriptase and an oligo-dT primer to generate full-length first-strand cDNA from the mRNA template. The cDNA was stored at −70°C until in vitro amplification of TCR genes was performed with a 5′ primer specific for the appropriate TCRβsubfamily19 and a 32P-labeled 3′ primer for the TCRβ constant region, which was common to all rearranged β-chain genes.19 The reverse primer was radioactively end-labeled in a solution consisting of 27 MBq (0.73 mCi)/mL γ32P]adenosine 5′-triphosphate (ATP) (Amersham), and 2.3 μM T4 polynucleotide kinase (9.6 × 102 U/mL) (Boehringer Mannheim, Mannheim, Germany). The polymerase chain reaction (PCR) reaction mix also contained the following components at the final concentrations listed here: 2.3 μM forward primer; 0.38 μM deoxy-ATP (dATP), d–cytidine 5′-triphosphate, d–ribothymidine 5′-triphosphate, and d–guanosine 5′-triphosphate (Boehringer Mannheim); a 1 × PCR-reaction buffer with either 1.5 mM or 3 mM MgCl2(Boehringer Mannheim); and 0.01 U Taq polymerase (Boehringer Mannheim). We added 5 μL PCR reaction mix to each well of a Thermo-fast 96-well plate (Advanced Biotechnologies, Epsom, Surrey, United Kingdom) containing 5 μL cDNA. The mixture was overlaid with 25 μL mineral oil (ICN). PCR amplification was performed in an Omni-e thermal cycler (Hybaid, Ashford, United Kingdom) by means of the following thermocycling protocol: initial denaturation at 95°C for 5 minutes; followed by 45 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute; with a final extension at 72°C for 7 minutes. Polyacrylamide gel electrophoresis was used to distinguish PCR products, which had been diluted 2-fold with sequencing gel–loading buffer, denatured for 5 minutes at 95°C in an Omni-e thermal cycler, and snap-chilled on ice. To allow an estimation of PCR bands sizes, γ32P]ATP–labeled molecular weight markers (Number VIII) (Boehringer Mannheim) were electrophoresed on the same gel. The PCR products were electrophoresed at 900 V for approximately 3 hours. Following electrophoresis, gels were transferred to 3-mm chromatography paper (Whatman International, Maidstone, United Kingdom), covered in plastic wrap (Huntsman, Castle Hill, Australia), and directly exposed to X-OMAT AR scientific imaging film (Eastman Kodak, Rochester, NY) overnight at −70°C, by means of intensifying screens (DuPont NEN, Denver, CO). Film was developed in a Curix 60 film processor (Agfa-Gevaert, Munich, Germany).
Sequencing of PCR products of specific TCRVβsubfamilies
To obtain PCR products of specific TCRVβsubfamilies for DNA sequencing, PCR was performed with the use of cDNA samples with Ready-to-go PCR beads (Amersham). The PCR amplification conditions were similar to those described above except that no radioactive label was used. Samples were taken from 3 patients (A, B, and C) who had been previously shown to have single, dual, and multiple dominant bands, respectively, on polyacrylamide gel electrophoresis. Direct sequencing of the PCR products was carried out by Supermac (Camperdown, Australia). Where direct sequencing failed, the Original TA Cloning Kit (Invitrogen, Carlsbad, CA) was used to clone the PCR products in plasmid vectors according to the manufacturer's protocol. The linearized vector supplied in this kit ends with single 3′deoxythymidine (dT) residue that allows PCR inserts to ligate efficiently with the vector, since the nontemplate-dependent activity of Taq polymerase adds a single deoxyadenosine (dA) to the 3′ ends of PCR products. A few isolated white transformants were picked individually and allowed to grow for 12 hours at 37°C in 5 mL Luria-Bertani medium containing 50 μg/mL ampicillin in a rotary shaking incubator at 225 rpm. The plasmid DNA was then purified with a NucleoSpin kit (Macherey-Nagel, Düren, Germany). DNA sequencing employing dye-terminator chemistry with M13 forward and reverse primers was performed on the extracted plasmid DNA. PCR products from TCRVβ age-matched controls were also direct sequenced (Supermac).
Estimation of T-cell proliferative index
Ficoll-separated cells from patients A to F were first labeled with nonconjugated antibodies specific for the TCRVβsubfamily known to be expanded in each individual. After a 20-minute incubation on ice, cells were washed and stained for a further 20 minutes with biotinylated antimouse immunoglobulin (Dako, Carpinteria, CA), except for anti-TCRVβ1 for which a biotinylated rabbit anti–rat immunoglobulin (Dako) was used. Cells were then washed and stained with streptavidin conjugated to Alexa-488 (Molecular Probes). Alternatively, to determine the proliferative index in total CD8 and CD4 populations in MM patients (no. = 8; patient A and patients J through P) and normal controls (no. = 4; healthy laboratory staff), cells were labeled initially with biotinylated anti-CD8 or anti-CD4 (Pharmingen), followed by streptavidin conjugated to Alexa-488, and then washed with PBS. The supernatant was removed and the pellet resuspended. While the cells were gently vortexed, 100 μL DNA-Prep lysing and permeabilizing reagent (Beckman Coulter, Fullerton, CA) and then 2 mL DNA-Prep Stain (Beckman Coulter) was added, and the sample was incubated for 15 minutes at room temperature. Cells were analyzed on a Coulter Epics XL flow cytometer. Multicycle for Windows (Phoenix Flow Systems, San Diego, CA) was used to analyze the DNA histograms.
CD95 surface staining
Ficoll-separated cells from 12 MM patients (patients A through I and N through P) were examined for expression of CD95 on total CD3+ T cells or TCRVβ subsets (both expanded and nonexpanded) by means of 3-color labeling. An aliquot of cells was stained with biotinylated anti-CD8 to allow the percentage of CD8 or CD4 cells within each TCRVβ subset to be calculated (Table 2). Propidium iodide (PI) was added to all samples before flow analysis. Cells were analyzed on a Coulter Epics XL flow cytometer. Normally, more than 99% of the cells in the lymphocyte gate were viable as assessed by PI. Biotinylated mouse IgG1 was used as the isotype control. For a 4-color labeling experiment, 5 MM patients (A through E) were examined for expression of CD95 on total CD3+ T cells and different TCRVβ subsets. For 4-color labeling, Ficoll-separated cells were stained with biotinylated anti-CD95 (Pharmingen), which was followed by a cocktail of FITC-conjugated anti-CD57, PE-conjugated anti-TCRVβ or anti-CD3, and streptavidin conjugated to Alexa-594 (Molecular Probes) and APC-conjugated anti-CD4 (Pharmingen). Stained cells were washed once and then fixed in 1% paraformaldehyde for at least 30 minutes before analysis by means of a FACStar Plus cytometer.
Median CD95 expression and proportion of CD8+cells within the CD57+ and CD57− compartments of individual TCRVβ subfamilies in 2 patients
| . | Vβ in CD3+ T cells, %* . | 3 SD above mean of age-matched controls . | CD8 in Vβ+cells, % . | CD95 expression in CD57+TCRVβ+ cells (MCF)† . | CD8 in CD57+ TCRVβ+ cells, %‡ . | CD95 expression in CD57−TCRVβ+ cells (MCF)2-153 . | CD8 in CD57− TCRVβ+ cells, %‡ . |
|---|---|---|---|---|---|---|---|
| Patient A | |||||||
| Vβ1 | 12.82-155 | 6.2 | 87.0 | 59 | 81.4 | 92 | 83.4 |
| Vβ2 | 5.0 | 14.5 | 69.0 | 54 | 84.9 | 174 | 32.3 |
| Vβ3 | 10.1 | 10.7 | 91.0 | 59 | 97.1 | 121 | 64.7 |
| Vβ7 | 1.7 | 4.7 | 69.0 | 80 | 72.7 | 161 | 43.1 |
| Vβ8 | 3.6 | 8.8 | 68.0 | 71 | 89.9 | 151 | 41.5 |
| Vβ13.6 | 2.5 | 4.5 | 81.0 | 75 | 99.6 | 178 | 21.3 |
| Vβ17 | 1.5 | 9.3 | 30.0 | 75 | 17.3 | 169 | 18.5 |
| Patient B | |||||||
| Vβ1 | 8.82-155 | 6.2 | 59.0 | 72 | 97.6 | 186 | 32.8 |
| Vβ2 | 13.0 | 14.5 | 22.0 | 78 | 18.8 | 170 | 16.1 |
| Vβ7 | 9.02-155 | 4.7 | 34.0 | 77 | 98.8 | 136 | 63.5 |
| Vβ8 | 7.5 | 8.8 | 23.0 | 70 | 62.0 | 199 | 16.6 |
| Vβ13.6 | 4.82-155 | 4.5 | 20.0 | 100 | 75.3 | 182 | 27.2 |
| Vβ16 | 1.8 | 2.5 | 23.0 | 87 | 16.7 | 178 | 18.9 |
| Vβ17 | 6.3 | 9.3 | 27.0 | 82 | 53.0 | 175 | 23.2 |
| . | Vβ in CD3+ T cells, %* . | 3 SD above mean of age-matched controls . | CD8 in Vβ+cells, % . | CD95 expression in CD57+TCRVβ+ cells (MCF)† . | CD8 in CD57+ TCRVβ+ cells, %‡ . | CD95 expression in CD57−TCRVβ+ cells (MCF)2-153 . | CD8 in CD57− TCRVβ+ cells, %‡ . |
|---|---|---|---|---|---|---|---|
| Patient A | |||||||
| Vβ1 | 12.82-155 | 6.2 | 87.0 | 59 | 81.4 | 92 | 83.4 |
| Vβ2 | 5.0 | 14.5 | 69.0 | 54 | 84.9 | 174 | 32.3 |
| Vβ3 | 10.1 | 10.7 | 91.0 | 59 | 97.1 | 121 | 64.7 |
| Vβ7 | 1.7 | 4.7 | 69.0 | 80 | 72.7 | 161 | 43.1 |
| Vβ8 | 3.6 | 8.8 | 68.0 | 71 | 89.9 | 151 | 41.5 |
| Vβ13.6 | 2.5 | 4.5 | 81.0 | 75 | 99.6 | 178 | 21.3 |
| Vβ17 | 1.5 | 9.3 | 30.0 | 75 | 17.3 | 169 | 18.5 |
| Patient B | |||||||
| Vβ1 | 8.82-155 | 6.2 | 59.0 | 72 | 97.6 | 186 | 32.8 |
| Vβ2 | 13.0 | 14.5 | 22.0 | 78 | 18.8 | 170 | 16.1 |
| Vβ7 | 9.02-155 | 4.7 | 34.0 | 77 | 98.8 | 136 | 63.5 |
| Vβ8 | 7.5 | 8.8 | 23.0 | 70 | 62.0 | 199 | 16.6 |
| Vβ13.6 | 4.82-155 | 4.5 | 20.0 | 100 | 75.3 | 182 | 27.2 |
| Vβ16 | 1.8 | 2.5 | 23.0 | 87 | 16.7 | 178 | 18.9 |
| Vβ17 | 6.3 | 9.3 | 27.0 | 82 | 53.0 | 175 | 23.2 |
CD95 expression results of the 2 multiple myeloma patients A and B have been graphed in Figure 5A.
MCF indicates median channel fluorescence.
The normal range of individual TCRVβsubfamilies was established by means of the age-matched controls.
For this column, the mean expression was 74; median, 75; and SD, 12.
The percentage of CD8+ cells in the CD57+ and CD57− subpopulations of each Vβ subset was calculated from the 3-color flow analysis for specific TCRVβ, CD8, and CD57.
For this column, the mean expression was 162; media, 172; and SD, 29.
Indicates expanded TCRVβ population.
Results
Overrepresentation of CD8+ cells within TCRVβ expansions in MM patients
We have previously reported that T-cell expansions, defined as those TCRVβ+ T-cell populations exceeding the mean + 3 SD of the percentage of Vβ+cells in peripheral blood T cells of normal controls, were found in 79% of patients with MM (no. = 38; mean age = 62 years), but in only 19% of normal controls (no. = 17; mean age = 45 years).5 In the present study, we included an age-matched control group (no. = 27; mean age = 66 years) and found that 63% (17 of 27 subjects) had at least one T-cell expansion, consistent with the normal age-dependent increase in T-cell expansions.20Since the T-cell expansions in MM patients had previously been shown to express predominantly CD85 whereas those in the normal control group of younger age did not (data not shown), we decided to make a more detailed comparison of the frequency and phenotype of T-cell expansions in patients with MM versus the age-matched controls. Data from the 27 age-matched controls in this study and those MM patients from the previous group5 for whom Vβ analysis had included CD8 status (22 of the 38) were reanalyzed after gating for either CD8+CD3+ or CD8−CD3+cells (Figure1). Given the small percentage of circulating CD4+CD8+ and CD4−CD8− T cells in human blood, CD8−CD3+ cells were composed predominantly of CD4+ T cells. The CD8+CD3+TCRVβ+ and CD8−CD3+TCRVβ+percentages were calculated as a percentage either of total CD3+ cells or of CD8+CD3+ or CD8−CD3+ T cells, respectively. The latter calculation was included to correct for the low CD4+ T-cell counts that are characteristic of many patients with MM and that may mask any CD8− expansions. TCRVβ+expansions were defined as those populations exceeding the mean + 3 SD of the percentage in the normal controls.
CD8 expression in T-cell expansions.
T-cell expansions from 27 age-matched donors were compared with 22 patients with MM. The data of patients with MM have been published previously5 and were reanalyzed and reported again here for comparison. Cells were gated for either CD8−CD3+ (panels A through D) or CD8+CD3+ T cells (panels E through H) before calculation of the percentage of cells expressing each Vβas a function of either total CD3+ T cells (panels A through B and E through F), CD8−CD3+ (panels C and D), or CD8+CD3+ (panels G and H). Horizontal bars represent 3 SD above the means for the age-matched control group. #The 21 Vβ families studied (left-right) were Vβ1, 2, 3, 5.1, 5.2, 5.3, 7, 8, 9, 11, 12, 13.1, 13.6, 14, 16, 17, 18, 20, 21.3, 22, and 23.
CD8 expression in T-cell expansions.
T-cell expansions from 27 age-matched donors were compared with 22 patients with MM. The data of patients with MM have been published previously5 and were reanalyzed and reported again here for comparison. Cells were gated for either CD8−CD3+ (panels A through D) or CD8+CD3+ T cells (panels E through H) before calculation of the percentage of cells expressing each Vβas a function of either total CD3+ T cells (panels A through B and E through F), CD8−CD3+ (panels C and D), or CD8+CD3+ (panels G and H). Horizontal bars represent 3 SD above the means for the age-matched control group. #The 21 Vβ families studied (left-right) were Vβ1, 2, 3, 5.1, 5.2, 5.3, 7, 8, 9, 11, 12, 13.1, 13.6, 14, 16, 17, 18, 20, 21.3, 22, and 23.
The new analysis indicated that CD8− expansions were present both in patients with MM and in age-matched controls (Table3). The slightly reduced frequency in MM (10 of 22 patients versus 17 of 27 controls) was not statistically significant (χ2 = 1.85, P > .1), and the number was unaffected by being calculated as a percentage of CD8−CD3+ cells rather than total CD3+ cells. In contrast, reanalysis of Vβfamily expression within CD8+ T cells indicated that CD8+ expansions in MM patients were significantly more frequent than in age-matched controls (Table4). A higher percentage of MM patients had at least one CD8+ T-cell expansion (χ2 = 8.6, P < .005), and the total number of expansions in the MM patients was also significantly higher (χ2 = 26.7, P < .005). Thus, in addition to the CD8+ expansions previously described,5MM patients showed the CD4+ expansions expected as a function of age. Thirteen of the 22 MM patients in this analysis (59%) had CD8+ expansions representing at least 5% of total CD3+ T cells, which is the estimated detection limit of the Southern blot technique previously used to demonstrate the prognostic significance of T-cell expansions in MM.9 In contrast, the CD8+ T-cell expansions, which represented more than 5% of total CD3+ T cells, were found in only 15% of normal age-matched controls. In summary, MM patients have a greater number of CD8+ expansions, and these constitute a significantly greater percentage of total T cells than the age-matched controls.
CD8− expansions in age-matched controls and multiple myeloma patients
| . | Age-matched controls . | Patients with MM3-150 . |
|---|---|---|
| No. subjects in study | 27 | 22 |
| No. subjects showing CD8− expansions3-151 | 9 | 7 |
| Total no. CD8− expansions3-151 | 17 | 10 |
| Mean no. CD8− expansions per positive subject3-151 | 1.9 | 1.4 |
| No. subjects showing CD8− expansions predicted to be detectable by Southern blot3-152 | 3 | 1 |
| Total no. CD8− expansions predicted to be detectable by Southern blot3-152 | 4 | 1 |
| No. CD8− expansions predicted to be detectable by Southern blot per positive subject3-152 | 1.3 | 1 |
| Total no. CD8− expansions3-151,3-153 | 18 | 10 |
| . | Age-matched controls . | Patients with MM3-150 . |
|---|---|---|
| No. subjects in study | 27 | 22 |
| No. subjects showing CD8− expansions3-151 | 9 | 7 |
| Total no. CD8− expansions3-151 | 17 | 10 |
| Mean no. CD8− expansions per positive subject3-151 | 1.9 | 1.4 |
| No. subjects showing CD8− expansions predicted to be detectable by Southern blot3-152 | 3 | 1 |
| Total no. CD8− expansions predicted to be detectable by Southern blot3-152 | 4 | 1 |
| No. CD8− expansions predicted to be detectable by Southern blot per positive subject3-152 | 1.3 | 1 |
| Total no. CD8− expansions3-151,3-153 | 18 | 10 |
Data were calculated as a function of total CD3+cells, except where noted otherwise.
MM indicates multiple myeloma.
Reanalyzed data of 22 patients with MM from previous publication.5
After gating for CD8−CD3+ T cells, expanded populations were those calculated as a function of total CD3+ cells and present at 3 SD or more above the mean percentage of Vβ+CD8− cells of the age-matched controls.
A 4% to 5% monoclonal population can be detected with the Southern blot technique.8 The size of monoclonal Vβ+ populations (predicted to be detectable by Southern blot) was estimated by subtracting the mean values for the particular Vβ from the percentage of CD8−CD3+ T cells, calculated as above. The mean values represent the percentage of polyclonal populations within individual TCRVβ subfamilies.
Calculated as a function of total CD8− cells.
CD8+ expansions in age-matched controls and multiple myeloma patients
| . | Age-matched controls . | Patients with MM4-150 . |
|---|---|---|
| No. subjects in study | 27 | 22 |
| No. subjects with CD8+ expansions4-151 | 16 | 21 |
| Total no. CD8+ expansions4-151 | 23 | 71 |
| Mean no. CD8+ expansions per positive subject4-151 | 1.4 | 3.4 |
| No. subjects with CD8+ expansions predicted to be detectable by Southern blot‡ | 4 | 13 |
| Total no. CD8+ expansions predicted to be detectable by Southern blot‡ | 5 | 25 |
| No. CD8+ expansions predicted to be detectable by Southern blot per positive subject‡ | 1.3 | 1.9 |
| Total no. CD8+expansions4-151,4-153 | 20 | 35 |
| . | Age-matched controls . | Patients with MM4-150 . |
|---|---|---|
| No. subjects in study | 27 | 22 |
| No. subjects with CD8+ expansions4-151 | 16 | 21 |
| Total no. CD8+ expansions4-151 | 23 | 71 |
| Mean no. CD8+ expansions per positive subject4-151 | 1.4 | 3.4 |
| No. subjects with CD8+ expansions predicted to be detectable by Southern blot‡ | 4 | 13 |
| Total no. CD8+ expansions predicted to be detectable by Southern blot‡ | 5 | 25 |
| No. CD8+ expansions predicted to be detectable by Southern blot per positive subject‡ | 1.3 | 1.9 |
| Total no. CD8+expansions4-151,4-153 | 20 | 35 |
Data were calculated as a function of total CD3+cells, except where noted otherwise.
MM indicates multiple myeloma.
Reanalyzed data of 22 patients with MM from previous publication.5
After gating for CD8+CD3+ T cells, expanded populations were those calculated as a function of total CD3+ cells and that present at 3 SD or more above the mean percentage of Vβ+CD8+ cells of the age-matched controls.
The size of monoclonal Vβ+populations (predicted to be detectable by Southern blot) was estimated by subtracting the mean values for the particular Vβ from the percentage of CD8+CD3+ T cells, calculated as above.
Calculated as a function of total CD8+ cells.
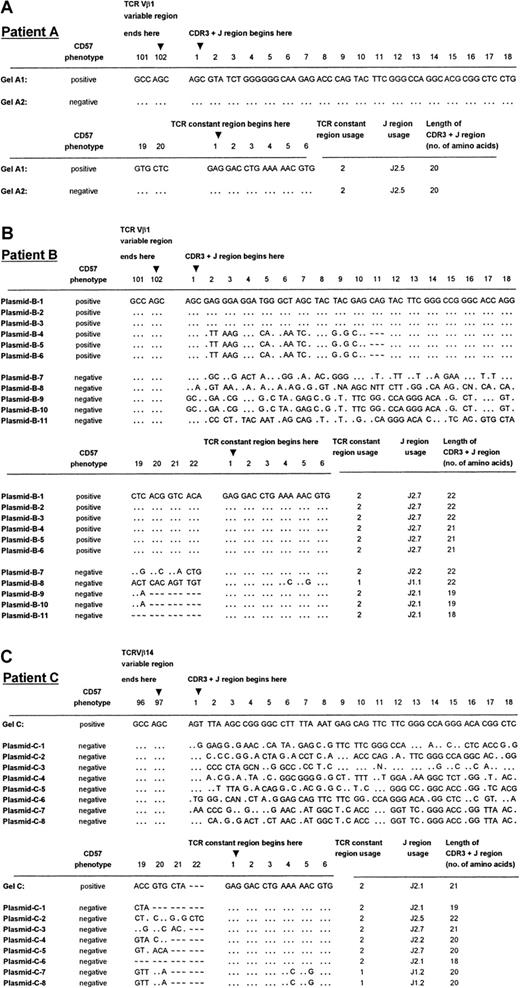
CDR3 length analysis suggests that the CD57+subset of expanded cytotoxic T cells is monoclonal or oligoclonal
We have previously shown that the phenotype of the expanded Vβ populations in MM was predominantly CD57+and CD28−, which differs significantly from the nonexpanded Vβ populations.5 The reactivity of T cells to specific anti-TCRVβ antibodies can represent either a true clonal expansion or a polyclonal response. Therefore, the clonality of sorted CD57+CD8+TCRVβ+ T cells from either MM patients or age-matched controls was compared with that of CD57−CD8+TCRVβ+ T cells by means of TCRVβ CDR3 length analysis and DNA sequencing of the variable region of the TCR. Cells were sorted by their CD57 phenotype because we have recently shown that expanded cytotoxic T cells contain intracytoplasmic perforin, and its expression is directly associated with the expression of CD57 in expanded Vβ populations.5
PCR was performed with cDNA prepared from the sorted cells, a 5′ primer specific for the appropriate TCRVβ subfamily, and a 32P-labeled 3′ primer from the TCRβconstant region. Figure 2A shows the length of TCRVβ CDR3 amplified from the CD8+CD57+ and CD8+CD57− T cells of 6 patients (patients A through F) with MM. A single band derived by PCR of the CD8+TCRVβ+CD57+ cDNA was obtained for 4 of the 6 patients examined (Figure 2A, lanes 1, 9, 13, and 15). One third of the patients in this study (patients B and F) showed 2 bands of equal intensity (Figure 2A, lanes 5 and 17). In the case of patient C, a second expanded population (Vβ17+) (Figure 2A, lane 11) showed multiple bands within the CD57+ subset. The CD57−sorted T cells generally contained multiple bands of nearly equal intensity (Figure 2A, lanes 4, 6, 8, 10, 16, and 18). For example, the TCRVβ1 and Vβ14 amplification products of the CD57−TCRVβ+CD8+sorted T cells of patients B and C, respectively, both consisted of 5 bands each separated by 3 nucleotides (Figure 2A, lanes 6, 10). However, in some cases, such as patient A, the CD57−sorted cells of an expanded TCRVβ also contained a single band (Figure 2A, lane 2). Similarly, the CD57− sorted cells of an expanded TCRVβ of patient C were oligoclonal (Figure 2A, lane 12). Therefore, although CD57− T cells were generally polyclonal, they may occasionally be oligoclonal. The monoclonality and biclonality of the CD57+ subset of expanded CD8+TCRVβ+ T cells were also observed in age-matched controls (Figure 2B; lanes 23, 25, and 27 showed a single band and lanes 19 and 29 showed 2 equally dominant bands).
TCRVβ CDR3 length analysis.
The PCR products (approximately 220 base pairs) and molecular markers were run on a 6% polyacrylamide gel, and the sizes of the radioactive PCR bands were determined by exposing an x-ray film to the gel. Expanded T cells were sorted according to their specific surface phenotype of TCRVβ+ and CD8+ and CD57 expression. Bands are lined up side by side for ease of comparison. The relative position of the PCR products of different TCRVβ families to each other is not the same as the actual gel. *Expanded TCRVβ populations are underlined. (A) CDR3 length analysis results from CD8+ T-cell expansions found in 6 patients with MM, together with CD8 and CD57 expression data. (B) Results of 6 age-matched normal controls for comparison. **Direct DNA sequencing has been performed on some of the PCR products and results are summarized.
TCRVβ CDR3 length analysis.
The PCR products (approximately 220 base pairs) and molecular markers were run on a 6% polyacrylamide gel, and the sizes of the radioactive PCR bands were determined by exposing an x-ray film to the gel. Expanded T cells were sorted according to their specific surface phenotype of TCRVβ+ and CD8+ and CD57 expression. Bands are lined up side by side for ease of comparison. The relative position of the PCR products of different TCRVβ families to each other is not the same as the actual gel. *Expanded TCRVβ populations are underlined. (A) CDR3 length analysis results from CD8+ T-cell expansions found in 6 patients with MM, together with CD8 and CD57 expression data. (B) Results of 6 age-matched normal controls for comparison. **Direct DNA sequencing has been performed on some of the PCR products and results are summarized.
Direct DNA sequencing and plasmid cloning confirmed the monoclonality or biclonality of the CD57+ expanded T-cell population
TCRVβ CDR3 PCR products of patients A (Vβ1+), B (Vβ1+), and C (Vβ14+) were sequenced to ensure that the distribution of bands observed was a true reflection of the clonality of the Vβ+ expansions. These patients were chosen because they had previously been shown to have single, dual, and multiple dominant bands, respectively, in CDR3 length analysis. In every case, PCR products that appeared as a single band gave unambiguous direct DNA sequencing data (Figure 2A, lanes 1 and 2; Figure 3A, Gel-A1, A2; Figure 2A, lane 9; Figure 3C, Gel-C), indicative of a dominant sequence within the PCR product. On the other hand, none of the TCRVβ+CD8+CD57− PCR products yielded readable sequence data except for patient A, whose CD57− PCR product indicated a monoclonal population (Figure 2A, lane 2; Figure 3A, Gel-A2).
TCRVβ DNA sequences of patients A through C.
Panels A, B, and C show the sequencing results of patients A, B, and C, respectively. DNA sequences were derived from direct sequencing of the CD57+ PCR products from patient A (labeled Gel-A1 in panel A) and patient C (labeled Gel-C in panel C). No readable sequence was obtained from direct sequencing of the CD57− PCR products, except in the case of patient A (labeled Gel-A2 in panel A). Plasmids B-1 to B-6 are 6 independent sequences obtained from 6 cDNA clones chosen at random from a library containing the CD57+ PCR product from patient B. Plasmids B-7 to B-11 are the independent sequences obtained from 5 cDNA clones from the CD57− PCR product from the patient shown in panel B. Plasmids C-1 to C-8 are 8 independent sequences obtained from 8 cDNA clones chosen at random from a library containing the CD57− PCR product from patient C (panel C). The sequences summarized in this Figure have been given the European Molecular Biology Laboratory (Heidelberg, Germany) nucleotide sequence database accession numbers: AJ276183 through AJ276184 for panel A, AJ276185 through AJ276195 for panel B, and AJ276196 throughAJ276204 for panel C. Keys: … identical nucleotides; ∼ ∼ ∼ absence of an amino acid at the relative position in the CDR3 sequence alignment.
TCRVβ DNA sequences of patients A through C.
Panels A, B, and C show the sequencing results of patients A, B, and C, respectively. DNA sequences were derived from direct sequencing of the CD57+ PCR products from patient A (labeled Gel-A1 in panel A) and patient C (labeled Gel-C in panel C). No readable sequence was obtained from direct sequencing of the CD57− PCR products, except in the case of patient A (labeled Gel-A2 in panel A). Plasmids B-1 to B-6 are 6 independent sequences obtained from 6 cDNA clones chosen at random from a library containing the CD57+ PCR product from patient B. Plasmids B-7 to B-11 are the independent sequences obtained from 5 cDNA clones from the CD57− PCR product from the patient shown in panel B. Plasmids C-1 to C-8 are 8 independent sequences obtained from 8 cDNA clones chosen at random from a library containing the CD57− PCR product from patient C (panel C). The sequences summarized in this Figure have been given the European Molecular Biology Laboratory (Heidelberg, Germany) nucleotide sequence database accession numbers: AJ276183 through AJ276184 for panel A, AJ276185 through AJ276195 for panel B, and AJ276196 throughAJ276204 for panel C. Keys: … identical nucleotides; ∼ ∼ ∼ absence of an amino acid at the relative position in the CDR3 sequence alignment.
Plasmid cloning and sequencing of the cDNA were performed on the PCR products of the 2 patients (B, C) in whom direct sequencing failed to provide interpretable sequencing results of the CDR3 regions in TCRVβ+CD8+CD57+ T cells. Cloning and sequencing of the PCR products from the CD57− sorted T cells gave an array of sequences, indicative of a polyclonal population (Figure 3B, plasmids B-7 to B-11; Figure 3C, plasmids C-1 to C-8). Cloning and sequencing of the CD57+ PCR product of the expanded CD8+ T-cell subset of patient B, which gave 2 bands of equal intensity in the CDR3 length polymorphism analysis, revealed 2 distinct sequences, each present in 3 of the 6 randomly chosen cDNA clones (Figure 3B, plasmids B-1 to B-6). These 2 TCRVβ1 clones differed by only 1 amino acid in length, consistent with the CDR3 length polymorphism analysis (Figure 2A, lane 5). The CD57− sorted cells of patient B revealed 4 different clones in 5 plasmid preparations, which differed in length by 1 to 4 amino acids. Furthermore, T-cell clones plasmids B-7 and B-8, which had the same CDR3 length, possessed unique sequences and used a different joining-β gene segment. The fact that the dominant clones of TCRVβ1+CD8+CD57+ were not identified in the corresponding CD57− sorted populations from patient B may have been due to the limited number of plasmid preparations analyzed. Of the 8 independent sequences obtained from 8 cDNA clones chosen at random from a library containing the CD8+TCRVβ14+CD57−PCR product of patient C, 8 clones that differed by 1 to 4 amino acids in length were identified (Figure 3C). Once again, the single dominant clone identified in the CD57+ sorted cells of patient C was not found in the CD57− subset.
The monoclonality of the CD57+ subsets of the expanded CD8+TCRVβ+ T cells in age-matched controls was shown by direct DNA sequencing of the PCR products showing a single band (Figure 2B, lanes 23, 25, and 27) or PCR products with a single dominant band plus other weaker bands (Figure 2B, lane 21). Neither PCR products with double bands nor multiple dominant bands shown in polyacrylamide gel electrophoresis (Figure 2B, lanes 19, 20, 22, and 26) gave an interpretable direct DNA sequencing result, indicating nonclonal populations.
The majority of the expanded T cells in MM patients are not proliferating
We have previously shown that the expanded T-cell clones of patients with MM remain stable over an 18-month period.5To examine the level of turnover in the expanded T-cell populations, we examined the proliferative index of T cells from the expanded TCRVβ subsets of patients with MM by measuring the proportion of cells in the S phase by means of a protocol previously established to measure cycling plasma cells in our laboratory.21 The proliferative index of the TCRVβ expanded cells from 6 MM patients (patients A through F) was compared with that of total CD8+ and CD4+ T cells in MM (no. = 8, patients A, J through P) and normal controls (no. = 4) (Figure 4). Only 0.1% to 1.8% of the TCRVβ expanded cells of MM patients were in the S phase, a percentage that did not differ significantly from the CD8+ and CD4+ T cells of MM patients or normal controls examined (ANOVA, P = .84). On the other hand, the TCRVβ expanded T cells and total CD8+ T cells of these MM patients retained the capacity to proliferate in a dose-related fashion in response to polyclonal stimuli such as PHA or concanavalin A. The percentage of cells in S phase increased within 24 hours of 15 μg/mL PHA stimulation from 1.6% to 11.1% in a dose-related fashion (Figure 4, far right column).
Proliferative index of the expanded TCRVβsubsets.
The proliferative index measured by the percentage of cells within S phase of expanded TCRVβ+ T cells in MM patients (far left column) and compared with that of the total CD8+ or total CD4+ T cells in MM patients (second and third columns from left) and normal controls (NCs, fourth and fifth columns from left). No significant difference was found among the groups (one-way analysis of variance [ANOVA],P = .84). The positive control was cultured CD8+ cells of patient D at 24 hours after stimulation with various amounts of phytohemagglutinin (PHA). *Far right column, shown as squares in order from top to bottom with the following amounts of PHA added to the 1-mL culture medium: 15 μg, 11.1%; 5 μg, 5.3%; 1.5 μg, 3.7%, and 0 μg, 1.6%.
Proliferative index of the expanded TCRVβsubsets.
The proliferative index measured by the percentage of cells within S phase of expanded TCRVβ+ T cells in MM patients (far left column) and compared with that of the total CD8+ or total CD4+ T cells in MM patients (second and third columns from left) and normal controls (NCs, fourth and fifth columns from left). No significant difference was found among the groups (one-way analysis of variance [ANOVA],P = .84). The positive control was cultured CD8+ cells of patient D at 24 hours after stimulation with various amounts of phytohemagglutinin (PHA). *Far right column, shown as squares in order from top to bottom with the following amounts of PHA added to the 1-mL culture medium: 15 μg, 11.1%; 5 μg, 5.3%; 1.5 μg, 3.7%, and 0 μg, 1.6%.
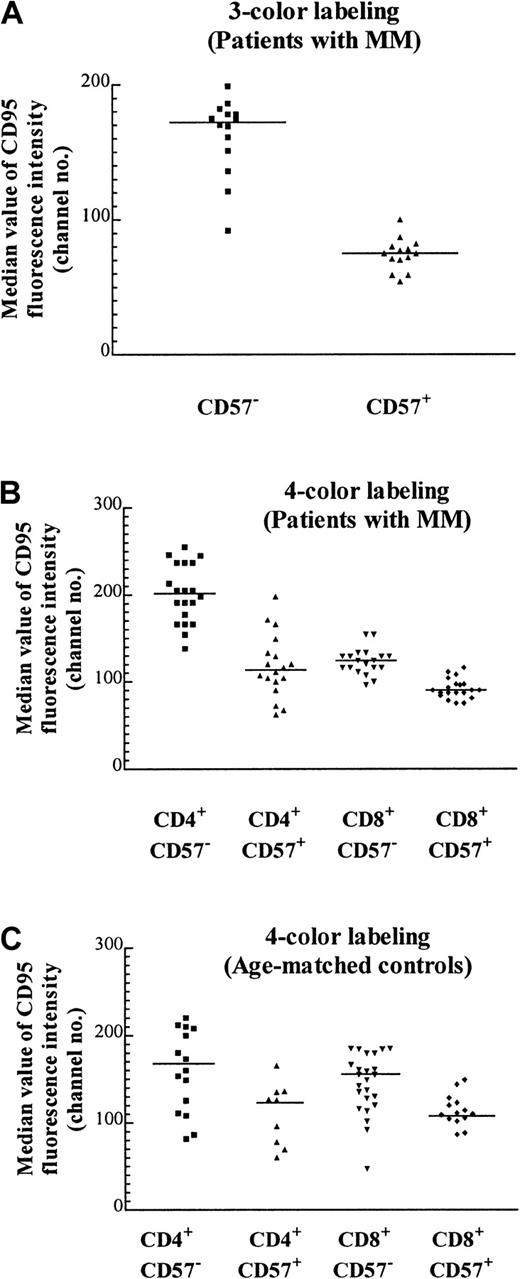
CD57+ T cells express CD95 at lower levels than their CD57− counterparts
We have previously noted that the proportion of T cells expressing the apoptotic marker Fas (CD95) tends to be lower in expanded populations.5 CD95 expression as determined by quantitative flow cytometry of the CD57+ Vβpopulations was approximately 2-fold lower than that of their CD57− counterparts (Figure5A; Table 2). Because CD95 expressed by the CD8+ subset of the TCRVβ+CD57− cells could not be definitively determined by a 3-color staining protocol, it was unclear whether the increase in the percentage of CD8+ cells within particular TCRVβ populations was responsible for the decrease in CD95 expression (eg, Table 2, Vβ1 of patient A and Vβ7 of patient B). To unambiguously examine the expression of CD95 on expanded CD57+ subsets, 4-color labeling was used so that the CD95 levels in each of the 4 subsets (namely, CD4+CD57−, CD4+CD57+, CD8+CD57−, and CD8+CD57+) could be determined. It was found that there was a 30% reduction of CD95 expression in CD8+CD57+ cells compared with their CD8+CD57− counterparts (Figure 5B, column 3 versus 4; P < .0001, Mann-Whitney test). The reduction in CD95 expression within the CD4+ compartment was even more marked (Figure 5B, column 1 versus 2; P < .0001, Mann-Whitney test). This suggests that although CD95 (Fas) may be involved in the accumulation of CD57+ T cells, an additional factor, such as chronic antigenic stimulation, could be more important in the determination of the fate of these cells. The median CD95 levels in these 4 subsets from patients with MM were similar to that of the age-matched normal controls (Figure 5C). The number of CD4+CD57+ cells was low in the age-matched controls, and in some cases the number was too low to include in the subset analysis.
Median CD95 expression on T-cell subsets with the use of 3- and 4-color labeling.
(A) Median value of CD95 fluorescence with the use of 3-color labeling on CD57+ and CD57− subsets of various expanded and nonexpanded TCRVβ+ subsets from 2 MM patients (patients A and B) performed on the same day (P < .0001, Mann-Whitney test, and Table 2). The staining pattern is representative of the results found in 12 MM patients examined. (B) Median CD95 expression on T cells of 5 MM patients (patients A through E) with the use of 4-color labeling. The CD57+ and CD57− subsets in either the CD4+ or the CD8+ subpopulations of the TCRVβ+ subpopulations are significantly different (P < .0001, Mann-Whitney test). (C) Median CD95 expression of T cells in 5 age-matched controls. The CD57+and CD57− subsets in either CD4+ or CD8+ subpopulations of the TCRVβ subsets are significantly different (P = .027 and .002 respectively, Mann-Whitney test).
Median CD95 expression on T-cell subsets with the use of 3- and 4-color labeling.
(A) Median value of CD95 fluorescence with the use of 3-color labeling on CD57+ and CD57− subsets of various expanded and nonexpanded TCRVβ+ subsets from 2 MM patients (patients A and B) performed on the same day (P < .0001, Mann-Whitney test, and Table 2). The staining pattern is representative of the results found in 12 MM patients examined. (B) Median CD95 expression on T cells of 5 MM patients (patients A through E) with the use of 4-color labeling. The CD57+ and CD57− subsets in either the CD4+ or the CD8+ subpopulations of the TCRVβ+ subpopulations are significantly different (P < .0001, Mann-Whitney test). (C) Median CD95 expression of T cells in 5 age-matched controls. The CD57+and CD57− subsets in either CD4+ or CD8+ subpopulations of the TCRVβ subsets are significantly different (P = .027 and .002 respectively, Mann-Whitney test).
Discussion
There is currently considerable interest in the development of immunotherapy as a novel treatment option for patients with myeloma and other malignancies. In myeloma, the unique tumor antigen, the idiotype, has been adopted as the immunizing agent. The potential value of immunotherapy in MM is based on a dual rationale. First, it has been postulated that plateau-phase disease may have an immunoregulatory basis,22 and second, our group has recently found that expanded T-cell clones with a cytotoxic phenotype (CD8+, CD57+, CD28−, and perforin positive) are present in a majority of MM patients5 and that their presence is associated with longer survival, consistent with an antitumor effect.5 We have also recently shown that patients whose myeloma cells carry the CD86 antigen (B7-2+) have both a poor prognosis and a lower number of expanded T-cell clones, suggesting that an immunological basis exists for the improved prognosis.23 In the belief that these expanded T-cell clones may possess anti-idiotypic specificity for myeloma cells, most studies to date have involved a strategy of idiotype immunization with the patients' own paraproteins. However, so far, only minimal antitumor responses have been reported with the use of either in vitro or in vivo markers, which raises questions about the true specificity of the expanded clones and has necessitated further analysis of their properties.24-29
In the present study, flow cytometric analysis of TCR Vβsubset distribution within circulating T cells indicated once again that MM patients and age-matched controls differed in the frequent occurrence of large CD8+ T-cell expansions in MM. It has been previously reported that in the healthy elderly both CD4+ and CD8+ expansions are seen.20 This observation has been confirmed in the present study and extended to include MM patients. The oligoclonality, low proliferative index, and lower CD95 expression of the CD57+subpopulations was seen in both MM and aged-matched normal control groups. These findings did not appear to be related to previous therapy administered to MM patients. Although 7 of 16 patients received chemotherapy within 6 months of this study, no patient had received chemotherapy within 2 weeks of the day of study, nor did we observe any difference in the results of CDR3 length analysis, proliferation study, and CD95 expression between those who received and those who did not receive chemotherapy. Four patients had autologous stem cell transplantation at least 2 years before their entry into this study. We have investigated the effect of transplantation on T-cell populations, and this will be reported elsewhere. In summary, we found that CD8+ T-cell expansions appeared shortly after the transplantation, but are all short-lived and most disappeared within 3 to 6 months after transplantation.
In the CDR length analysis study, the extracted mRNA encodes those TCRβ chains that are expressed as proteins on the surface of sorted T cells, and only in-frame rearrangements are represented. Hence, the PCR products from a polyclonal population form a ladder at 3-nucleotide intervals, corresponding to differences of 1 amino acid in the length of the CDR3 region. Furthermore, the intensity of each band is proportional to the initial number of TCRVβtranscripts with that particular CDR3 length. In this report, we analyzed the CD57+ and CD57− subpopulations within expanded TCRVβ+CD8+ cells and demonstrated that the CD57+ subpopulations are generally clonal, whereas the corresponding CD57− cells are frequently polyclonal. In a study of healthy subjects that used a similar approach based on sorting for CD28+ and CD28− cells, the CD8+CD28− T cells were found to be dominated by relatively few clones.30 However, the expanded T-cell clones in MM patients may represent up to 25% of total CD3 cells, whereas such clones in healthy subjects compose only a small percentage of total T cells30 and have been postulated to reflect a response to long-standing viral infection.13
Using the Southern blot technique, we previously reported expanded clonal T cells in 32% of MM patients (no. = 119).9 This is a lower percentage than in the cohort of MM patients in this study, in which T-cell expansions composing more than 5% of total CD3+ cells were seen in 59% of patients. This discrepancy could be due to a number of factors: (1) the sensitivity of the Southern blot technique previously used may have been lower than the 5% limit estimated by van Dongen and Wolvers-Tettero8; (2) total T-cell counts of patients may have differed in the 2 study times; and (3) the presence of more than one TCR within the T-cell expansions, due either to biclonality within the CD57+subset (as demonstrated in one third of MM patients in this study) or polyclonality within the CD57− population, may have led to an overestimate of the number of T-cell expansions representing more than 5% of total T cells.
The CD57+ clonal T cells had a low rate of turnover, as demonstrated by S-phase analysis, and expressed relatively lower levels of the apoptotic marker CD95 than their CD57−counterparts. This finding provides an explanation for the accumulation of CD8+CD57+ T cells and is consistent with a previous report that they are not susceptible to spontaneous or activation-induced apoptosis in vitro.16,31 Further evidence in support of this concept comes from an in vivo study in mice, which revealed that cells with the highest levels of CD95 expression are preferentially deleted.32
After prolonged stimulation and proliferation, CD8+CD28+ cells tend to lose expression of CD28.33 Thus, the previous demonstration by our group that the expanded TCRVβ+CD8+ cells in patients with myeloma have reduced levels of CD28 is suggestive of chronic antigenic stimulation.5 In theory, this could have been due either to a persistent viral infection in a compromised host or to an ongoing antitumor response. However, no correlation was demonstrated between the presence of viral infections, such as CMV and hepatitis, and expanded T-cell clones in a cohort of MM patients (no. = 40) previously studied (unpublished observations, September 2000), although one patient did have a transient increase in the TCRVβ+CD8+ cells associated with a recurrent herpes infection. In the current study, 7 of 27 age-matched controls showed no evidence of CD8+ or CD8− T-cell expansion. Although all these donors were negative for HIV and hepatitis B antibodies, 5 of them had anti-CMV antibodies. This suggests that CMV infection in the elderly is not necessarily associated with T-cell expansion. A role for human herpes virus 8 in the pathogenesis of myeloma remains controversial, and clonal expansion of T cells is not a feature of patients with Kaposi sarcoma.34-38 Moreover, expanded TCRVβ+CD8+ T-cell populations in patients with MM can persist for many years, consistent with chronic stimulation by a tumor-associated antigen.5
In our previous study,5 we showed that the expression of perforin was directly associated with that of CD57 in the expanded TCRVβ populations as well as in peripheral blood lymphocytes in general. It was suggested that CD57 is not a direct marker of cytotoxicity but may be related to a state of activation. Similar coexpression of perforin and CD57 has also been reported in T-cell expansions in patients with Hodgkin disease.39
The identity of possible tumor-associated antigens in myeloma is still unknown. Circulating idiotype remains an attractive candidate owing to its persistence throughout the course of the disease and the fact that it represents a unique tumor antigen. Idiotype is a possible antigen, and anti-idiotypic peptide responses that lead to tumor protection have been demonstrated in artificial transgenic models,40-46but evidence of anti-idiotype CD4+-dependent or CD8+-dependent responses in MM remains controversial.24-29 Idiotype-specific cytotoxic T lymphocytes in MM capable of lysing autologous primary tumor cells have been recently reported.47
Having shown that monoclonality or oligoclonality is a property of the CD57+CD8+ T-cell subset, we aim to identify whether these CD8+ expanded T-cell clones are really tumor-specific and, if so, which specific tumor antigen they recognize. Some plasma cell-specific antigens, such as HM1.2448 and CD138 (syndecan-1),49 50 have been previously identified and may be potential targets for immunotherapy. Knowledge of the specificity of the CD57+CD8+ T-cell clones may also allow us to design immunization regimens for MM patients who have failed to make spontaneous antitumor responses.
We wish to thank Joseph Webster for expert technical assistance in the flow-sorting work on FACStar Plus and Dr Robert Brink for his kind gift of the Original TA Cloning Kit.
Supported by the Foundation IV fund and the International Myeloma Foundation (D.M.-Y.S.) and the Academy of Finland (M.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Daniel M.-Y. Sze, Institute of Haematology, Royal Prince Alfred Hospital, Camperdown, NSW, Australia 2050; e-mail:d.sze@centenary.usyd.edu.au.



![Fig. 4. Proliferative index of the expanded TCRVβsubsets. / The proliferative index measured by the percentage of cells within S phase of expanded TCRVβ+ T cells in MM patients (far left column) and compared with that of the total CD8+ or total CD4+ T cells in MM patients (second and third columns from left) and normal controls (NCs, fourth and fifth columns from left). No significant difference was found among the groups (one-way analysis of variance [ANOVA],P = .84). The positive control was cultured CD8+ cells of patient D at 24 hours after stimulation with various amounts of phytohemagglutinin (PHA). *Far right column, shown as squares in order from top to bottom with the following amounts of PHA added to the 1-mL culture medium: 15 μg, 11.1%; 5 μg, 5.3%; 1.5 μg, 3.7%, and 0 μg, 1.6%.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/9/10.1182_blood.v98.9.2817/4/m_h82111715004.jpeg?Expires=1763489279&Signature=eYi26wnXgasqrZmAQHqa5C3Jy91WZGG61WDio2RF2pbVDYP9GS3Bvv7i08DAgEufEGCZwrgQgJBDADwe049y78d3aL2W0jgZxfgS7IJYsILXymPXGJKV3VqMsmnT6yTX3q-Vf6XW1fwNxzxe8Ptg7h95-uJYgq96fTYgXaOGpekQiyVBn08bz1Okz6narCgCZO~k-4YozH-Sp6LmYUUILEjAY2GTAIHtQfu5EonM~QSSPGCp-fZGV7XV~YLn~899p1iDE1ZEMmeHJzU~Y5tgyYwtM2NoeGbxD64M0qfaAI3McH7bcqeOg0~t41zl582RJQ8yX3LyWi6-FcyEFzguxQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal