The ataxia–telangiectasia mutated (ATM) gene codifies for a protein critically involved in the cellular response to DNA damage. ATM alterations have been observed in some sporadic lymphoproliferative disorders. The recurrent 11q22-23 deletions found in mantle cell lymphoma (MCL) suggest that ATM could be inactivated in these lymphomas. In this study, ATM gene alterations and protein expression were examined in 20 and 17 MCL tumor specimens, respectively. Previously, these patients had been examined forp53 and p14ARF gene status and analyzed by comparative genomic hybridization. Nine patients had 11q22-23 losses. Eight ATM gene mutations were detected in 7 patients. These alterations were 3 missense mutations in the phosphatidylinositol-3 kinase (PI-3K) domain and 5 truncating mutations, including 3 frameshifts, a nonsense mutation, and a substitution of the initial methionine. All truncating mutations were associated with lack of protein expression. Somatic origin was demonstrated in 3 mutations, whereas one mutation was carried heterozygously in the patient germ line. Chromosomal imbalances were significantly higher in typical MCL with ATM inactivation (7.8 ± 1.3) than in tumors with the wild-type gene (3 ± 1.1) (P = .001). Moreover, tumors with bi-allelic ATM alteration were associated with 3q gains (P = .015) and frequent extranodal involvement (P = .049).ATM gene alterations were not related to the histologic variant of the tumors, p53/p14ARF gene status, survival, or other clinicopathologic features of the patients. These findings indicate that ATM gene mutations in MCL are mainly truncating or missense mutations involving the PI-3K domain, and that may play a role in the pathogenesis of a subset of these tumors with increased numbers of chromosomal imbalances.
Introduction
Mantle cell lymphoma (MCL) is a lymphoproliferative disorder characterized by the t(11;14) (q13;q32) translocation, which leads to the rearrangement and overexpression of the cyclinD1 gene.1,2 However, the tumorigenic and transforming potential of cyclin D1 in experimental models is relatively limited, and it requires the cooperation of other oncogenic factors such as c-myc.3 Additional alterations in the tumor suppressor genes p16INK4a andp53 have been described in aggressive variants of MCL, suggesting that these genes may cooperate with cyclin D1overexpression in the progression of these lymphomas.4-7Classical cytogenetic and comparative genomic hybridization (CGH) studies have shown a high number of recurrent chromosomal alterations in MCL, indicating that other genes may be involved in the pathogenesis of these tumors.8-10 One of the most frequent secondary chromosomal aberrations in MCL is the loss of the 11q22-23 region, where the ataxia–telangiectasia mutated (ATM) gene is located.11 12
Mutations in the ATM gene are responsible for the ataxia–telangiectasia (AT) syndrome, a rare autosomal recessive disorder characterized by progressive cerebellar ataxia, ocular telangiectasia, immunodeficiency, high sensitivity to ionizing radiation, and predisposition to lymphoid malignancies.13AT cells show chromosomal instability, telomere shortening, and defects in response to ionizing radiation and radiomimetic drugs.14 Mutations and deletions in the ATMgene have also been found in a variety of sporadic neoplasias, including T-prolymphocytic leukemia (T-PLL)15-17 and B-cell chronic lymphocytic leukemia (B-CLL).18,19 More recently,ATM mutations have been identified in MCL, mainly associated with 11q22-23 deletions.20 However, the incidence of these mutations in tumors with no alterations in chromosome 11 and the possible relation between ATM inactivation and morphologic variants of tumors and the clinicopathologic characteristics of patients are unknown.
ATM gene encodes for a serine-threonine kinase belonging to the phosphatidylinositol-3 kinase (PI-3K) family. This enzyme plays a central role in signaling pathways activated by DNA damage.21-23 Different studies have now identified a number of ATM targets including c-abl, p53, Chk-2, Nbs-1, and BRCA-1.21,24-27 Particularly, p53 seems to participate in the ATM regulation of the G1/S checkpoint activated by DNA damage. Thus, in response to different DNA damaging agents, ATM phosphorylates p53, promoting its stabilization and transcriptional activation and leading to cell cycle arrest, DNA repair, or apoptosis.26 28p53 Gene inactivation has been recognized as a frequent phenomenon in the pathogenesis of aggressive variants of lymphoproliferative disorders including MCL. However, the possible relation between p53 and ATM inactivation in the development and progression of human tumors is not well known.
The aim of this study was to analyze the role of ATM gene alterations in the pathogenesis of MCL and possible relationships with genetic, clinical, and pathologic characteristics of the tumors. Our findings indicate that ATM gene inactivation is a relatively frequent phenomenon in these lymphomas, mainly occurring by truncating mutations and nucleotide substitutions in the PI-3K domain. ATM aberrations are independent of p53 gene status, and they are associated with a significantly higher number of chromosomal imbalances in typical variants of MCL.
Materials and methods
Tumor selection
Twenty MCL tumor specimens were obtained from the Department of Pathology of the Hospital Clinic, University of Barcelona (Spain) on the basis of the availability of frozen samples for molecular studies. These specimens consisted of 12 typical and 8 blastoid MCL tumors.1 Immunophenotype was analyzed using immunohistochemistry on tissue sections or cell suspensions by flow cytometry. All samples were positive for cyclin D1 expression by Northern blot analysis, immunohistochemistry analysis, or both. All these tumors had been previously examined for mutations and deletions of the p53 gene and the INK4a/ARF locus, and chromosomal imbalances were also analyzed by CGH.9 29
RNA extraction and reverse transcription–polymerase chain reaction
Total RNA was obtained from frozen tissues using guanidine isothiocyanate extraction and cesium chloride gradient centrifugation. Complementary DNA was synthesized from 1 μg total RNA using random hexamers (TaqMan reverse transcription reagents; Applied Biosystems, Warrington, United Kingdom) and following the manufacturer's specifications. The ATM complete coding region was amplified by nested polymerase chain reaction (PCR) in 8 partly overlapping portions, ranging in size from 0.9 to 1.6 kb. Reactions were performed using the primers and PCR conditions previously described, with minor modifications.30 Briefly, PCR amplifications of fragments 1 through 5 and fragment 7 were carried out using a PCR program with a touchdown profile, with final temperatures ranging from 58°C to 54°C. Fragments 6 and 8 were amplified following the originally proposed profiles.30 All the amplifications were carried out in a 2400 Perkin-Elmer thermocycler (Norwalk, CT).
Restriction endonuclease fingerprinting and sequencing analyses
The whole ATM coding region was screened for mutations using restriction endonuclease fingerprinting (REF) analysis and direct sequencing in 17 MCL specimens, following previously described protocols.30 31 In each gel, wild-type DNA from reactive tonsils was included to determine the normal restriction patterns of the analyzed fragments. Direct sequencing of positive reverse transcription–PCR products was performed by cycle sequencing dRhodamine or BigDye terminator chemistry (Applied Biosystems). Sequencing reactions were run on a Perkin-Elmer ABI-377 automated sequencer. All mutations were confirmed by sequencing both strands. In 3 additional samples with normal ATM protein expression and 11q CGH profile, only the PI-3K domain was analyzed by direct sequencing.
Germline studies
To analyze the germline status of the ATM gene, constitutional normal DNA was obtained in 6 specimens. In 3, DNA was extracted from peripheral blood granulocytes separated by density gradient centrifugation. In the remaining samples, constitutional DNA was obtained from normal tissue microdissected from frozen tissue sections. Microdissection was carried out using the laser pressure catapulting technique of the Robot-microbeam system (P.A.L.M. GmbH, Benried, Germany). DNA extraction from microdissected samples was performed as described elsewhere.32 Possible contamination of these normal DNA samples by tumor cells was ruled out by analysis of the immunoglobulin heavy chain gene, bcl-1 rearrangement, or both.33
PCR amplifications of the ATM gene were performed using a hemi-nested strategy with primers flanking the genomic region in which the mutations were located in the tumor complementary DNA (Table1). The first PCR amplification conditions were 35 cycles at 94°C for 1 minute, 55°C to 60°C for 1 minute, and 72°C for 1.5 minutes. The amplification profile of the second PCR round was the same, with an annealing temperature ranging from 60°C to 65°C depending on the particular amplification. Finally, direct sequencing using the same PCR primers was performed.
Primers used in germline studies
| Exon . | Mutation . | Primers . |
|---|---|---|
| Forward: 5′-TTGTGCCTTTGACCAGAATGTGC-3′ | ||
| Exon 4 | 67 CT | Reverse: 5′-GGGTTACTAATCACACATTTCAAGG-3′ |
| Forward nested: 5′-TGACCAGAATGTGCCTCTAATTG-3′ | ||
| Forward: 5′-CAAATACAAGCTGAAAACTTTGGC-3′ | ||
| Exon 5 | Δ73-76 | Reverse: 5′-ACCTAAAAACAGCATCCCAATTC-3′ |
| Forward nested: 5′-TGACCAGAATGTGCCTCTAATTG-3′ | ||
| Exon 12 | Δ1564- | Forward: 5′-CAAATACAAGCTGAAAACTTTGGC-3′ |
| 1565 | Reverse: 5′-CTCGGCCAAACAAGAAACGCATC-3′ | |
| Forward nested: 5′-GCTGAAAACTTTGGCTTACTTGGA-3′ | ||
| Forward: 5′-CCACACCCGGCCTAAAGTTGTAG-3′ | ||
| Exon 57 | 8150 AT | Reverse: 5′-AAGCAACCTCACTGTACATGTCTG-3′ |
| Forward nested: 5′-GGTGGACCACACAGGAGAATATG-3′ |
| Exon . | Mutation . | Primers . |
|---|---|---|
| Forward: 5′-TTGTGCCTTTGACCAGAATGTGC-3′ | ||
| Exon 4 | 67 CT | Reverse: 5′-GGGTTACTAATCACACATTTCAAGG-3′ |
| Forward nested: 5′-TGACCAGAATGTGCCTCTAATTG-3′ | ||
| Forward: 5′-CAAATACAAGCTGAAAACTTTGGC-3′ | ||
| Exon 5 | Δ73-76 | Reverse: 5′-ACCTAAAAACAGCATCCCAATTC-3′ |
| Forward nested: 5′-TGACCAGAATGTGCCTCTAATTG-3′ | ||
| Exon 12 | Δ1564- | Forward: 5′-CAAATACAAGCTGAAAACTTTGGC-3′ |
| 1565 | Reverse: 5′-CTCGGCCAAACAAGAAACGCATC-3′ | |
| Forward nested: 5′-GCTGAAAACTTTGGCTTACTTGGA-3′ | ||
| Forward: 5′-CCACACCCGGCCTAAAGTTGTAG-3′ | ||
| Exon 57 | 8150 AT | Reverse: 5′-AAGCAACCTCACTGTACATGTCTG-3′ |
| Forward nested: 5′-GGTGGACCACACAGGAGAATATG-3′ |
Protein extraction and Western blot analysis
Nuclear protein extracts were obtained from 17 MCL samples in which additional frozen tissue was available. Cryostat frozen sections were lysed in ice-cold buffer containing 10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.1% Nonidet P-40, 0.5 mM dithiothreitol, 2 μg/mL leupeptin, 5 μg/mL aprotinin, and 0.5 mM phenylmethylsulfonyl fluoride for 15 minutes. After centrifugation at 3500 rpm, the precipitated nuclei were lysed in a buffer containing 80 mM Tris-HCl (pH 6.8), 2% sodium dodecyl sulfate, and 10% glycerol. Eighty micrograms nuclear protein was run per lane on a 5% sodium dodecyl sulfate–polyacrylamide gel and electroblotted to a nitrocellulose membrane (Amersham, Buckinghamshire, United Kingdom). Membranes were blocked by overnight incubation in 5% dry milk and 0.1% Tween-20 at 4°C. Blocked membranes were then incubated with the polyclonal antibody anti-ATM Ab3 (Oncogene Research, Boston, MA) overnight at 4°C, washed with phosphate-buffered saline 0.1% Tween-20, and incubated with a sheep anti–rabbit secondary antibody conjugated to horseradish peroxidase (Amersham). Polyclonal antibody antipoly ADP ribose polymerase (PARP) (Oncogene Research) was used as a loading control in all cases. After washing, antibody binding was detected by chemiluminescence detection procedures according to the manufacturer's recommendations (ECL; Amersham). Intensities of the ATM proteins were normalized to the PARP band signal.
Statistical analysis
Tumors were grouped according to histologic subtype andATM gene status (wild-type and 1 or 2 affected ATM alleles). The following clinicopathologic variables were recorded and included in the analysis: age, sex, histologic subtype, performance status, stage (Ann Arbor), extranodal involvement (0 or 1 vs 2 or more sites), lactate dehydrogenase serum level, International Prognostic Index, response to therapy (complete or partial vs failure), peripheral blood involvement, length of survival from diagnosis, number of chromosomal imbalances, and different chromosomal aberrations. Comparison between the number of chromosomal imbalances detected by CGH and the status of ATM alleles was performed using the Kruskal-Wallis test. Associations between ATM inactivation and the presence of a particular clinicopathologic parameter were evaluated with the 2-tailed Fisher exact test. Probability of survival was calculated using the method of Kaplan and Meier,34 and different curves were statistically compared by means of the log-rank test.35Level of significance was set at .05 for all analyses.
Results
Mutational analysis
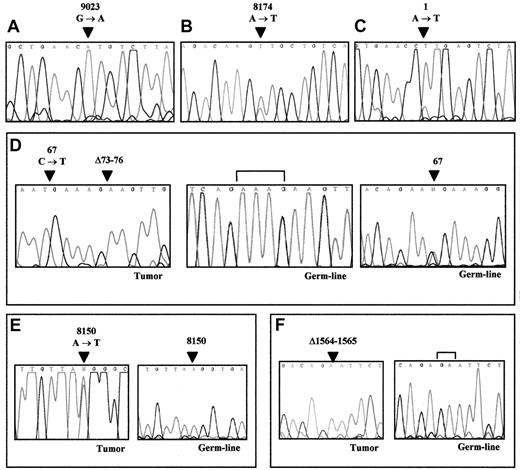
The entire coding region of the ATM gene was initially examined in 17 MCL samples. Nine had been shown by CGH analysis to exhibit 11q losses, including the 11q22-23 region. In this group, 6ATM gene mutations were identified in 5 tumors (Table2). Four of these mutations (in samples 1, 7, and 8) predicted premature termination of the protein lacking the PI-3K domain. Two additional alterations (samples 2 and 6) were missense mutations (9023 G→A and 8174 A→T, respectively) leading to an amino acid change in conserved residues of the PI-3K domain. Truncating mutations included an insertion and a small deletion causing frameshift changes and creating stop codons, a nonsense mutation, and a substitution of the initial methionine (Table 2). In sample 1, 29 nucleotides from the intron 28 sequence were inserted at position 4182 of the mRNA, causing a frameshift in the open-reading frame predicting a protein truncation 17 amino acids after the insertion point. This insertion had been previously recognized in T-PLL.15 The inserted sequence corresponds to a fragment of the intron 28 flanked by potential acceptor and donor splicing sites.36 As in the previously described T-PLL sample, no alterations in the genomic sequence were found. Tumor 8 showed a C→T transition at codon 67 creating a stop codon at R23, exhibiting an additional downstream 4-bp deletion (Δ73-76) that caused the generation of a second stop codon. The lack of a normal allele in the REF analysis and sequencing plot and the 11q loss present in this tumor suggest that both mutations were in the same allele. Normal DNA from this patient could be obtained from peripheral blood granulocytes. The 67C→T transition was present in heterozygosity in the germline. However, the second mutation was only present in the tumor sample, indicating a somatic origin. The third truncating mutation in these tumors was observed in specimen 7. This MCL showed the nucleotide change 1A→T, which resulted in the substitution of the initial methionine codon preventing protein translation. All truncating mutations were associated with a total absence of protein expression on Western blot analysis (see below).
ATM studies in 20 MCL tumors
| Sample . | MCL variant . | Genetic analysis . | Gene product . | Protein expression . | CGH . | p53 and p14 ARF . | |||
|---|---|---|---|---|---|---|---|---|---|
| Polymorphisms . | Mutations . | Germline . | 11q . | N . | |||||
| 1 | T | — | 4182ins29 | NA | FS1349STOP | Absent | DEL | 6 | WT |
| 2 | T | — | 9023 GA | NA | R3008H | Low | DEL | 9 | p14 del |
| 3 | T | (1)(2)(4) | WT | — | WT | Absent | DEL | 8 | WT |
| 4 | T | — | WT | — | WT | NA | DEL | 8 | WT |
| 5 | T | (2)(4) | WT | — | WT | NA | DEL | 5 | WT |
| 6 | T | (4) | 8174 AT | NA | D2725V | NA | DEL | 8 | WT |
| 7 | B | (4) | 1 AT | NA | M(Start)1L | Absent | DEL | 8 | WT |
| 8 | B | (4) | 67 CT | Present | R23STOP | Absent | DEL | 9 | WT |
| Δ73-76 | NP | ||||||||
| 9 | B | (1)(3)(4) | WT | — | WT | Low | DEL | 9 | WT |
| 10 | T | (4) | WT | — | WT | Normal | WT | 4 | p53 mut |
| 11 | B | (4)(5) | WT | — | WT | Normal | WT | 20 | p53 mut |
| 12 | B | (4) | 8150 AT | NP | K2717M | Low | WT | 12 | p53 del |
| 13 | B | (4) | Δ1564-1565 | NP | FS564STOP† | Absent | WT | 9 | p53 del |
| 14 | T | (1)(4) | WT | — | WT | Normal | WT | 4 | WT |
| 15 | T | (1) | WT | — | WT | Normal | WT | 3 | WT |
| 16 | T | (1)(4) | WT | — | WT | Normal | WT | 3 | WT |
| 17 | T | (1)(4)(6) | WT | — | WT | Normal | WT | 3 | WT |
| 18 | T | — | WT* | — | WT | Normal | WT | 1 | WT |
| 19 | B | — | WT* | — | WT | Normal | WT | 7 | WT |
| 20 | B | — | WT* | — | WT | Normal | WT | 4 | WT |
| Sample . | MCL variant . | Genetic analysis . | Gene product . | Protein expression . | CGH . | p53 and p14 ARF . | |||
|---|---|---|---|---|---|---|---|---|---|
| Polymorphisms . | Mutations . | Germline . | 11q . | N . | |||||
| 1 | T | — | 4182ins29 | NA | FS1349STOP | Absent | DEL | 6 | WT |
| 2 | T | — | 9023 GA | NA | R3008H | Low | DEL | 9 | p14 del |
| 3 | T | (1)(2)(4) | WT | — | WT | Absent | DEL | 8 | WT |
| 4 | T | — | WT | — | WT | NA | DEL | 8 | WT |
| 5 | T | (2)(4) | WT | — | WT | NA | DEL | 5 | WT |
| 6 | T | (4) | 8174 AT | NA | D2725V | NA | DEL | 8 | WT |
| 7 | B | (4) | 1 AT | NA | M(Start)1L | Absent | DEL | 8 | WT |
| 8 | B | (4) | 67 CT | Present | R23STOP | Absent | DEL | 9 | WT |
| Δ73-76 | NP | ||||||||
| 9 | B | (1)(3)(4) | WT | — | WT | Low | DEL | 9 | WT |
| 10 | T | (4) | WT | — | WT | Normal | WT | 4 | p53 mut |
| 11 | B | (4)(5) | WT | — | WT | Normal | WT | 20 | p53 mut |
| 12 | B | (4) | 8150 AT | NP | K2717M | Low | WT | 12 | p53 del |
| 13 | B | (4) | Δ1564-1565 | NP | FS564STOP† | Absent | WT | 9 | p53 del |
| 14 | T | (1)(4) | WT | — | WT | Normal | WT | 4 | WT |
| 15 | T | (1) | WT | — | WT | Normal | WT | 3 | WT |
| 16 | T | (1)(4) | WT | — | WT | Normal | WT | 3 | WT |
| 17 | T | (1)(4)(6) | WT | — | WT | Normal | WT | 3 | WT |
| 18 | T | — | WT* | — | WT | Normal | WT | 1 | WT |
| 19 | B | — | WT* | — | WT | Normal | WT | 7 | WT |
| 20 | B | — | WT* | — | WT | Normal | WT | 4 | WT |
Only the kinase domain was analyzed by direct sequencing.
Absence of the normal wild-type allele in the sequencing analysis, suggesting a microdeletion.
Polymorphisms appear in parentheses as follows: (1) R2486R; (2) D1853N; (3) Q476Q; (4) N750K; (5) C532Y; (6) P1054R. 11q indicates 11q status by CGH; N, number of chromosomal imbalances by CGH; T, typical; B, blastoid; WT, wild-type; NA, not available; and NP, not present.
In the 8 MCL specimens without 11q deletions by CGH, ATMgene mutations were detected in 2. Sample 12 showed a nucleotide change (8150 A→T) leading to an amino acid substitution located in a conserved residue of the PI-3K domain. Sample 13 exhibited a 2-bp deletion at nucleotide 1563 that caused a frameshift change and truncation of the protein at position 1604. A signal of the normal allele was not observed in the REF gel and sequencing plot, suggesting that this tumor may have a microdeletion of the paired allele that was not detected in the CGH analysis (Figure1). Sequencing analysis of normal DNA in these 2 samples showed that the ATM gene mutations detected were not present in the germline, indicating that they were acquired during tumor development.
ATM sequencing analysis of REF-positive MCL.
Arrows point to nucleotide positions affected by mutations. (A) Missense mutation in sample 2, resulting in R3008H. (B) Missense mutation in sample 6, resulting in D2725V. (C) Substitution of the first methionine in sample 7, resulting in M1L. (D) Nonsense mutation (R23stop) and a downstream 4-bp deletion in sample 8 (left). Germline DNA sequencing of the same patient showing the absence of the deletion (center) and the presence of the R23stop mutation in heterozygosity (right). Brackets span over the deleted nucleotides. (E) Heterozygous missense mutation resulting in K2717M in sample 12 (left). The germline DNA of the same patient (right) shows a normal sequence demonstrating the somatic acquired origin of this mutation in the tumor. (F) Sample 13 showing a 2-bp deletion resulting in FS564stop with no evidence of the normal allele (left). Germline DNA sequencing of the same patient (right), demonstrating the somatic acquired origin of this deletion in the tumor sample. Brackets span the deleted nucleotides.
ATM sequencing analysis of REF-positive MCL.
Arrows point to nucleotide positions affected by mutations. (A) Missense mutation in sample 2, resulting in R3008H. (B) Missense mutation in sample 6, resulting in D2725V. (C) Substitution of the first methionine in sample 7, resulting in M1L. (D) Nonsense mutation (R23stop) and a downstream 4-bp deletion in sample 8 (left). Germline DNA sequencing of the same patient showing the absence of the deletion (center) and the presence of the R23stop mutation in heterozygosity (right). Brackets span over the deleted nucleotides. (E) Heterozygous missense mutation resulting in K2717M in sample 12 (left). The germline DNA of the same patient (right) shows a normal sequence demonstrating the somatic acquired origin of this mutation in the tumor. (F) Sample 13 showing a 2-bp deletion resulting in FS564stop with no evidence of the normal allele (left). Germline DNA sequencing of the same patient (right), demonstrating the somatic acquired origin of this deletion in the tumor sample. Brackets span the deleted nucleotides.
Because all ATM gene aberrations in the previous 17 tumors were truncating mutations that led to lack of protein expression or nucleotide substitutions in the PI-3K domain, we expanded the protein analysis and the mutational study of the PI-3K domain in 3 additional tumors with no 11q deletions. Western blot analysis showed normal levels of ATM protein expression in all tumors (see below), suggesting that they did not have truncating mutations (Table 2). In addition, no mutations were identified in the PI-3K domain.
Polymorphic variants
In the mutational analysis of this series of MCL, various polymorphic changes were also identified (Table 2). The D3003N/Q3031Q polymorphism37 and the change A554T, not previously recognized as an ATM polymorphic variant, were detected in all tumor specimens, normal DNA from the same patients, and 10 additional DNA samples from Spanish blood donors. These findings indicate that the changes may represent the wild-type ATM allele in our geographic area. Because these changes were present in all patients, they are not included in Table 2. The polymorphic variant D1853N was present in this series at a lower prevalence (8%) than that previously recognized in the healthy population (16%).38 The 3161 C→G, P1054R nucleotide change found in one MCL specimen had previously been described as a polymorphic variant.39 Two nucleotide changes that did not cause amino acidic substitutions (1428 A→G, Q476Q and 7458 G→A, R2486R) were identified in 7 tumor specimens. Two additional nucleotide substitutions, N750K and C532Y, were identified in 13 (77%) and 1 (6%) tumors, respectively. Both changes were also identified in the respective normal DNA of the patients and in healthy blood donors with allele frequencies similar to those of tumor patients (0.9 and 0.05, respectively). Interestingly, the N750K variant had been recognized as a possible ATM mutation in one MCL tumor.20 However, no normal DNA from the same patient could be examined. The findings in our study indicate that this change is a polymorphic variant frequently found in the healthy population.
Protein expression analysis
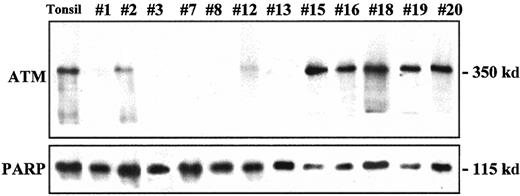
ATM protein expression was examined in 17 tumors. Five showed a complete absence of ATM protein (Figure2). Four of these lymphomas (specimens 1, 7, 8, 13) had truncating mutations and also showed concordant absence of the paired allele in the CGH analysis (specimens 1, 7, 8) and sequencing studies (specimens 1, 7, 8, 13). No gene mutations were detected in an additional tumor (specimen 3) with no protein expression in spite of sequencing the full coding region. Three tumors showed relatively low levels of protein expression. Two of these had an 11q deletion by CGH (specimens 2 and 9). The third tumor (specimen 12) had a missense mutation in the PI-3K domain, but the CGH analysis did not show an 11q loss. Low levels of ATM protein in this tumor suggested the presence of an additional truncating mutation or microdeletion in the paired allele that was not detected in the mutational or CGH studies. Relative normal levels of protein expression were observed in the remaining specimens (Table 2).
ATM protein expression analysis in MCL.
Relatively high levels of ATM protein expression were detected in samples 15, 16, 18, 19, and 20, similar to those observed in normal tonsil. Partial loss (samples 2, 12) or complete absence (samples 1, 3, 7, 8, 13) of protein expression was detected in several tumors. Expression of PARP in the same samples used as a loading control is shown in the lower panel.
ATM protein expression analysis in MCL.
Relatively high levels of ATM protein expression were detected in samples 15, 16, 18, 19, and 20, similar to those observed in normal tonsil. Partial loss (samples 2, 12) or complete absence (samples 1, 3, 7, 8, 13) of protein expression was detected in several tumors. Expression of PARP in the same samples used as a loading control is shown in the lower panel.
ATM inactivation and clinicopathologic features
ATM gene and ATM protein alterations were detected at a relatively similar incidence in typical (4 of 12, 33%) and blastoid (4 of 8, 50%) MCL variants. In addition, p53 andp14ARF gene alterations had been previously analyzed in this series.29 Four showed p53 gene alterations, including 2 mutations and 2 homozygous deletions. One showed a homozygous deletion of the INK4a/ARF locus. Tumors with homozygous deletions of the p53 gene and theINK4/ARF locus also had mutations of the ATMgene. However, no ATM alterations were detected in the 2 additional tumors with p53 mutations (Table 2).
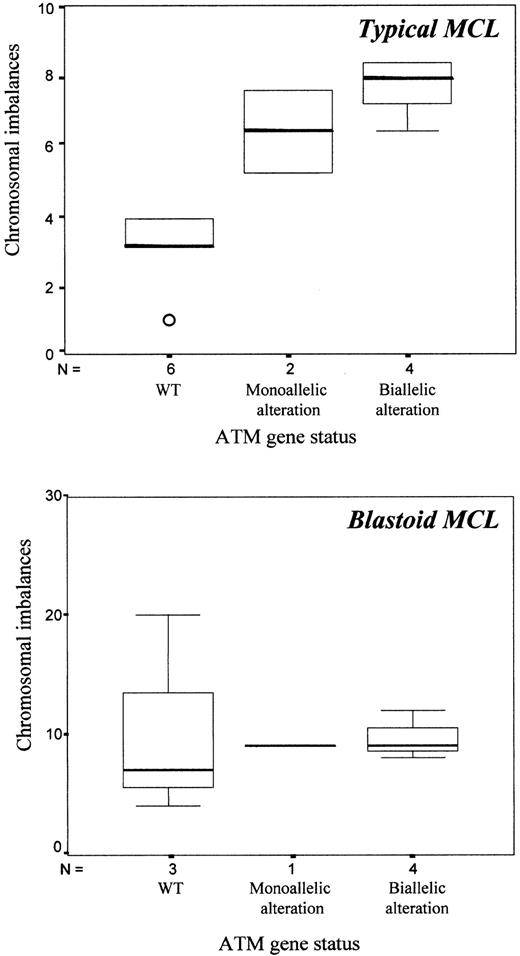
To determine the possible relation between ATM gene alterations and number of chromosomal imbalances, tumors were grouped according to the status of the ATM gene. Thus, MCL with no gene mutations or allelic losses and normal protein expression were considered tumors with wild-type ATM (samples 10, 11, 14-20). Tumors with biallelic alterations of the ATM gene included those with gene mutations associated with losses of the paired allele (samples 1, 2, 6-8, 12, 13). Sample 3, showing complete absence of protein expression on Western blot analysis, was also included in this group. Finally, tumors in which no gene mutations were detected on REF analysis but in which the CGH study showed 11q losses were considered to have monoallelic alterations (samples 4, 5, 9). Typical MCL with the wild-type ATM gene had significantly lower numbers of chromosomal imbalances (mean, 3; SD, 1.1) than tumors with inactivation of both alleles (mean, 7.8; SD, 1.3) (P = .001). The 2 typical MCL tumors with monoallelic ATM alterations had an intermediate number of CGH alterations (mean, 6.5) (Figure3). However, all blastoid MCL had a high number of chromosomal imbalances with no differences between samples with wild-type ATM (mean, 10.4; SD, 8.5), monoallelic (n = 9), or biallelic ATM alterations (mean, 9.5; SD, 1.7) (Figure 3).
ATM alterations and chromosomal imbalances in typical and blastoid MCL.
Chromosomal imbalances were significantly higher in typical MCL with ATM alterations than in samples with wild-type ATM (P = .001) (top). No differences were observed in blastoid variants (bottom).
ATM alterations and chromosomal imbalances in typical and blastoid MCL.
Chromosomal imbalances were significantly higher in typical MCL with ATM alterations than in samples with wild-type ATM (P = .001) (top). No differences were observed in blastoid variants (bottom).
Additionally, biallelic ATM alterations were associated with the presence of the +3q chromosomal abnormality (biallelic altered, 87% vs wild-type 22%; P = .015). Moreover, a statistical trend was observed with the +12q abnormality (biallelic altered, 60% vs wild-type, 0%; P = .082). Biallelic ATM inactivation was also associated with the presence of extranodal involvement (2 or more sites affected) (altered, 62% vs wild-type, 11%;P = .049). The same significant variables were obtained when the 3 tumors with monoallelic ATM alteration were included in the group of tumors with biallelic ATM alteration.
Median survival time in this series was 48 months (95% CI, 40-56 months). Median survival time when tumors had typical histology was 70 months, whereas it was 24 months for the blastoid variants. No statistical differences in survival were observed when grouped according to the ATM gene status.
Discussion
In this study, we have analyzed ATM gene alterations and ATM protein expression in a series of 12 typical and 8 blastoid variants of MCL. All these tumors had been previously characterized for the status of the p53 and ARFgenes.29 Chromosomal imbalances had been examined by CGH.9 ATM alterations were detected in 8 (40%) tumors, including 6 of 9 (67%) lymphomas with 11q losses and 2 of 11 (18%) tumors in which the CGH analysis showed a normal chromosome 11 profile. However, protein analysis and sequencing plots suggested that the paired allele in these 2 tumors was also inactivated. ATM alterations were slightly more frequent in blastoid (50%) than in typical (33%) variants, but the differences were not statistically significant. These findings indicate that ATM inactivation is a frequent phenomenon in MCL. The number of these alterations in typical variants suggests that they may be a relatively initial event in the development of these tumors. ATM gene and ATM protein alterations have been previously recognized in other lymphoproliferative disorders, including 46% to 67% T-PLL and 19% to 34% B-CLL.15-19,40 More recently, Schaffner et al20 have identified ATMmutations in 7 (100%) MCL with 11q22-23 deletions and in 2 of 5 (40%) MCL with no chromosome 11q losses. Although the number of ATM alterations in our study was relatively lower, these observations together confirm the role of ATM inactivation in the pathogenesis of MCL.
Most ATM mutations in T-PLL are missense mutations clustering in the PI-3K domain or are truncating mutations leading to the absence or early termination of the protein.15-17 In contrast, ATM mutations in B-CLL are mainly missense changes outside the kinase domain.18,19,40 In our study, 3 missense mutations were detected in conserved positions of the PI-3K domain, and 4 additional changes led to absence or early truncation of the protein. One additional tumor had a complete absence of protein expression, but no structural gene alterations were found. In the previous study on MCL, the mutations detected were mainly truncating changes and one missense substitution in the PI-3K domain. Two additional missense changes (N750K and E2423G) were located in other regions of the gene. However, the possible polymorphic significance of these nucleotide substitutions could not be assessed.20 In our study, we found that one of these changes (N750K) was present in 77% of the tumors, in normal DNA of the patients, and in a healthy population with a high allele frequency (0.9), indicating that this nucleotide substitution is a relatively common polymorphism. These observations confirmed that ATM inactivation in MCL follows a pattern similar to that for T-PLL, with truncating mutations and changes clustering in the kinase domain. Our observations differ from the findings in B-CLL, in which most mutations are missense substitutions distributed in different areas of the gene.
All mutations detected in our study predicted a potential inactivation of the protein function. Two of the 3 missense mutations in the PI-3K domain occurred at R3008 and D2725, 2 conserved amino acid positions that have mutated in B-CLL, T-PLL, and one MCL, suggesting that they may be crucial spots in protein inactivation.17,20,40 The third mutation, K2717M, has not previously been recognized but also involves a highly conserved amino acid in different members of the PI-3K family.17 The remaining ATM mutations found in our series were frameshift or nonsense mutations leading to absent proteins or truncating proteins. Two of these mutations have been initially recognized in patients with T-PLL (sample 1) and AT (sample 7).15,41 In this latter, however, the nucleotide substitution was identified in the first position whereas in the former it was in the second nucleotide.41 Sample 8 showed 2 simultaneous truncating mutations, apparently in the same allele. Interestingly, multiple structural lesions in one allele of theATM gene have also been observed in several patients with T-PLL.17 The significance of these multiple changes in theATM gene is unclear.
In this MCL series, ATM protein expression was concordant with the status of the gene and the presence of allelic losses in virtually all samples. Thus, tumors with truncating mutations and 11q loss by CGH showed a complete absence of protein expression, suggesting that these mutations prevented translation or generated unstable products. Samples with allelic losses associated with missense mutations or wild-type genes in the paired allele expressed low levels of protein. Normal protein signal was observed in samples with no apparent alterations of the gene. Only 2 samples (samples 3 and 12) showed discordance between protein expression and gene status. The ATM protein in these 2 samples was absent and low, respectively. However, sample 3 showed an 11q loss, but no mutations were detected in the remaining allele. Similarly, sample 12 with no 11q loss showed a missense mutation only in one allele. REF analysis is considered of high sensitivity for the detection of gene mutations,31 but it is possible that occasional truncating alterations or microdeletions were unidentified in these tumors.
Germline ATM mutations have been detected in patients with B-CLL,18,37 suggesting a possible role of ATM in the genetic predisposition of this disorder. In the current series, 2 tumor mutations were not present in the normal DNA of the patients (samples 12 and 13), indicating their somatic origin. Interestingly, in sample 8, 1 of the 2 simultaneous mutations (R23stop) was carried in heterozygosity in the germline of the patient. However, the tumor sample of this patient had lost the wild-type allele and had acquired a second 4-bp deletion downstream of the constitutional mutation. Germline heterozygous ATM gene mutations have been estimated to be present in approximately 0.5% to 5% of the general population.42 43 Whether these mutations may cause significant genetic predisposition to the development of malignancies is still debatable.
One of the targets of the ATM protein function is p53.26,28,44 Inactivation of p53 is a frequent phenomenon in aggressive MCL and other lymphoproliferative disorders. However, the possible relation between the genetic alterations of these 2 genes in human tumors is unknown. In T-PLL, in which ATM inactivation is relatively common, no p53 mutations have been detected, suggesting that ATM and p53 could have an alternative pathogenetic role.17 However, in the current study, p53mutations were found either in tumors with ATM inactivation or in tumors with wild-type ATM, suggesting that these 2 alterations may act independently in the development of MCL.
The ATM gene plays a central role in the cellular response to DNA damage. Human and murine cells deficient in the ATMgene present an increasing number of chromosomal abnormalities and genetic instability.14 A possible association betweenATM gene alterations and chromosomal aberrations in human tumors has not been previously explored. In this study, we have observed a significantly higher number of chromosomal imbalances in typical MCL with ATM gene alterations than in tumors with wild-type ATM, suggesting that ATM inactivation may favor increasing chromosomal instability in these lymphomas. The fact that blastoid variants have a high number of chromosomal imbalances independent ofATM gene status suggests that other genes may be involved in the pathogenesis of these variants and in their chromosomal instability.
In conclusion, our findings indicate that the ATM gene is frequently inactivated in MCL. The relatively similar number of alterations in typical and blastoid variants suggests that ATM may be involved in early steps of tumor development. ATM gene mutations are mainly truncating or, alternatively, missense mutations in conserved residues of the PI-3K domain, and they seem independent of p53 alterations. The significant association between ATM inactivation and higher number of chromosomal imbalances in typical MCL suggests a possible role of these aberrations in increasing chromosomal instability in these tumors.
We thank Iracema Nayach and Olga Luna for excellent technical assistance. Sequencing analysis was performed using the Serveis Cientı́fico-Tècnics of the University of Barcelona.
Supported by the Comision Interministerial de Ciencia y Tecnologia (CICYT) SAF 99/20, European Commission contract QLRT-1999-30687, FEDER 1FD97-1678, and CIRIT, Generalitat de Catalunya 2000SGR118.
E.C. and L.H. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Elias Campo, Laboratory of Pathology, Hospital Clinic, Villarroel 170, 08036 Barcelona, Spain; e-mail:campo@medicina.ub.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal