Acquired mutations of the PIG-A gene result in the hemolysis characteristic of paroxysmal nocturnal hemoglobinuria (PNH). Although the etiology of the mutation(s) is unclear, mutable conditions have been suggested by the coexistence of multiple clones with different mutations of PIG-A and by the appearance of leukemic clones in patients with PNH. This study sought to test this hypothesis by examining the frequency of hypoxanthine-guanine phosphoribosyl transferase (HPRT)gene mutations, identified by both resistance to 6-thioguanine (6-TG) and gene analysis. T-cell colonies resistant to 6-TG formed in methylcellulose culture were found in 8 (67%) of 12 PNH patients and 3 (18%) of 17 age-matched healthy volunteers (P < .02, Fisher exact probability test). The incidence of resistant colonies ranged from 40 to 367 (mean 149, × 10−7) in the 8 patients and from 1 to 16 (mean 7, × 10−7) in the 3 healthy donors. Thus, theHRPT gene mutated more frequently in patients with PNH than in healthy controls (P < .02, Mann-Whitney test). Analysis of bone marrow cells supported these findings. Like the PIG-A mutations in PNH, the HPRT mutations were widely distributed in the coding regions and consisted primarily of base deletions. Unlike PNH cells, 6-TG–resistant cells expressed CD59, indicating that the HPRT mutations did not occur in PNH clones. No correlation was noted between HPRT mutation frequency and content of therapy received by the patients. It is concluded that in PNH patients, conditions exist that favor the occurrence of diverse somatic mutations in blood cells.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired stem cell disorder characterized by intravascular hemolysis, venous thrombosis, frequent episodes of infection, intractable bone marrow (BM) failure, and rarely, the development of leukemia.1,2 Among clinical manifestations, only the molecular events leading to hemolysis have been clarified.3-5 PNH cells do not produce glycosylphosphatidylinositol (GPI); they lack a series of GPI-linked membrane proteins, including complement-regulatory molecules such as decay-accelerating factor (DAF) and CD59, and are eventually susceptible to complement. Therefore, affected erythrocytes undergo the complement-mediated intravascular hemolysis characteristic of PNH. The membrane defects are easily detected by flow cytometry and have been used to diagnose PNH.6-8 The gene responsible for these membrane defects, PIG-A, has been cloned and shown to act in the synthesis of N-acetylglucosaminyl phosphatidylinositol at the first stage of GPI biosynthesis.9,10 Inactivating mutations ofPIG-A have been detected in all reported PNH patients from Asia, Europe, and North America.10 This uniformity of the responsible gene in PNH is attributed to the localization ofPIG-A, mapped to the X chromosome at p22.1.11 APIG-A mutation in one allele suffices to cause a GPI deficiency because of X chromosome inactivation, whereas all other autosomal GPI synthesis genes require mutations in both alleles to manifest a defect.
Of major interest is the etiology of the mutations ofPIG-A in patients with PNH.12-15 There are at least 2 possibilities: one is spontaneous mutation at the rate found in healthy individuals. Indeed, PIG-A mutations were detected in non-PNH patients with lymphoma who had undergone therapy with antibody to a GPI-linked membrane protein on lymphocytes16as well as, albeit at extremely low levels, in healthy individuals.17,18 Another possibility is that there are mutable conditions that facilitate somatic mutations. Molecular analysis showed that in PNH, PIG-A mutations occurred randomly without mutation hot spots.10,12PIG-Amay be genetically unstable. This hypothesis is supported by the presence of multiple clones with different PIG-A mutations in the same PNH patient.19-21 The erythrocyte glycophorin A locus has also been reported as another case of increased mutations in PNH.13 Furthermore, PNH often gives rise to other abnormal clones of myelodysplastic syndromes (MDSs), and some rare patients develop leukemia.22-24 Thus, it is possible that mutations are not limited to specific genes or clones; rather, they may be random events. Based on these considerations, we hypothesized that PNH patients have a hematopoietic environment that allows the emergence of abnormal clones with distinct somatic mutations.
Acquired somatic mutations can be measured on the basis of mutation frequency at the X-linked single-gene locus encoding the purine salvage enzyme hypoxanthine-guanine phosphoribosyl transferase(HPRT).25-32 The survival of cells in culture with 6-thioguanine (6-TG) is a sensitive and specific indicator of the acquisition of HPRT gene mutations by these cells. In the present study, we attempted to identify the mutable conditions by quantitating the mutation frequency of HPRT and by characterizing the nature of HPRT mutations in peripheral blood T lymphocytes from patients with PNH.31,33 34 This analysis could lead to a better understanding of the etiology ofPIG-A mutations, the mechanism(s) underlying a selective expansion of PNH clones leading to the clinical manifestations characteristic of PNH, and the progression to leukemia in some PNH patients.
Patients, materials, and methods
Clinical profiles and laboratory data of the 12 PNH patients are shown in Table 1. Of these, 8 patients had de novo PNH and 4 had aplastic anemia (AA)–PNH syndrome. There were 6 females and 6 males; their ages ranged from 22 to 77 years. PNH was diagnosed on the basis of acquired intravascular hemolysis negative for Coombs test, detection of complement-sensitive erythrocytes by Ham acidified serum test, and detection by flow cytometry of blood cells negative for GPI-linked membrane proteins such as DAF and CD59.35 None of the patients manifested elevated serum uric acid levels or signs or symptoms seen in hereditaryHPRT gene abnormalities. The controls were 17 age-matched healthy volunteers. There were 4 light cigarette smokers: one in the PNH group and 3 in the control group. In addition, a 21-year-old male with ataxia telangiectasia (AT), known for a genetic instability, was also enrolled.36
Patient profiles
| Patient no., age/sex . | Diagnosis . | Duration* . | Leukocytes (109/L) . | Hemoglobin (g/dL) . | Therapy . | T cells % CD59− . | BM cells† % DAF−/CD59− . |
|---|---|---|---|---|---|---|---|
| 1, 24/M | PNH | 7 | 3.5 | 7.4 | Prednisolone | 17 | ND |
| 2, 31/M | PNH | 10 | 3.8 | 8.0 | Cyclosporine, G-CSF | 11 | 35 |
| 3, 52/F | PNH | 7 | 3.0 | 7.5 | None | 17 | ND |
| 4, 61/M | PNH | 7 | 3.5 | 11.7 | None | 6 | ND |
| 5, 63/M | PNH | 14 | 6.0 | 6.1 | Prednisolone Methenolone acetate | 5 | ND |
| 6, 66/F | PNH | 19 | 2.9 | 11.9 | Methenolone acetate | 36 | ND |
| 7, 75/F | PNH | 3 | 5.0 | 10.1 | Prednisolone | 6 | 53 |
| 8, 77/M | PNH | 6 | 4.9 | 13.3 | Prednisolone Methenolone acetate | 1 | ND |
| 9, 22/F | AA-PNH | 4 | 7.2 | 12.6 | Cyclosporine, G-CSF | 5 | 23 |
| 10, 59/M | AA-PNH | 5 | 3.5 | 14.0 | Methenolone acetate | 1 | ND |
| 11, 62/F | AA-PNH | 0.2 | 3.2 | 6.8 | Cyclosporine, G-CSF | 1 | 6 |
| 12, 68/F | AA-PNH | 7 | 3.9 | 9.8 | Prednisolone | 2 | 67 |
| Patient no., age/sex . | Diagnosis . | Duration* . | Leukocytes (109/L) . | Hemoglobin (g/dL) . | Therapy . | T cells % CD59− . | BM cells† % DAF−/CD59− . |
|---|---|---|---|---|---|---|---|
| 1, 24/M | PNH | 7 | 3.5 | 7.4 | Prednisolone | 17 | ND |
| 2, 31/M | PNH | 10 | 3.8 | 8.0 | Cyclosporine, G-CSF | 11 | 35 |
| 3, 52/F | PNH | 7 | 3.0 | 7.5 | None | 17 | ND |
| 4, 61/M | PNH | 7 | 3.5 | 11.7 | None | 6 | ND |
| 5, 63/M | PNH | 14 | 6.0 | 6.1 | Prednisolone Methenolone acetate | 5 | ND |
| 6, 66/F | PNH | 19 | 2.9 | 11.9 | Methenolone acetate | 36 | ND |
| 7, 75/F | PNH | 3 | 5.0 | 10.1 | Prednisolone | 6 | 53 |
| 8, 77/M | PNH | 6 | 4.9 | 13.3 | Prednisolone Methenolone acetate | 1 | ND |
| 9, 22/F | AA-PNH | 4 | 7.2 | 12.6 | Cyclosporine, G-CSF | 5 | 23 |
| 10, 59/M | AA-PNH | 5 | 3.5 | 14.0 | Methenolone acetate | 1 | ND |
| 11, 62/F | AA-PNH | 0.2 | 3.2 | 6.8 | Cyclosporine, G-CSF | 1 | 6 |
| 12, 68/F | AA-PNH | 7 | 3.9 | 9.8 | Prednisolone | 2 | 67 |
ND indicates not determined.
Years from initial diagnosis.
Bone marrow mononuclear cells negative for both DAF and CD59.
In vitro colony assay
Lymphocytes of peripheral blood (patients, 20 mL; controls, 200 mL) were obtained with informed consent from all participants in this study, using Ficoll-Paque (Amersham Pharmacia Biotech AB, Uppsala, Sweden) by the methods described elsewhere.8 Aliquots of cells were plated with and without 6-TG. Briefly, in the absence of 6-TG, cells were plated in 4 culture dishes (35-mm dishes; Becton Dickinson, Franklin Lakes, NJ) at a concentration of (2 × 105 cells/1 mL semisolid culture medium per dish) and incubated to form colonies at 37°C under 5% CO2. The medium was RPMI 1640 supplemented with 0.9% methylcellulose (Fisher Scientific, Fair Lawn, NJ), 10% fetal calf serum (FCS; JRH Bioscience, Lenexa, KS), 200 ng/mL interleukin-2 (IL-2; Shionogi, Osaka, Japan), and 5 μg/mL phytohemagglutinin (PHA; Seikagaku, Tokyo, Japan). In parallel, all the rest of the cells were also cultured under the same conditions as described above except for the presence of 5 μg/mL 6-TG (Wako, Osaka, Japan) in order to detect 6-TG–resistant colonies, ie, to select colonies with HPRT mutations. After 14 days, the T-cell colonies were counted under a microscope. The incidence of 6-TG–resistant T-cell colonies was determined by the ratio of the number of colonies formed in the presence and absence of 6-TG. To characterize the colonies by flow cytometry and by HPRTmutation analysis, colonies were transferred to 96-well, round-bottom microtiter plates and expanded in RPMI 1640 medium containing 20% FCS, 200 ng/mL IL-2, and 5 μg/mL PHA for an additional 14 days at 37°C under 5% CO2. In addition to T cells, immature lymphocyte-depleted BM cells were obtained from 5 PNH patients and 4 healthy controls by methods described previously.37 Then, 2 × 105 BM cells were incubated in 1 mL semisolid culture medium per 35-mm dish at 37°C under 5% CO2. The medium was Iscoves modified Dulbecco medium containing 1.0% methylcellulose, 30% FCS, and a set of recombinant human cytokines such as 50 ng/mL stem cell factor, 10 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF), 10 ng/mL IL-3, and 3 U/mL erythropoietin (Methocult GF H4434, Stem Cell Technologies, Vancouver, Canada). After 14-day culture, erythroid burst-forming unit (BFU-E), granulocyte-macrophage colony-forming unit (CFU-GM), CFU-G, CFU-M, and CFU-GEMM (granulocyte-erythrocyte-megakaryocyte-macrophage) colonies were observed. As in the T-cell colony assay, 6-TG (5 μg/mL) was used to detect mutant colonies.
Flow cytometry
Peripheral blood granulocytes were isolated with dextran sulfate as described previously.38 Lymphocytes and granulocytes were analyzed by flow cytometry described previously.39 In brief, blood cells were incubated with a mouse monoclonal antibody (mAb) to DAF or CD59 and labeled with fluorescein isothiocyanate (FITC)–conjugated mAb to mouse IgG (Zymed, San Francisco, CA). Lymphocytes were further labeled with phycoerythrin (PE)–conjugated mAb to CD2 (Pharmingen, San Diego, CA). BM mononuclear cells were also incubated with anti-DAF mAb and labeled with PE-conjugated anti–mouse IgG (Dako, Glostrup, Denmark) and then with FITC-conjugated anti-CD59 mAb. Labeled cells were analyzed with a flow cytometry analyzer (Epics, Beckman Coulter, Fullerton, CA).
Mutation analysis of HPRT transcripts by reverse transcription–polymerase chain reaction
To confirm and identify the HPRT mutations that explain the 6-TG resistance, we determined the sequence of theHPRT mRNA of cells forming 6-TG–resistant colonies using reverse transcription–polymerase chain reaction (RT-PCR).31 In brief, total RNA was isolated using a kit with a silica-gel membrane (RNeasy, Qiagen, Hilden, Germany) and converted to cDNA with a random primer kit (ReverTra, Toyobo, Osaka, Japan) according to the manufacturer's instructions. The coding region of the cDNA was then amplified by PCR with 0.5 U recombinant Taq DNA polymerase (ExTaq, Takara, Tokyo, Japan), 200 μM deoxynucleoside triphosphate in a buffer (20 μL; 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin), and 10 pmol of a sense primer (5′-CCTCCTCTGCTCCGCCACCG-3′) and an antisense primer (5′-GACTCCAGATGTTTCCAAAC-3′). After initial denaturation at 94°C for 4 minutes, DNA amplification was performed with a PCR thermocycler (Astec, Fukuoka, Japan) for 30 cycles; each cycle consisted of denaturation at 94°C for 30 seconds, annealing at 58°C for 45 seconds, and extension at 72°C for 60 seconds. The products were further amplified by a nested PCR using a sense primer (5′-CCACCGGCTTCCTCCTCCTGA-3′) and an antisense primer (5′-GTTTCCAAACTCAACTTGAATTCTCATC-3′) under the same conditions as the first PCR. The PCR products were analyzed by 1.5% agarose gel electrophoresis and ethidium bromide staining. Nucleotide sequences of PCR products were determined by the automated fluorescence sequence method using a DNA sequencing kit containing rhodamine-labeled dye terminators and a DNA sequencer (ABI PRISM 377, Applied Biosystems, Foster City, CA).40
Statistical analysis
The Fisher exact probability test and Mann-Whitney test were used to determine significance levels. P values less than .05 were considered significant.
Results
Detection of 6-TG–resistant colonies
Figure 1 shows T-cell colony formation in semisolid methylcellulose cultures with and without 6-TG. In the absence of 6-TG, each plate yielded many colonies. The number of colonies per plate ranged from 1200 to 13 600 (mean 8000) in the 12 PNH patients and from 2500 to 19 100 (mean 10 300) in the 17 controls. Colony formation was less in PNH than control (Mann-Whitney test, P < .05). In contrast, colony formation was extremely poor in the presence of 6-TG. Resistant colonies were found in 8 (67%) of 12 PNH patients and in only 3 (18%) of 17 healthy controls (Fisher exact probability test, P < .02, Table2). In the 11 individuals positive for 6-TG–resistant colonies, the ratio of T-cell colonies per plate in the presence (b in Table 2) and absence (a in Table 2) of 6-TG was considered to be an indicator of the incidence of resistant T-cell clones. According to this assessment, the ratio ranged from 40 to 367 (mean 149, × 10−7) in the 8 PNH patients and from 1 to 16 (mean 7, × 10-7) in the 3 healthy donors. The ratio was 43 (× 10−7) in the patient with AT, who had an established genetic instability. The ratio given by our assay is low, judging from the report of Purow et al.14 The discrepancy may reflect the difference in the methods used for HPRTmutation analysis. In any event, the incidence of 6-TG–resistant T-cell clones was markedly higher in the 8 patients with PNH (21-fold increase) than in the 3 controls (Mann-Whitney test,P < .02). No close correlation was found between the incidence of 6-TG–resistant T cells and factors such as content of therapy and duration of suffering (years from initial diagnosis of PNH) listed in Table 1. Next, we examined the colonies formed by BM hematopoietic progenitor cells obtained from 5 of the 8 PNH patients who had 6-TG–resistant T-cell colonies and from 4 healthy volunteers. The major colonies were primarily BFU-E and CFU-GM. In the absence of 6-TG, the number of BM colonies per plate ranged from 31 to 262 (mean 94) in the PNH patients and from 423 to 1130 (mean 709) in the healthy controls. Poor colony formation in PNH has been reported and explained by a decrease in BM progenitor cells and their poor response to hematopoietic cytokines.17 Despite the presence of fewer colonies in PNH patients than in the controls in the absence of 6-TG, 6-TG–resistant colonies were detected in 2 of the 5 PNH patients but in none of the 4 healthy controls. In those 2 patients (patients 9 and 12), the incidence of 6-TG–resistant BM colonies was 496 and 1493 (× 10−6), respectively. In PNH patients, the frequency of 6-TG–resistant colonies was markedly higher in the BM cells than the peripheral T cells.
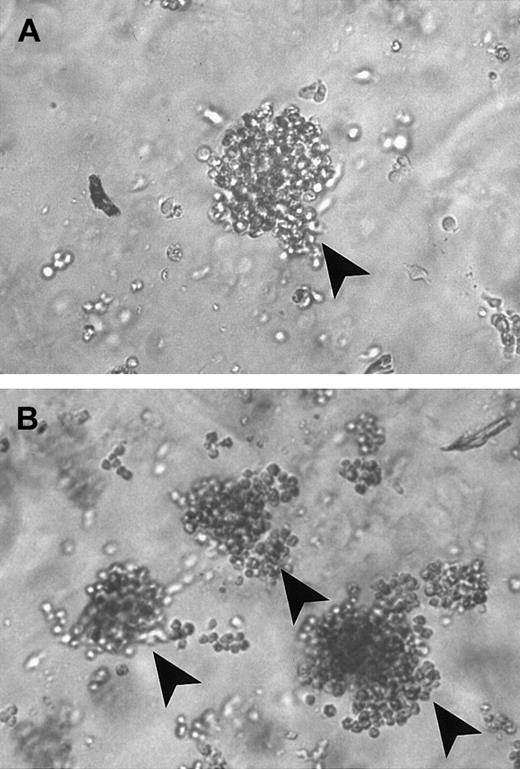
T-cell colonies of representative case of PNH.
(A) In the presence of 6-TG. (B) In the absence of 6-TG. Arrows indicate individual colonies.
T-cell colonies of representative case of PNH.
(A) In the presence of 6-TG. (B) In the absence of 6-TG. Arrows indicate individual colonies.
Incidence of 6-TG–resistant T-cell colonies
| . | In the absence of 6-TG . | In the presence of 6-TG . | (b)/(a)* (/107) . | ||||
|---|---|---|---|---|---|---|---|
| Plates . | Colonies (±SD) × 102 . | Colonies per plate (a) × 102 . | Plates . | Colonies (±SD) . | Colonies per plate (b) . | ||
| Patients | |||||||
| 2† | 4 | 200 ± 120 | 50 | 15.3 | 0.3 ± 0.5 | 0.020 | 40 |
| 3 | 4 | 185 ± 81 | 46 | 10.7 | 0.3 ± 0.5 | 0.028 | 61 |
| 4 | 4 | 347 ± 140 | 87 | 5.3 | 0.3 ± 1.0 | 0.057 | 66 |
| 6 | 4 | 47 ± 17 | 12 | 16.0 | 0.7 ± 1.0 | 0.044 | 367 |
| 7 | 4 | 298 ± 116 | 75 | 18.3 | 1.7 ± 1.4 | 0.093 | 124 |
| 9 | 4 | 436 ± 180 | 109 | 16.8 | 2.0 ± 1.7 | 0.119 | 109 |
| 11 | 4 | 451 | 113 | 16 | 1 | 0.063 | 56 |
| 12 | 4 | 165 ± 95 | 41 | 13.3 | 2.0 ± 1.0 | 0.150 | 366 |
| AT | 4 | 362 | 90 | 76 | 3 | 0.039 | 43 |
| Healthy donors‡ | |||||||
| 1 | 4 | 584 ± 201 | 146 | 133.7 | 0.3 ± 0.6 | 0.002 | 1 |
| 2† | 4 | 375 ± 63 | 94 | 66.0 | 1.0 ± 0.6 | 0.015 | 16 |
| 3 | 4 | 459 ± 50 | 114 | 106.7 | 0.3 ± 0.6 | 0.003 | 3 |
| . | In the absence of 6-TG . | In the presence of 6-TG . | (b)/(a)* (/107) . | ||||
|---|---|---|---|---|---|---|---|
| Plates . | Colonies (±SD) × 102 . | Colonies per plate (a) × 102 . | Plates . | Colonies (±SD) . | Colonies per plate (b) . | ||
| Patients | |||||||
| 2† | 4 | 200 ± 120 | 50 | 15.3 | 0.3 ± 0.5 | 0.020 | 40 |
| 3 | 4 | 185 ± 81 | 46 | 10.7 | 0.3 ± 0.5 | 0.028 | 61 |
| 4 | 4 | 347 ± 140 | 87 | 5.3 | 0.3 ± 1.0 | 0.057 | 66 |
| 6 | 4 | 47 ± 17 | 12 | 16.0 | 0.7 ± 1.0 | 0.044 | 367 |
| 7 | 4 | 298 ± 116 | 75 | 18.3 | 1.7 ± 1.4 | 0.093 | 124 |
| 9 | 4 | 436 ± 180 | 109 | 16.8 | 2.0 ± 1.7 | 0.119 | 109 |
| 11 | 4 | 451 | 113 | 16 | 1 | 0.063 | 56 |
| 12 | 4 | 165 ± 95 | 41 | 13.3 | 2.0 ± 1.0 | 0.150 | 366 |
| AT | 4 | 362 | 90 | 76 | 3 | 0.039 | 43 |
| Healthy donors‡ | |||||||
| 1 | 4 | 584 ± 201 | 146 | 133.7 | 0.3 ± 0.6 | 0.002 | 1 |
| 2† | 4 | 375 ± 63 | 94 | 66.0 | 1.0 ± 0.6 | 0.015 | 16 |
| 3 | 4 | 459 ± 50 | 114 | 106.7 | 0.3 ± 0.6 | 0.003 | 3 |
Each value represents the mean of 3 independent experiments, except the patients with PNH (no. 11) and AT, who were analyzed once.
Pairwise difference between patients with PNH and healthy donors was significant (P < .02, Mann-Whitney test32).
Cigarette smokers.
1, 26-year-old female; 2 and 3, 40- and 69-year-old males.
Mutations of the HPRT gene in 6-TG–resistant colonies
The resistance of cells to 6-TG is an indicator of HPRTmutations. To verify the mutations and to identify the nature of the mutations in our assays, we determined the sequence ofHPRT cDNA of the cells forming colonies in the presence of 6-TG (Table 3). All of the clones manifested distinct mutations in the HPRT gene, confirming the 6-TG resistance and the clonality of each colony. In PNH patients, the mutations occurred at various sites: exons 2, 3, 6, 7, and 8. The types of cDNA changes were also heterogeneous: a single-base change, deletions of a whole exon, and the deletion and insertion of a large number of bases. In a single clone of patient 7, whose 2 mutant clones were analyzed, both a deletion and an insertion coexisted. The predicted size of this peptide indicated truncated form (Table 3), supporting the hypothesis that 6-TG resistance is due to functional impairment of the HPRT enzyme. Analysis of a BM-cell colony of patient 12 showed consistent results: the deletion of 202 bases in exon 3, resulting in a small peptide of 150 amino acids. The patient with AT showed a deletion of 66 bases in exon 4, resulting in a peptide of 196 amino acids, smaller than the normal peptide of 218 amino acids. In contrast to various deletions found in patients with PNH and the AT patient, 2 single-base substitutions were noted in healthy donors 2 and 3 (Table 3). In healthy donor 3, a base change in exon 6 induced premature translation termination, resulting in a truncated form of 151 amino acids. A base change in healthy donor 2 involved the same amino acid as in PNH patient 7. The single-base substitutions, GC-to-AT transitions, and mutation at base 197 detected in our healthy controls (Table 3) have frequently been demonstrated in spontaneous HPRT mutations in vivo.27,29 We detected no mutations in HPRT cDNA prepared from T-cell and BM-cell colonies formed in the absence of 6-TG in the individuals listed in Table 3. Figure 2 shows a schematic presentation of the HPRT mutations detected by the cDNA analysis. Molecular analysis revealed that HPRTmutations spread to virtually all exons, indicating the absence of mutation hot spots.31 The coexistence of multiple clones with different mutations of the HPRT gene in a single patient (patient 7) is similar to the phenomenon observed inPIG-A mutations.19-21 Patient 9 also had 3 different HPRT mutant clones when analyzed by RT-PCR (data not shown).
HPRT mutations in 6-TG–resistant colonies
| . | Site . | cDNA change . | Predicted size of protein3-150 . |
|---|---|---|---|
| Patients with PNH | |||
| 2 | Exon 8 | Deletion of whole exon | 182 amino acids |
| 7 | Exon 3 | Base change (198T>G) | 218 amino acids (Cys66Trp) |
| Exons 2 and 6 | 49-base insertion (exon 2) 62-base deletion (exon 6) | 26 amino acids | |
| 9 | Exon 7 | Deletion of whole exon | 165 amino acids |
| 123-151 | Exon 3 | 202-base deletion | 150 amino acids |
| Patient with AT | Exon 4 | 66-base deletion | 196 amino acids |
| Healthy donors | |||
| 2 | Exon 3 | Base change (197G>A) | 218 amino acids (Cys66Tyr) |
| 3 | Exon 6 | Base change (444C>T) | 151 amino acids (Gln152X) |
| . | Site . | cDNA change . | Predicted size of protein3-150 . |
|---|---|---|---|
| Patients with PNH | |||
| 2 | Exon 8 | Deletion of whole exon | 182 amino acids |
| 7 | Exon 3 | Base change (198T>G) | 218 amino acids (Cys66Trp) |
| Exons 2 and 6 | 49-base insertion (exon 2) 62-base deletion (exon 6) | 26 amino acids | |
| 9 | Exon 7 | Deletion of whole exon | 165 amino acids |
| 123-151 | Exon 3 | 202-base deletion | 150 amino acids |
| Patient with AT | Exon 4 | 66-base deletion | 196 amino acids |
| Healthy donors | |||
| 2 | Exon 3 | Base change (197G>A) | 218 amino acids (Cys66Tyr) |
| 3 | Exon 6 | Base change (444C>T) | 151 amino acids (Gln152X) |
218 amino acids in healthy controls.
Bone marrow progenitor cell colony.
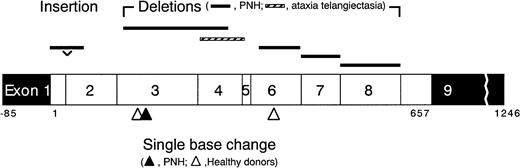
Schematic presentation of the
HPRT mutations found in 6-TG–resistant T- and BM-cell colonies. Open boxes show open reading frame (657 bases) consisting of exon 1 through 9, and closed boxes show noncoding regions.
Schematic presentation of the
HPRT mutations found in 6-TG–resistant T- and BM-cell colonies. Open boxes show open reading frame (657 bases) consisting of exon 1 through 9, and closed boxes show noncoding regions.
Expression of GPI-anchored proteins in 6-TG–resistant colonies
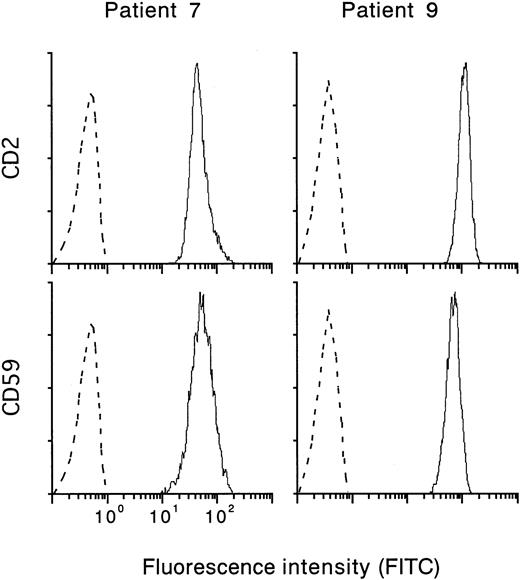
To characterize the T-cell colonies resistant to 6-TG, we examined the expression of GPI-anchored membrane proteins such as CD59 and DAF that are deficient in PNH cells. Figure3 shows flow cytometry results obtained in representative patients 7 and 9. The 6-TG–resistant colonies consisted of a single population of cells positive for CD2 (a T-cell marker) and CD59. Our findings that HPRT mutations did not occur within PNH clones indicate that mutations of more than one gene were involved and that they were not limited to a specific clone in PNH.
Expression of CD59 and CD2 on 6-TG–resistant T-cell colonies.
Dotted lines show nonspecific background staining with isotype-matched mouse Ig.
Expression of CD59 and CD2 on 6-TG–resistant T-cell colonies.
Dotted lines show nonspecific background staining with isotype-matched mouse Ig.
Discussion
Resistance to 6-TG—that is, the survival of cells cultured in the presence of 6-TG—is a good indicator for HPRT gene mutation(s).27,29,30,33,41 We used this indicator and the in vitro colony formation by individual T-cell clones to identify T cells with HPRT mutations and assessed the mutation frequency in patients with PNH. In the absence of 6-TG, T cells from healthy donors and PNH patients formed many colonies. In the presence of 6-TG, approximately 7 per 107 colonies from the controls survived, a frequency that may reflect the incidence of spontaneousHPRT mutations in our assay.31,32,41 As expected, sequence analysis of cDNA showed HPRT gene mutations in the 6-TG–resistant colonies. We detected T-cell colonies with HPRT mutations more frequently in PNH than in control: in 8 of 12 PNH patients and 3 of 17 healthy donors. The incidence of 6-TG–resistant T-cell colonies was also higher (21-fold increase) in the 8 patients than in the 3 controls. The markedly high incidence of resistant colonies in patients 6 and 12 may be partly ascribable to clonal expansion that occurred in a cell that happened to contain anHPRT mutation.41 Analysis of BM hematopoietic colonies obtained from 5 PNH patients and 4 healthy donors corroborated the increased frequency of HPRTmutations. Thus, we conclude that the HPRT gene more frequently mutated in the PNH patients than in the healthy individuals.
Sequence analysis of HPRT cDNA characterized theHPRT gene mutations. Despite the use of a reliable kit for RNA isolation, the mutation was determined in only 7 of 12 individuals positive for 6-TG–resistant colonies. One explanation for this is impaired expression of HPRT due to the mutation(s).41 As expected, HPRT mutations identified in 6-TG–resistant colonies from one donor were not found in colonies from the same donor that were formed in the absence of 6-TG, indicating somatic mutations of the HPRT gene. Most of the predicted HPRT proteins were small and truncated, supporting the idea of 6-TG resistance. Interestingly, the mutations were heterogeneous and widely distributed in the coding regions, indicating that they occurred at random sites. Therefore, there is no mutation cluster region or mutation hot spot within the HPRT gene. The mutations were large base deletions, a large base insertion, and a single-base substitution in PNH patients; only single-base substitutions were detected in the healthy controls. The nature of the acquired HPRT mutations found in our healthy individuals accord well with that in the published literature.27,29Judging from the difference in the HPRT gene mutation spectrum between PNH patients and healthy controls, it is then conceivable that different mutagens operated in the 2 groups, because the spectrum should be similar if spontaneous mutation alone were involved. In patient 7, 2 clones with 3 mutations ofHPRT were detected, indicating that several differentHPRT mutant clones may coexist in a single patient. These observations suggest that mutable conditions are present in PNH patients. In a current report, somatic mutations of PIG-Afound in 62 PNH patients consisted of deletions of a few bases in 28 and single-base substitutions in 20 of the patients.10No mutation hot spot(s) were detected. The coexistence in the same PNH patient of multiple clones with different mutations ofPIG-A has also been reported.19-21 In our PNH patients, the nature of the HPRT mutations was similar to the reported PIG-A mutations except for the size of the base deletion(s). We therefore posit that in PNH, both HPRT andPIG-A genes are exposed to common mutable conditions. Flow cytometry results indicated that HPRT mutations occurred in clones different from PNH clones. Thus, we conclude that in our PNH patients there existed a hematopoietic environment favorable to the emergence of multiple clones with different mutations in more than one gene.
The present study yields essentially the same analyses as that of Purow et al14 and supports the existence of mutable conditions, as has been suggested by the analysis of glycophorin13 and the detection of leukemic clones and MDS clones in patients with PNH.23 For further characterization of these conditions, analysis of patients with aplastic anemia and MDS, disorders that share the emergence of mutant clones and intractable BM failure with PNH, is necessary.24,42,43 The hypothesis of mutable conditions will help to elucidate the etiology of PIG-Amutations in PNH clones,12-15 the mechanism(s) of PNH clone expansion (by either survival or growth advantage) leading to clinical manifestations,10,12,37 and the mechanism(s) underlying the rare progression to leukemia in patients with PNH.23 24
We thank Toshio Suda, Nobuyuki Takakura, and Hiroshi Maeda of Kumamoto University for their advice and help.
Supported by grants from the Sagawa Foundation for Promotion of Cancer Research and the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Submitted March 6, 2001; accepted August 31, 2001.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hideki Nakakuma, Second Dept of Internal Medicine, Kumamoto University School of Medicine, Honjo 1-1-1, Kumamoto 860-8556, Japan; e-mail: nakakuma@kaiju.medic.kumamoto-u.ac.jp

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal