Follicular lymphomas (FLs) are neoplastic counterparts of normal germinal center (GC) B cells. FLs are characterized by t(14;18) with deregulation of the Bcl-2 (BCL2) gene. The presence of t(14;18) and overexpression of Bcl-2 is necessary, but not sufficient, to cause this disease. An array containing 588 complementary DNAs (cDNAs) was used to compare the gene expression between GC B cells and FL cells. To specifically monitor genes expressed in normal GC B and FL cells and not the entire tissue compartment, normal and malignant B cells were purified from tissues. Using the array, 37 genes were up-regulated and 28 were down-regulated in FL cells as compared to normal GC B cells. The expression level of each differentially expressed gene was verified by quantitative polymerase chain reaction. Following these studies 24 genes were up-regulated and 8 genes down-regulated with a P value less than .1. Included among the genes that were up-regulated in FLs were cell cycle regulator proteins CDK10, p120, p21CIP1, and p16INK4A; transcription factors/regulators Pax-5 and Id-2, which are involved in normal B-cell development; and genes involved in cell-cell interactions, tumor necrosis factor, interleukin-2Rγ (IL-2Rγ), and IL-4Rα. Among the genes that were down-regulated in FLs wereMRP8 and MRP14, which are involved in adhesion. Interestingly, several of these genes are localized within chromosomal regions already described to be altered in FLs. These findings provide a basis for future studies into the pathogenesis and pathophysiology of FL and may lead to the identification of potential therapeutic targets as well as antigens for immunotherapeutic strategies.
Introduction
Follicular lymphomas (FLs) are characterized by the t(14;18) (q32;q21) translocation in virtually all patients. This translocation involves the Bcl-2 gene (BCL2) on chromosome 18 juxtaposed with the immunoglobulin heavy chain (IGHG1) joining region on chromosome 14 (Bcl-2/IgH).1 This leads to deregulation of theBCL2 gene, with overproduction of the antiapoptotic Bcl-2 protein. A substantial body of evidence supports the hypothesis that the Bcl-2/IgH translocation is necessary, but not sufficient, to cause FL. Compelling evidence comes from studies of transgenic mice that bear a Bcl-2/IgH fusion gene to mimic the t(14;18) translocation in humans. These mice developed polyclonal B-cell hyperplasia in lymph nodes and spleen with IgM/IgD+ mature B cells, but not a monoclonal lymphoproliferative disorder.2,3 Similar to the progression seen in human FL, a minor subset of these transgenic mice go on to develop a monoclonal diffuse large B-cell lymphoma that is associated with a rearranged c-Myc (MYC) gene in about 50% of cases.3
Further evidence that the Bcl-2/IgH translocation alone is not causative for follicular non-Hodgkin lymphoma (NHL) is that B cells with t(14;18) are present in a substantial number of healthy individuals. In peripheral blood, t(14;18)+ cells have been reported in 8% to 88% of healthy subjects using polymerase chain reaction (PCR). Similarly using fluorescein in situ hybridization (FISH), B cells with t(14;18) have been detected in hyperplastic lymphoid tissues (tonsil, lymph nodes) of 12% to 54% of healthy individuals.4-6 Similar to the risk of FL increasing with age, the detection of t(14;18) in peripheral blood of healthy humans appears to increase with age.7 Cells with t(14;18)+ have also been detected by PCR in peripheral blood of patients with localized FL who are in long-term remission (> 10 years) after radiation therapy.8 One interpretation of these finding is that these cells lack other genetic changes necessary for transformation into recurrent lymphoma.
One approach to understand neoplastic transformation is by comparing gene expression of normal cells to their malignant counterparts. The development of complementary DNA (cDNA) array technology allows one to simultaneously quantify the expression of a very large number of genes.9-18 The application of DNA array technology to studies of human malignancies includes identifying the gene profile of malignancies of different cell types or lineages; genes involved in pathogenesis, progression, and metastasis; genetically determined risk groups; genes involved in resistance; and ultimately potential molecular targets of therapy. To date, gene profiling studies of NHL have identified clinically relevant subgroups, related histologic subgroups to normal cellular counterparts, and molecules that are specific to certain disease entities.10 13 In the present report, we have used a cDNA microarray to compare the expression of 588 genes in FL cells with their normal cellular counterparts, germinal center (GC) B cells isolated from hyperplastic tonsils. The levels of differentially expressed genes were verified by real-time quantitative reverse transcription PCR (RT-PCR). Our data suggest that there is differential expression of a large number of genes between normal GC B cells and FL cells. Among these are transcription factors, cell cycle regulators, signal transduction molecules, and molecules involved in cell-cell interactions. These studies may provide further understanding of the pathogenesis and pathophysiology of FL.
Materials and methods
Cells
Single-cell suspensions were prepared from tissues after mincing with a scalpel blade in the presence of collagenase IV and DNAse I, followed by passage through a fine wire mesh as previously described.19 Mononuclear cells were then isolated by Ficoll-Hypaque (Pharmacia, Uppsala, Sweden) density gradient centrifugation. Normal GC B cells were enriched from single-cell suspensions of tonsils by immunomagnetic bead depletion of T cells, monocytes, natural killer (NK) cells, naive B cells, and non-GC B cells using anti-CD4, -CD8, -CD14, -CD56, -IgD, and -CD44, respectively.19 FL cells were isolated from single-cell suspensions from lymph nodes or spleen that were histologically involved with FL. These populations of cells were depleted of T cells, monocytes, and NK cells as described above. Normal GC B cells were obtained from tonsils from 6 different individuals and FL cells isolated from 6 individuals with relapsed FL who had not received any therapy for at least 6 months before biopsy. All samples were obtained according to appropriate Human Protection Committee validation and informed patient consent. Following immunomagnetic bead treatment, a small aliquot of cells was phenotyped by flow cytometry and the remainder quickly frozen before RNA extraction.
Flow cytofluorometric analysis and antibodies
Cells were washed once in phosphate-buffered saline (PBS) and resuspended in PBS containing 1% bovine serum albumin (BSA; w/v; PBS-A). Immunophenotyping was performed as previously described19 using monoclonal antibodies (mAbs) directed against CD4, CD8, CD14, CD20, CD38, CD44, CD56, IgD, κ or λ light chains (Southern Biotechnology Associates, Birmingham, AL). An irrelevant isotype-matched control mAb was used in all experiments. The secondary antibody was a phycoerythrin R–conjugated goat F(ab′)2 antimouse IgG (Southern Biotechnology Associates) diluted at 2 μg/mL. Flow cytometry was carried out within 2 hours using an EPICS XL (Beckman-Coulter, Hialeah, FL). Cells were gated using forward versus side scatter to exclude dead cells and debris.
Poly(A)+ RNAs extraction and cDNA labeling
Cells were resuspended in TRIzol reagent (Gibco BRL, Grand Island, NY) according to the manufacturer's procedures. Contaminating DNA was removed by treatment with RNAse-free DNase I (Boehringer Mannheim, Indianapolis, IN) as described in the manufacturer's manual (Clontech, Palo Alto, CA). After 1 hour of incubation, the samples were extracted with phenol/chloroform/isoamylalcohol (25:24:1) and chloroform/isoamyl-alcohol (24:1). Total RNAs were finally precipitated with 1:5 (v/v) 7.5 M ammonium acetate and 1:2.5 (v/v) 100% ethanol. Poly(A)+ RNA was purified using Oligotex messenger RNA (mRNA) isolation kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The cDNA probes were synthesized either from 1 μg mRNA extracted from normal GC B cells or FL cells by reverse transcription according to the Atlas cDNA Expression Array manufacturer's protocol (Clontech) using 35 μCi (1.295 KBq) γ[32P]dATP (3.000 Ci/mmol; 111 TBq/mmol) (NEN, Boston, MA).
Probe purification and hybridization
Reverse transcribed products were purified using CHROMA SPIN-200 DEPC-H2O columns as described by the manufacturer (Clontech). Incorporation of 32P into the probes was determined after counting an aliquot in a liquid scintillation counter (Packard, Meridien, CT). The fractions corresponding to the peak were pooled and denatured (Clontech). The Atlas cDNA Expression Array membranes containing 588 cDNAs (Clontech) were prehybridized in ExpressHyb buffer (Clontech) containing 100 μg/mL heat-denatured salmon sperm (Sigma, St Louis, MO) for 3 hours in a rotating oven at 68°C. The denatured probes were added directly into the hybridization buffer and further incubated for 18 hours at 68°C. The membranes were washed 4 times 30 minutes at 68°C with 2 × standard sodium citrate (SSC)–1% sodium dodecyl sulfate (SDS), twice 30 minutes at 68°C with 0.1 × SSC-0.5% SDS, and finally at room temperature for 5 minutes in 2 × SSC. Hybridizations were performed in duplicate. Duplicate measurements were analyzed for reproducibility. Quantitation was assessed and transformed by the logarithm (base 10). The ratios of the measurements had a median of 0.036; 17% of the measurements were either above 0.10 or below −0.10 on the log scale, suggesting that 83% of the genes could be replicated with no more than 10% error.
Image acquisition and DNA array analysis
The membranes were exposed to phosphorimager screens at room temperature for 2 days (Molecular Dynamics, Sunnyvale, CA). The screens were scanned on a Storm 860 Phosphorimager (Molecular Dynamics) at a resolution of 50 μm. Scans were imported and arrayed using the AtlasImage 1.5 software (Clontech). The arrays from the 6 different normal GC B-cell samples were averaged after normalization of their intensities using the sum method as described in the AtlasImage 1.5 user manual. The same procedure was applied to the arrays from the 6 different FL samples. Using this methodology β-actin (GenBank accession no. X00351, Human Genome Organization (HUGO) nomenclature ACTB) and ribosomal protein S9 (GenBank accession no. U14971, HUGO nomenclature RPS9) were equally expressed in all the samples. Two composite arrays were generated, respectively. These 2 composite arrays were finally compared using the AtlasImage 1.5 software. Intensity of the genes ranged from 0 to 65 280 arbitrary units (AU). For a given gene, positive ratio corresponded to the normalized value of the intensity of a gene in FL cells divided by the normalized value in normal GC B cells. Negative ratios are the negative value of the ratio of the normalized gene intensities of normal GC B cells divided by the one of FL cells. Ratios greater than or equal to 2 and less than or equal to −2 were identified for further investigation. To include genes for which the value was below background level (ie, 0 for the AtlasImage 1.5 software), we took into consideration differences in intensities (FL minus GC) greater than or equal to 2000 and less than or equal to −2000 AU. All the expressed genes that did not meet these criteria are available athttp://dfciwww.dfci.harvard.edu/AO/Freedman_Lab/cDNA_array/not_different_expressed.html.
Real-time quantitative PCR analysis
Real-time quantitative RT-PCR analysis was performed using an ABI PRISM 7700 Sequence Detection System instrument using the SYBR Green I dye (PE Biosystems, Foster City, CA). Briefly, direct detection of PCR product was monitored by measuring the increase in fluorescence caused by the binding of SYBR Green I dye to double-stranded DNA by the Sequence Detection System directly into the reaction tube. The sequence detector software 1.7 calculates the threshold cycle number (CT) when signals reach 10 times the SD of the baseline. It was previously demonstrated that the calculated CT values are a quantitative measurement for the mRNA levels of various genes tested.20 21 DNase I-treated total RNA (1 μg) was reverse transcribed using the Advantage RT-for-PCR kit (Clontech) following the manufacturer's instructions, resuspended in 100 μL final volume, and aliquoted to avoid freezing and thawing procedures. RT reaction (2.5 μL), or water as control, was amplified in triplicate by real-time PCR in a final volume of 50 μL using the SYBR Green Master Mix reagent at a final concentration of 1 × (PE Biosystems). The appropriate primers were designed with the Primer Express 1.0 software (PE Biosystems). To ensure that any DNA contamination was removed by DNAse I treatment, real-time RT-PCR was performed on non–reverse-transcribed RNA. No amplification was observed in these conditions for all 65 genes that were differentially expressed. Due to the nonspecific binding of SYBR Green I to double-stranded DNA, the optimal primer concentrations were defined after verifying that no amplification was observed in the no template controls (NTCs) for a given set of forward and reverse primer concentrations. To ensure the specificity of the reaction, the size of the PCR product for each gene was verified by gel electrophoresis. The sequences and the concentrations of the primers are available athttp://dfciwww.dfci.harvard.edu/AO/Freedman_Lab/cDNA_array/sequences.html. Conditions were as follows: 1 cycle at 50°C for 2 minutes, 1 cycle at 95°C for 10 minutes, 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute. Validation experiments to verify the efficiencies of amplification of the primers of target and reference genes were approximately equal.
For each sample, CT values for β-actin and ribosomal protein S9 genes were generated for normalization purposes. For each run of each sample, a correction factor was calculated by dividing by the minimum β-actin and ribosomal protein S9 values, and then averaging the adjusted control values for the sample. This value was used to correct each sample for differences in RNA content.
Statistical analysis
Genes were identified from the Clontech array as described above. Results of quantitative RT-PCR were assessed by the 2-sample t test; nominal P values are presented without correction for multiple comparisons.
Results
Cell surface analysis of purified GC B cells and FL cells
To try to better understand the differences in gene expression between normal GC B cells and FL cells we used purified cell populations. Single mononuclear cell suspensions of tonsil and tissues biopsies involved with FL contain significant numbers of T cells, NK cells, and monocytes and in the case of tonsil, naive and memory B cells. To limit the interference of any genes from the microenvironment in our study, we used immunomagnetic bead depletion to remove these cells from our preparations. Normal GC B cells were purified from single-cell suspensions of tonsils. The immunophenotype of GC B cells has been previously described to be CD20+/CD38+and IgD−/CD44−.19,22 Following immunomagnetic bead depletion of T cells, monocytes, NK cells, and IgD+/CD44+ cells, the population of cells were phenotyped and analyzed by flow cytometry. The preparation of GC B cells was more than 98% CD20+/CD38+, with no detectable CD4+, CD8+, CD56+, CD14+, CD44+, and IgD+ cells. Similarly, immunomagnetic depletion of T cells, NK cells, and monocytes was performed with single-cell suspensions of tissues involved with FL. Cell surface marker analysis of FL cells showed a monoclonal population CD20+ B cells.23-25
cDNA array analysis
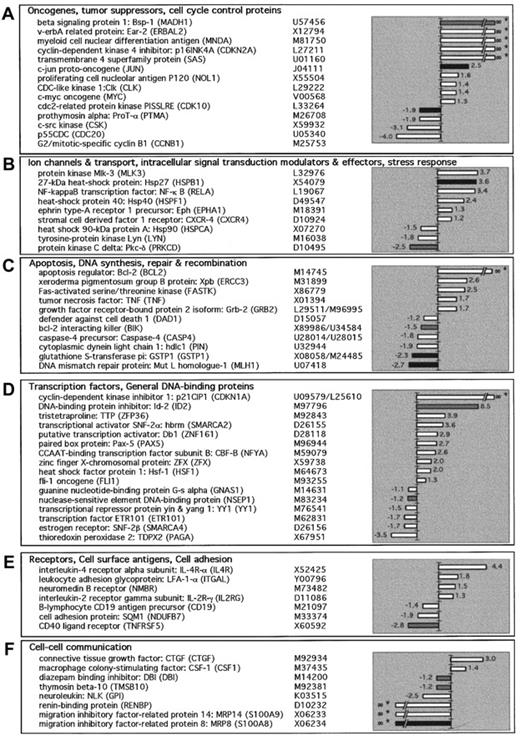
In an attempt to further understand differences between FL and normal GC B cells, we compared the gene expression between these 2 cell types by cDNA microarray. In the Clontech array the randomly selected cDNA represented all known sequences spotted onto a nylon membrane as amplified products of 200 to 600 base pairs (bp) in length. These fragments have been carefully selected from gene regions that lack the poly-A tail, repetitive elements, or other highly homologous sequences. The array is organized in 6 different quadrants each containing one or several gene families. The families include oncogenes, tumor suppressors, cell cycle control proteins (Figure1A); ion channels and transport, intracellular signal transduction modulators and effectors, stress response proteins (Figure 1B); apoptosis, DNA synthesis, repair and recombination proteins (Figure 1C); transcription factors, general DNA-binding proteins (Figure 1D); receptors, cell surface antigens, cell adhesion (Figure 1E); and cell-cell communication factors (Figure 1F).
Gene expression patterns in FLs.
Gene expression profiles generated from 6 healthy individuals (GC B cells) and 6 patients with relapsed FL were analyzed using cDNA arrays. The mRNA from purified cells were reverse transcribed and hybridized as described in “Materials and methods.” The 6 arrays obtained after hybridization of 6 GC B-cell probes were averaged, as well as 6 arrays from 6 FL-cell probes to create 2 composite arrays, respectively. Finally, after global normalization and subtraction of the background, the 2 composite arrays were compared using the AtlasImage 1.5 software. Genes were selected if ratios were more than 2 or if differences were more than 2000. Positive values indicate that the transcript is more abundant in FL than GC cells and negative values the opposite. Shades of gray indicate expression level of a gene (AU), black bars represent expression level more than 10 000, gray more than 5000, and white more than 2000. Also listed along side the gene names is the GenBank accession number. The genes are grouped according to their function as determined from the literature.
Gene expression patterns in FLs.
Gene expression profiles generated from 6 healthy individuals (GC B cells) and 6 patients with relapsed FL were analyzed using cDNA arrays. The mRNA from purified cells were reverse transcribed and hybridized as described in “Materials and methods.” The 6 arrays obtained after hybridization of 6 GC B-cell probes were averaged, as well as 6 arrays from 6 FL-cell probes to create 2 composite arrays, respectively. Finally, after global normalization and subtraction of the background, the 2 composite arrays were compared using the AtlasImage 1.5 software. Genes were selected if ratios were more than 2 or if differences were more than 2000. Positive values indicate that the transcript is more abundant in FL than GC cells and negative values the opposite. Shades of gray indicate expression level of a gene (AU), black bars represent expression level more than 10 000, gray more than 5000, and white more than 2000. Also listed along side the gene names is the GenBank accession number. The genes are grouped according to their function as determined from the literature.
Radiolabeled probes for hybridization were derived from 2 sources of mRNA: GC B cells derived from the tonsils of 6 healthy individuals and FL cells isolated from tissues of 6 patients with relapsed disease. Six arrays obtained after hybridization of the 6 GC B-cell probes were averaged as well as the 6 arrays from 6 FL probes to create 2 composite arrays, respectively. Finally, after global normalization and subtraction of the background, the 2 composite arrays were compared. The expression of actin and S9 were constant in all 12 samples analyzed. Figure 1 summarizes the genes for which the ratio of FL to GC was more than 2 or less than −2, or for which the difference was more than 2000 or less than −2000. Of the 588 genes in the cDNA array, 65 genes were differentially expressed in FL cells as compared to normal GC B cells, using the above-described criteria. Of the 65 genes, 37 (57%) were increased in FL, whereas 28 (43%) were decreased. The list of genes that were expressed by either FL or GC B cells but did not meet these criteria are listed athttp://dfciwww.dfci.harvard.edu/AO/Freedman_Lab/cDNA_array/not_different_expressed.html.
Real-time quantitative RT-PCR analysis
Although differences in gene expression between FL and normal GC B cells were observed with the cDNA array, it was critical to verify these results using another methodology. We monitored the mRNA level of all genes noted to be differentially expressed in the Atlas array, according to the criteria described above by real-time quantitative RT-PCR using SYBR Green 1.20,26 We also monitored Bcl-2, which is increased in the vast majority of FL as compared to normal GC B cells27 as well as 2 housekeeping genes (β-actin and ribosomal protein S9). Results representative of real-time quantitative PCR are shown Figure 2 for Bcl-2, β-actin, and ribosomal protein S9. The CT value at which the fluorescence reaches the threshold is higher for GC than for FL cells, which is reflective of a lower mRNA content for Bcl-2 in GC B cells. The CT values obtained for GC or FL for β-actin as well as ribosomal S9 protein are the same, indicating that the mRNA content was identical in these 2 samples. The NTCs remained below the threshold, showing that no amplification was observed without template.
Real-time RT-PCR quantification for Bcl-2, β-actin, and S9 ribosomal protein mRNAs levels.
Real-time quantitative RT-PCR was performed with appropriate primers for Bcl-2 (A), β-actin (B), and S9 ribosomal protein (C) using SYBR Green I dye. Amplification results were visualized using the Sequence Detector 1.7 software. These are representative plots showing increasing fluorescence (Δ Rn) detected through cycle 40 on normal GC B cell or FL cDNA samples. NTC represents the negative control amplification (using H20 as template). CT represents the threshold cycle at which fluorescence is first detected above background. A higher CT value is indicative of lower mRNA levels (more PCR cycles required to exceed background).
Real-time RT-PCR quantification for Bcl-2, β-actin, and S9 ribosomal protein mRNAs levels.
Real-time quantitative RT-PCR was performed with appropriate primers for Bcl-2 (A), β-actin (B), and S9 ribosomal protein (C) using SYBR Green I dye. Amplification results were visualized using the Sequence Detector 1.7 software. These are representative plots showing increasing fluorescence (Δ Rn) detected through cycle 40 on normal GC B cell or FL cDNA samples. NTC represents the negative control amplification (using H20 as template). CT represents the threshold cycle at which fluorescence is first detected above background. A higher CT value is indicative of lower mRNA levels (more PCR cycles required to exceed background).
Genes increased in FL
Besides the increase of Bcl-2 in FL, relatively few genes have been shown to be increased as compared to normal B cells. Among these genes are those for Pax-5 (PAX5), TNF (TNF) andCXCR4 (SDF-1 receptor).28-30 To confirm the differential expression of the 37 genes that were increased in FL cells, we performed real-time quantitative RT-PCR analysis. The CT values were normalized for each gene in all of the samples obtained from the GC B and FL cells as described in “Materials and methods.” The data were analyzed using a 2-sample t test. As seen in Table1, 24 genes are listed, 21 that were found to be increased in FL using the array (P < .1), and 3 genes whose expression was decreased in FL by the array. The difference between the mean of the CT value for FL and the mean of the CT value for GC was calculated (Δ mean CT). These 24 genes are listed in descending order of their level of expression (Δ mean CT). As reported previously, approximately 2.5 Δ CT values correspond to a 10-fold increase.26 Consistent with prior reports, we found that Bcl-2, TNF, and Pax-5 were all overexpressed in FL cells.1,13,28 29 In FL cells, several genes encoding for cell cycle regulator proteins were overexpressed including CDK10, p120 (NOL1), p21CIP1 (CDKN1A), and p16INK4A (CDK2A). Considering that FL cells are often in G0/G1, it is of interest that regulators of G1 phase p21CIP1 and p16INK4A were overexpressed. A large number of transcription factors and DNA binding proteins were preferentially expressed in FL cells including Bsp-1 (MADH1); Id-2 (ID2); c-Jun (JUN); v-erbA (ERBAL2); ZFX (ZFX); Db1 (ZNF161); Pax-5; SNF2-α (SMARCA2); and Hsf-1 (HSF1). There is evidence that Id-2 and Pax-5 are both important in B-cell differentiation. A set of genes that are involved in microenvironmental interactions including cytokines and cytokines receptors were also overexpressed in FL cells including TNF, IL-4Rα, IL-2Rγ. Three genes (YY1, Lyn, cyclin B1 [CCNB1]), which had decreased expression in FL cells using the cDNA array, were overexpressed by quantitative RT-PCR with Pvalues of less than .02, .08, and .03, respectively.
Gene expression analysis by real-time quantitative RT-PCR (genes increased in FL)
| Gene . | HUGO . | CTGC ± SEM . | CTFL ± SEM . | ΔCT . | P . |
|---|---|---|---|---|---|
| Bsp-1* | MADH1 | 29.2 ± 0.7 | 22.8 ± 1.6 | 6.4 | .0001 |
| Bcl-2* | BCL2 | 26.1 ± 0.9 | 20.2 ± 1.4 | 5.9 | .0001 |
| Mlk-3* | MLK3 | 36.1 ± 2.9 | 31.5 ± 2.5 | 4.6 | .04 |
| p21CIP1* | CDKN1A | 26.5 ± 0.7 | 23.4 ± 1.2 | 3.1 | .002 |
| Hsp27 | HSPB1 | 28.1 ± 1.1 | 25.4 ± 1.7 | 2.7 | .04 |
| Hsp40 | HSPF1 | 24.8 ± 0.2 | 22.2 ± 2.1 | 2.6 | .04 |
| TNF | TNF | 21.8 ± 0.8 | 19.3 ± 1.5 | 2.5 | .02 |
| Id-2* | ID2 | 27.3 ± 1.0 | 24.8 ± 1.3 | 2.4 | .02 |
| c-Jun | JUN | 24.5 ± 0.9 | 22.2 ± 1.9 | 2.3 | .05 |
| Ear-2* | ERBAL2 | 26.5 ± 0.4 | 24.4 ± 1.1 | 2.1 | .005 |
| YY1 | YY1 | 28.4 ± 0.8 | 26.3 ± 1.2 | 2.1 | .02 |
| CDK10* | CDK10 | 26.3 ± 0.6 | 24.7 ± 0.7 | 1.6 | .01 |
| ZFX | ZFX | 28.6 ± 0.6 | 27.1 ± 0.9 | 1.5 | .03 |
| IL-4R-α | IL4R | 18.1 ± 0.2 | 16.6 ± 0.4 | 1.5 | .0004 |
| Db1 | ZNF161 | 25.2 ± 1.0 | 23.8 ± 0.7 | 1.3 | .09 |
| IL-2R-γ* | IL2RG | 22.5 ± 0.7 | 21.2 ± 0.5 | 1.3 | .05 |
| Xpb* | ERCC3 | 24.0 ± 0.3 | 22.9 ± 0.7 | 1.1 | .02 |
| CCN1B* | CCN1B | 28.6 ± 0.8 | 26.5 ± 1.3 | 1.1 | .03 |
| Pax-5 | PAX5 | 23.0 ± 0.3 | 21.7 ± 1.0 | 1.1 | .08 |
| Lyn* | LYN | 21.5 ± 0.5 | 20.5 ± 0.6 | 1.0 | .08 |
| SNF2-α* | SMARCA2 | 23.3 ± 0.2 | 22.3 ± 1.0 | 1.0 | .08 |
| p16INK4A* | CDKN2A | 22.1 ± 0.1 | 21.1 ± 0.8 | 0.9 | .09 |
| p120* | NOL1 | 21.4 ± 0.5 | 20.7 ± 0.5 | 0.7 | .08 |
| Hsf-1* | HSF1 | 19.7 ± 0.2 | 19.1 ± 0.1 | 0.6 | .004 |
| Gene . | HUGO . | CTGC ± SEM . | CTFL ± SEM . | ΔCT . | P . |
|---|---|---|---|---|---|
| Bsp-1* | MADH1 | 29.2 ± 0.7 | 22.8 ± 1.6 | 6.4 | .0001 |
| Bcl-2* | BCL2 | 26.1 ± 0.9 | 20.2 ± 1.4 | 5.9 | .0001 |
| Mlk-3* | MLK3 | 36.1 ± 2.9 | 31.5 ± 2.5 | 4.6 | .04 |
| p21CIP1* | CDKN1A | 26.5 ± 0.7 | 23.4 ± 1.2 | 3.1 | .002 |
| Hsp27 | HSPB1 | 28.1 ± 1.1 | 25.4 ± 1.7 | 2.7 | .04 |
| Hsp40 | HSPF1 | 24.8 ± 0.2 | 22.2 ± 2.1 | 2.6 | .04 |
| TNF | TNF | 21.8 ± 0.8 | 19.3 ± 1.5 | 2.5 | .02 |
| Id-2* | ID2 | 27.3 ± 1.0 | 24.8 ± 1.3 | 2.4 | .02 |
| c-Jun | JUN | 24.5 ± 0.9 | 22.2 ± 1.9 | 2.3 | .05 |
| Ear-2* | ERBAL2 | 26.5 ± 0.4 | 24.4 ± 1.1 | 2.1 | .005 |
| YY1 | YY1 | 28.4 ± 0.8 | 26.3 ± 1.2 | 2.1 | .02 |
| CDK10* | CDK10 | 26.3 ± 0.6 | 24.7 ± 0.7 | 1.6 | .01 |
| ZFX | ZFX | 28.6 ± 0.6 | 27.1 ± 0.9 | 1.5 | .03 |
| IL-4R-α | IL4R | 18.1 ± 0.2 | 16.6 ± 0.4 | 1.5 | .0004 |
| Db1 | ZNF161 | 25.2 ± 1.0 | 23.8 ± 0.7 | 1.3 | .09 |
| IL-2R-γ* | IL2RG | 22.5 ± 0.7 | 21.2 ± 0.5 | 1.3 | .05 |
| Xpb* | ERCC3 | 24.0 ± 0.3 | 22.9 ± 0.7 | 1.1 | .02 |
| CCN1B* | CCN1B | 28.6 ± 0.8 | 26.5 ± 1.3 | 1.1 | .03 |
| Pax-5 | PAX5 | 23.0 ± 0.3 | 21.7 ± 1.0 | 1.1 | .08 |
| Lyn* | LYN | 21.5 ± 0.5 | 20.5 ± 0.6 | 1.0 | .08 |
| SNF2-α* | SMARCA2 | 23.3 ± 0.2 | 22.3 ± 1.0 | 1.0 | .08 |
| p16INK4A* | CDKN2A | 22.1 ± 0.1 | 21.1 ± 0.8 | 0.9 | .09 |
| p120* | NOL1 | 21.4 ± 0.5 | 20.7 ± 0.5 | 0.7 | .08 |
| Hsf-1* | HSF1 | 19.7 ± 0.2 | 19.1 ± 0.1 | 0.6 | .004 |
For each of the 6 individuals and the 6 patient samples, a real-time quantitative RT-PCR was performed for all the genes presented in Figure 2. The mean values are shown for GC (CTGC) ± SEM and FL (CTFL) ± SEM as well as the difference between the means (ΔCT). Note that a lower CTvalue is indicative of a higher mRNA content. A paired ttest was performed and only the genes for which the P value is less than .1 are presented here.
Confirmed in validation sample.
Genes decreased in FL
There is relatively little information on genes that are down-regulated in FL cells, as compared to normal B cells. We confirmed by RT-PCR the decreased expression in 7 of the 28 genes noted to be down-regulated in FL cells using the array (P < .1), whereas in 21 genes the decrease noted using the array was not confirmed by RT-PCR. One gene (LFA1), which was increased in FL by array, was found to be decreased by RT-PCR. Table 2 shows these 8 genes in descending order of their level of expression in GC B cells (Δ mean CT). The majority of genes are involved in microenvironmental or cell-cell interactions includingMRP14; MRP8; CD40; thymosin β 10 (TMSB10); and DBI (DBI). Consistent with the low proliferative index of FL cells, the cell cycle regulator p55CDC (CDC20) had decreased expression in the FL cells.
Gene expression analysis by real-time quantitative RT-PCR (genes decreased in FL)
| Gene . | HUGO . | CTGC ± SEM . | CTFL ± SEM . | ΔCT . | P . |
|---|---|---|---|---|---|
| MRP14* | S100A9 | 22.1 ± 2.1 | 30.1 ± 2.2 | −7.9 | .0006 |
| MRP8* | S100A8 | 22.5 ± 2.5 | 22.5 ± 2.9 | −6.6 | .005 |
| TDPX2* | PAGA | 22.6 ± 0.7 | 22.6 ± 0.5 | −1.7 | .01 |
| p55CDC* | CDC20 | 24.4 ± 0.6 | 24.4 ± 1.1 | −1.6 | .05 |
| CD40 | TNFRSF5 | 23.0 ± 0.5 | 23.7 ± 0.3 | −0.7 | .07 |
| Thymosin β-10* | TMSB10 | 14.9 ± 0.5 | 15.5 ± 0.4 | −0.7 | .08 |
| DBI | DBI | 18.3 ± 0.4 | 19.0 ± 0.4 | −0.7 | .03 |
| LFA-1 | ITGAL | 25.3 ± 0.8 | 25.8 ± 0.8 | −0.5 | .06 |
| Gene . | HUGO . | CTGC ± SEM . | CTFL ± SEM . | ΔCT . | P . |
|---|---|---|---|---|---|
| MRP14* | S100A9 | 22.1 ± 2.1 | 30.1 ± 2.2 | −7.9 | .0006 |
| MRP8* | S100A8 | 22.5 ± 2.5 | 22.5 ± 2.9 | −6.6 | .005 |
| TDPX2* | PAGA | 22.6 ± 0.7 | 22.6 ± 0.5 | −1.7 | .01 |
| p55CDC* | CDC20 | 24.4 ± 0.6 | 24.4 ± 1.1 | −1.6 | .05 |
| CD40 | TNFRSF5 | 23.0 ± 0.5 | 23.7 ± 0.3 | −0.7 | .07 |
| Thymosin β-10* | TMSB10 | 14.9 ± 0.5 | 15.5 ± 0.4 | −0.7 | .08 |
| DBI | DBI | 18.3 ± 0.4 | 19.0 ± 0.4 | −0.7 | .03 |
| LFA-1 | ITGAL | 25.3 ± 0.8 | 25.8 ± 0.8 | −0.5 | .06 |
Real-time quantitative RT-PCR was performed as described in Figure 2 for all the genes presented in Figure 1. The mean CT values are shown for (CTGC) ± SEM and FL (CTFL) ± SEM as well as the difference between the means (ΔCT). Note that a higher CT is indicative of a lower mRNA content. A paired t test was performed and only the genes for which the P value is less than .1 are presented here.
Confirmed in validation sample.
Validation sample
We conducted a small validation study of the 32 genes that were significantly different in their expression levels. Three additional FL samples were studied by RT-PCR only. Of the 24 genes found to be overexpressed in FL cells by RT-PCR, 15 were confirmed to be overexpressed in FL cells compared to GC cells in this small validation sample. Of the 8 genes that had decreased expression in FL cells, 5 were confirmed to be significantly decreased in the 3 additional cases. An asterisk in Tables 1 and 2 indicates these genes.
Discussion
Follicular lymphomas have been hypothesized to be neoplastic transformations of normal GC-B cells. This was supported first by morphology, and then by virtue of the expression of various cell surface antigens.31 32 However the finding that FL cells possess in essentially all cases the Bcl-2/IgH rearrangement and overexpression of Bcl-2 protein provided clear distinction from normal GC B cells. In the present report we have compared the gene expression profile of FL cells to normal tonsillar GC B cells. Based on the evidence that Bcl-2/IgH rearrangement and overexpression of Bcl-2 is necessary but not sufficient to induce FL, we hypothesized that additional alterations in gene expression may contribute to the malignant phenotype. Our objective was to identify genes that were differentially expressed in the malignant cells and their normal cellular counterparts using a cDNA array and confirm expression using real-time quantitative RT-PCR as a second platform. For a limited number of genes the results of quantitative PCR differed from the array data. This was unexpected, but emphasizes the need to confirm array data using a second methodology. We identified a series of genes that were either overexpressed or underexpressed in FL cells as compared to normal GC B cells. A number of these genes are known to be important in normal and malignant B cells and may help explain certain aspects of the pathophysiology of FL. Because all patients had relapsed disease, our data may not apply to FL cells isolated from untreated patients. Furthermore, it is possible that some of the alterations in gene expression were related to prior cytotoxic therapy. The localization of certain of these genes at sites of known chromosomal abnormalities in FL may provide further evidence for their role in the malignant transformation.
The genes that had the greatest Δ CT values and the most statistically significant differences in expression wereBCL2, BSP-1, MRP8, andMRP14. Observing overexpression of Bcl-2 in FL cells served as a control for this study. Bsp-1, also known as Smad1, is involved in transforming growth factor-β (TGF-β) and bone morphogenetic proteins (BMPs) signaling.33,34 It is unknown what function Bsp-1 has in B cells, although TGF-β has effects on human and murine B cells. The major effect of TGF-β is inhibiting DNA synthesis and inducing cell cycle arrest, and in some B-cell lines, apoptosis.35 The overexpression of Bcl-2 in FL cells may prevent apoptosis as a consequence of TGF-β. In the B-cell line RL, which has t(14;18), TGF-β treatment leads to an increase in hypophosphorylated Rb and a decrease in mutant p53 levels.36 In murine B cells, TGF-β caused G1arrest through up-regulation of p27Kip1, whereas in other cell types TGF-β up-regulated p21Cip (CDKN1A).35,37 Overexpression of Bsp-1 may promote increased sensitivity of FL cells to effects of TGF-β and contribute to the hypoproliferative nature of these cells. MRP8 and MRP14, also known as migration inhibitory factor–related proteins 8 and 14, are members of the S100 family, which are reported to be expressed in myelomonocytic cells.38 These proteins associate with the cytoskeleton, but are also secreted products. MRPs regulate adhesion through β2 integrin39; however, their precise function remains unknown. Expression in normal GC B cells, but not in FL cells, may be important in the adhesion of normal GC cells. In addition the low level of MRP8 and MRP14 in FL cells may explain the relative absence of monocytes in neoplastic GCs as compared to that found in normal GCs.40
Several transcription factors/regulators and DNA binding proteins were overexpressed in FL cells. Among these were Pax-5 and Id-2, both of which have important roles in B-cell development and differentiation.29,41-44 Pax-5, also known as B-cell–specific activator protein (BSAP), is a DNA binding protein. Pax-5 expression is restricted to B cells, the testis in adults, and the brain and liver in the fetus. In B-cell ontogeny, Pax-5 mRNA is expressed from the pro–B-cell stage through mature B cells, decreasing in plasma cells. Mice lacking Pax-5 have B-cell maturation arrest at the pre–B-cell stage. Pax-5 regulates expression of several B-cell–specific genes including CD19, the tyrosine kinase Blk, and pre-B-cell–specific surrogate light chain genes.45 Pax-5 localizes to 9p13 and has deregulated expression in lymphoplasmacytic lymphomas and some cases of diffuse large cell lymphoma with t(9;14).46,47 Our studies are consistent with a previous report that the majority of cases of FL overexpress Pax-5 protein.29 It is unclear what role Pax-5 plays in FL; however, overexpression in splenic B cells and cell lines enhances proliferation and modulates p53 expression.48 Similar to Pax-5, Id-2 has been shown to be important in B-cell development. Id proteins are helix-loop-helix transcription regulators that play an important role in hematopoiesis.49 Id proteins bind to other helix-loop-helix–bearing transcription factors, inhibiting their function.50 In mice, Id-2 is expressed in pro-B cells and decreases during differentiation.51 In normal peripheral blood mononuclear cells, Id-2 mRNA levels decrease with mitogen stimulation.50 Id-2 plays a critical role in lymphoid tissue development because Id-2−/− mice lack lymph nodes and Peyer patches, but have normal splenic architecture with T cells and B cells.45 Overexpression of Id-2 causes a block in normal murine T-cell differentiation.52 Therefore, the overexpression of Id-2 in FL cells may regulate proliferation or contribute to the maturation arrest in these tumor cells. Identification of the target genes that are regulated by Pax-5 and Id-2 in FL cells may provide important insights into understanding the growth pattern and maturation arrest of these cells.
The FL cells reside in a GC microenvironment within lymphoid tissues and in the marrow in association with other lymphohematopoietic and stromal cells.53 Evidence indicates that interactions of FL cells with components of the microenvironment modulate the growth and survival of the malignant cells.54 FL cells had increased mRNA expression of 2 cytokine receptors: IL-2Rγ and IL-4Rα. The IL2RG chain mediates signaling from IL-2 as well as IL-4, IL-7, and IL-15.55-57 Furthermore, there are often large numbers of T cells expressing CD40L (CD154), IL-2, and IL-4 in normal and malignant GCs.58,59 Survival of FL cells in vitro is enhanced by IL-4 in association with stimulation through CD40.19,60 Therefore IL-4 production by T cells and overexpression of the IL-4 receptor on FL cells may be important for their viability in vivo. Increased numbers of CD40L+ T cells in malignant GCs may be able to compensate for the lower expression of CD40 on FL cells as compared to normal GC B cells.13 As reported previously, TNF mRNA was overexpressed by FL cells.28 TNF can serve as an autocrine growth factor for normal B cells, although TNF has not been shown to stimulate proliferation of FL cells in vitro. TNF family members and their receptors play a central role in the generation and maintenance of the structure of lymphoid tissues, the development of GCs, and follicular dendritic cells (FDCs).61 A potentially important function of FL-derived TNF may be in the creation and maintenance of the tumor cell microenvironment, and functional FDCs, which may assist in the support of tumor cell viability and their recruitment within specific areas of the body.
One problem with comparing gene expression of normal and neoplastic tissues is the difficulty of determining whether the signal is originating from the tumor cells or from associated normal cells. Within normal tonsils and tissue involved with FL, there are many normal cellular constituents (ie, fibroblasts, monocytes, T, NK, endothelial cells, and FDCs). Although the microenvironment in FL may in fact differ from that of normal lymphoid tissues, in this study we only sought to examine gene expression in the B cells. The GC B cells used in this study were representative of a total population, rather than separated into centroblasts or centrocytes. The phenotype of the GC B cells used was CD38+, CD44− , and surface IgD−. Within the total population, the expression of CD77 is associated with the more proliferative centroblasts, whereas CD38+ GC B cells that lack CD77 are more reflective of centrocytes.62 CD77 has been reported to have variable expression on FL cells.63 However, despite the potential heterogeneity of different tonsillar B-cell preparations, when the gene expression of the various samples of GC B cells was examined, they were remarkably homogeneous. The finding that cell cycle regulatory proteins that are involved in G1 arrest, p21CIP1 and p16INK4A, were overexpressed in FL cells is consistent with their low proliferative nature.64 Moreover, p55CDC, which is also required for normal cell division, was underexpressed in FL cells.13 65An alternative interpretation is that the GC B cells we purified were predominantly centroblasts with a high proliferation rate and low levels of these CDK inhibitors.
Follicular lymphomas have been compared to normal GC B cells using oligonucleotide microarrays.13 Seventeen of the 32 genes we confirmed by RT-PCR to be differentially expressed were present on the microarray used by Alizadeh et al.13 Despite a different technical approach, a number of the genes that we found to be overexpressed in FL cells were also increased in the array analysis by Alizadeh et al. These include BCL2, BSP1, JUN, IL4R,IL-2Rγ, Lyn, and SNF-2a. In addition, 2 genes (p55CDC, CD40), which we found to have decreased expression in FL cells, were also decreased in the study of Alizadeh et al.13 In contrast, 2 genes that we found to be increased in FL cells were decreased in the Alizadeh13 study (p16INK4A and CCN1B).
In addition to the t(14;18) translocation, additional cytogenetic abnormalities have been described in FL.66-72 Additional chromosomal abnormalities involving, for example, p53, play a central role in the progression of FL. These additional abnormalities are likely to be important in the development of the disease, because Bcl-2/IgH is necessary but not sufficient for tumor development. These changes include gains and losses of chromosomal material that can either lead to an increased or a decreased expression of other genes. In this study we found a number of genes that were deregulated and localized to sites of known genetic abnormalities observed in FL (Table3). For example, p21CIP1 and TNF, assigned to 6p21.1 and 6p21.3, respectively, are located within high-level DNA amplifications on band 6p21.71MRP8, MRP14, and TDPX2 genes are located within commonly seen chromosome 1 breakpoints 1q21-22 and 1p22-36, respectively.66 69
Chromosomal localization of differentially expressed genes in FL
| Gene ID . | HUGO ID . | Chromosomal localization . | Known genetic abnormalities in FL . | References . |
|---|---|---|---|---|
| Genes increased | ||||
| Bcl-2 | BCL2 | 18q21 | 18q21 | 1 |
| p21CIP1 | CDKN1A | 6p21.1 | 6p21 | 71 |
| TNF | TNF | 6p21.3 | 6p21 | 71 |
| c-Jun | JUN | 1p32-p31 | 1p31-36 | 70 |
| Hsf-1 | HSF1 | 8q24.3 | 8q24 | 71 |
| XPB | XPB | 2q21 | 2q21 | 69 |
| Mlk-3 | MLK3 | 11q13.1-q13.3 | 11q13-25 | 67 |
| Hsp27 | HSP27 | 7q | 7 | 67 |
| Pax-5 | PAX5 | 9p13 | 9p13 | 67 |
| Genes decreased | ||||
| MRP8/14 | S100A8/9 | 1q12-q22 | 1q21-23 | 66,69 |
| TDPX2 | PAGA | 1p34.1 | 1p22-36 | 66,67,69 |
| Gene ID . | HUGO ID . | Chromosomal localization . | Known genetic abnormalities in FL . | References . |
|---|---|---|---|---|
| Genes increased | ||||
| Bcl-2 | BCL2 | 18q21 | 18q21 | 1 |
| p21CIP1 | CDKN1A | 6p21.1 | 6p21 | 71 |
| TNF | TNF | 6p21.3 | 6p21 | 71 |
| c-Jun | JUN | 1p32-p31 | 1p31-36 | 70 |
| Hsf-1 | HSF1 | 8q24.3 | 8q24 | 71 |
| XPB | XPB | 2q21 | 2q21 | 69 |
| Mlk-3 | MLK3 | 11q13.1-q13.3 | 11q13-25 | 67 |
| Hsp27 | HSP27 | 7q | 7 | 67 |
| Pax-5 | PAX5 | 9p13 | 9p13 | 67 |
| Genes decreased | ||||
| MRP8/14 | S100A8/9 | 1q12-q22 | 1q21-23 | 66,69 |
| TDPX2 | PAGA | 1p34.1 | 1p22-36 | 66,67,69 |
In conclusion, we have identified a number of genes that are differentially expressed in FL cells and their normal counterparts, GC B cells, using a combination of gene array technology and quantitative PCR analysis. These studies focused on only the normal and neoplastic B cells by using highly purified cells without contaminating cells from the associated microenvironment. Several of the genes identified are involved in regulating the cell cycle, and their overexpression in FL cells is consistent with their low proliferative state. The observation that a number of transcription factors overexpressed by FL cells, particularly Pax-5 and Id-2, have a central role in normal B-cell development, may lead to better understanding of the regulation of other genes involved in the pathogenesis and pathophysiology of this disease. In addition to insights into the biology of FL, comparing gene expression in normal B cells and FL cells may lead to the identification of potential targets as well as antigens for immunotherapeutic strategies.
Supported in part by National Institutes of Health grants CA55207 and CA66996, and the Norman Hirschfield Foundation. H.H. was supported by the Cure for Lymphoma Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Arnold S. Freedman, Department of Adult Oncology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail:arnold_freedman@dfci.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal