B-cell chronic lymphocytic leukemia (B-CLL) is a heterogeneous disease involving more than one molecular mechanism that leads to the transformation of CD5+ B cells at either the pregerminal or postgerminal center stage of differentiation. It was previously demonstrated that ataxia telangiectasia mutated (ATM) gene mutations can occur in B-CLL and cause a defect in the p53 pathway. Here the role of ATM mutations in the pathogenesis of B-CLL is addressed. Of 50 B-CLL tumors with fully analyzedATM and TP53, 16 had ATM mutations. Six of 50 B-CLLs showed mutations in TP53 and the remaining 28 tumors had wild-type ATM or TP53. No tumor had both ATM and TP53 mutations. Remarkably, all 16 ATM mutant B-CLLs showed the absence of somatic variable region heavy chain hypermutation indicating a pregerminal center cell origin and a common pathogenesis for these tumors. Furthermore, in 5 of the 16 B-CLLs, ATMmutation preceded the transformation stage of differentiation. At the cellular level, ATM mutant tumors exhibited a deficient ATM-dependent p53 response to gamma irradiation, failure to up-regulate TRAIL-R2, a downstream target that links irradiation-induced p53 response with apoptosis, and an inability to repair induced chromosome breaks. Mantle cell lymphoma (MCL) is also of pregerminal center origin and ATMmutations are frequent in this malignancy. It is concluded that ATM is likely to play an important role at the pregerminal center stage and a model is proposed where loss of ATM function during B-cell ontogeny drives B-CLL tumorigenesis in pregerminal B cells by a dual defect in p53 damage response and repair of chromosome breaks.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) is the most common leukemia in the western world. It is a heterogeneous disease with variable phenotypic and biologic features.1Two distinct subsets of B-CLL have recently been defined by the stage of B-cell differentiation in which they are transformed.2,3 B-CLL can develop either in B cells that have not entered the germinal center and, consequently, do not show the presence of VH somatic hypermutation or, from postgerminal center B cells that show frequent changes within their consensus VH sequences.4 B-CLLs with and without VH somatic hypermutation have different clinical outcomes and it was suggested that the VH status of B-CLL tumors reflects different pathogenic mechanisms involved in the development of this malignancy.2 3 It is conceivable that these differences are caused by the involvement of different molecular events.
Patients with inherited inactivation of both alleles of the ataxia telangiectasia mutated (ATM) gene show a high predisposition for the development of leukemia and lymphoma and hypersensitivity to agents causing DNA double-strand breaks (DSBs).5 The ATM protein has been proposed to function in multiple biochemical pathways linking the recognition and repair of DNA DSBs to downstream cellular processes, such as cell cycle regulation and programmed cell death.6 Some of the biologic functions of ATM are exerted through p53 phosphorylation and activation of the p53 pathway.6 Although there is some overlap in the spectrum of DNA damage to which ATM and p53 respond, ATM is restricted to response to DNA DSBs and is particularly important in lymphoid development during which there is frequent processing of such breaks. There is a predominance of T-cell over B-cell tumors in A-T individuals, with chromosome translocations involving T-cell receptor (TCR) genes contributing to the development of T-cell tumors.5ATM also has a role, through chromosome translocation, in the pathogenesis of sporadic T prolymphocytic leukemia (T-PLL).7-9 The mechanism of development of B-cell tumors in A-T individuals is largely unknown.
We showed recently that ATM mutations are frequent in sporadic B-CLL10 and that they can cause a p53 defect in response to irradiation11 in the absence ofTP53 mutations. However, despite the observation thatATM mutations can be observed in B-CLL tumors,10-13 the relationship between ATM inactivation and pathogenesis of sporadic B-CLL is still unclear. In the present study we have addressed the possibility that inactivation of theATM gene represents a molecular event that gives rise to a distinct subset of sporadic B-CLL tumors with a common pathogenesis. We show that ATM mutations are restricted to B-CLL with pregerminal center cell characteristics. We provide evidence that in these tumors, ATM inactivation occurs independently ofTP53 gene alterations and that functional loss of ATM determines a distinctive cellular response to DNA damage. Our results suggest a role for ATM in preventing both the accumulation of cells carrying DNA damage during B-cell differentiation and the consequent development of tumors at the pregerminal center cell stage.
Patients, materials, and methods
Patients
Peripheral blood was obtained after fully informed consent from 50 unselected patients with classical CD5+ and CD23+ B-CLL. There were 12 females and 38 males in our group of B-CLL patients. The median age of patients with ATMmutant tumors was 72.5 years (range, 39-90 years), whereas the median age in the group of patients with ATM wild-type tumors was 72.6 (range, 40-92 years). All B-CLL patients were followed for at least 2 years; the longest follow-up was 15 years. The median follow-up was 5.7 years (range, 3-10 years) for patients with ATMmutant B-CLL tumors and 7.6 years (range, 2-15 years) for patients withATM wild-type B-CLL tumors.
Stage of disease was defined according to National Cancer Institute (NCI) criteria.14 Four patients withATM mutant B-CLL were classified as in early disease (stage A) and 12 were classified as being at either stage B or C. Among patients with ATM/TP53 wild-type tumors, 14 were classified as stage A and 14 as stage B or C. Five patients with TP53mutant tumors were at either stage B or C at the time of analysis (Tables 1,2, and3). Nine of 16 patients withATM mutant B-CLL, 11 of 28 patients with ATM/TP53wild-type B-CLL, and 5 patients with TP53 mutant B-CLL were treated prior to collection and subsequent analysis of material (Tables1-3).
Genotypic and phenotypic features of B-CLL tumors withATM mutations
| Tumor . | ATM protein . | ATM mutation base (amino acid change) . | Germline/ somatic . | TP53 gene . | p53/MDM2/p21 induction . | Nearest VH/D/J family . | % Homology . | Karyotype . | Stage . | Th . |
|---|---|---|---|---|---|---|---|---|---|---|
| B-CLL1 | A | 1058delGT | G | WT | D | V1-02 | 100 | 45XY,-13,der(21)t(13;21) | B | C |
| G5464A(E1822Q) | Acq | D21-9 | (q12;p10)/46XY,add(4)(p1)/ | |||||||
| 46XY | ||||||||||
| B-CLL33 | A | G8600GA (G2867E) | Acq | WT | D | V3-11 | 100 | 46XX,del11q23 | C | E/C |
| Pre-l | ||||||||||
| LOH across ATM | D-39 | |||||||||
| B-CLL34 | A | 2114insA | Acq | WT | D | V3-23 | 99.6 | 46XY,der6;t(6;15)(p23;q14); | C | C |
| Pre-l | D3-16 | der18;t(8;18)(q11;p11) | ||||||||
| 4393insA | Acq | |||||||||
| B-CLL2 | A | G1048T(A350T) | Acq | WT | ND | V1-02 | 100 | ND | C | C/F |
| T1055C(I352T) | Acq | D3-22 | ||||||||
| B-CLL4 | R | G6820A (A2274K) | G | WT | D | V1-69 | 100 | ND | A | No |
| 6277delC | Acq | Dns/J6 | ||||||||
| B-CLL35 | R | A8266T(K2756stop) | G | WT | D | V3-33 | 99.4 | 46XX | B | No |
| D3-3 | ||||||||||
| B-CLL7 | R | G8084C (G2695A) | Acq | WT | D | V3-72 | 100 | 46XY,der(8)add(8)(p23)add | C | C/F/ |
| G7351A (A2451T) | Acq | D2-2 | (8)(q24),del(13)(q14q22)/ | CHOP | ||||||
| J4 | 46XY | |||||||||
| B-CLL36 | N | A8839C (T2947S) | ND | WT | D | V1-69 | 100 | 46XY,del11q23 | C | No |
| LOH across ATM | D2-21 | |||||||||
| B-CLL37 | N | T6055G (Y2019D) | ND | WT | D | V3-48 | 100 | ND | C | C |
| D21-10 | ||||||||||
| BCLL38* | N | C1300T (P434S) | ND | WT | D‡ | V1-46 | 99.3 | 46XY | C | C |
| D3-10 | ||||||||||
| B-CLL39 | N | A6295C(H2099Y) | ND | C1004T | D‡ | V1-69 | 100 | 46XY,del13q14 | B | C |
| (silent) | D3-10 | |||||||||
| B-CLL40 | R | G3556T(E1186stop) | ND | WT | D‡ | V4-59 | 98.9 | ND | C | C |
| D5-12 | ||||||||||
| B-CLL6† | R | 3994ins190 | Acq | WT | N | V3-73 | 100 | ND | A | No |
| B-CLL41 | R | 4829delG | ND | WT | N | V1-46 | 99.1 | ND | B | No |
| Dns | ||||||||||
| B-CLL42 | R | 7511del62 | Acq | WT | N | V3-33 | 99.7 | ND | A | No |
| D3-10 | ||||||||||
| BCLL5 | R | 3910del7 | Acq | WT | N | V3-23 | 99.6 | 46XYdel13q(q12;q32) or | A | No |
| Dns | (q14;q34) |
| Tumor . | ATM protein . | ATM mutation base (amino acid change) . | Germline/ somatic . | TP53 gene . | p53/MDM2/p21 induction . | Nearest VH/D/J family . | % Homology . | Karyotype . | Stage . | Th . |
|---|---|---|---|---|---|---|---|---|---|---|
| B-CLL1 | A | 1058delGT | G | WT | D | V1-02 | 100 | 45XY,-13,der(21)t(13;21) | B | C |
| G5464A(E1822Q) | Acq | D21-9 | (q12;p10)/46XY,add(4)(p1)/ | |||||||
| 46XY | ||||||||||
| B-CLL33 | A | G8600GA (G2867E) | Acq | WT | D | V3-11 | 100 | 46XX,del11q23 | C | E/C |
| Pre-l | ||||||||||
| LOH across ATM | D-39 | |||||||||
| B-CLL34 | A | 2114insA | Acq | WT | D | V3-23 | 99.6 | 46XY,der6;t(6;15)(p23;q14); | C | C |
| Pre-l | D3-16 | der18;t(8;18)(q11;p11) | ||||||||
| 4393insA | Acq | |||||||||
| B-CLL2 | A | G1048T(A350T) | Acq | WT | ND | V1-02 | 100 | ND | C | C/F |
| T1055C(I352T) | Acq | D3-22 | ||||||||
| B-CLL4 | R | G6820A (A2274K) | G | WT | D | V1-69 | 100 | ND | A | No |
| 6277delC | Acq | Dns/J6 | ||||||||
| B-CLL35 | R | A8266T(K2756stop) | G | WT | D | V3-33 | 99.4 | 46XX | B | No |
| D3-3 | ||||||||||
| B-CLL7 | R | G8084C (G2695A) | Acq | WT | D | V3-72 | 100 | 46XY,der(8)add(8)(p23)add | C | C/F/ |
| G7351A (A2451T) | Acq | D2-2 | (8)(q24),del(13)(q14q22)/ | CHOP | ||||||
| J4 | 46XY | |||||||||
| B-CLL36 | N | A8839C (T2947S) | ND | WT | D | V1-69 | 100 | 46XY,del11q23 | C | No |
| LOH across ATM | D2-21 | |||||||||
| B-CLL37 | N | T6055G (Y2019D) | ND | WT | D | V3-48 | 100 | ND | C | C |
| D21-10 | ||||||||||
| BCLL38* | N | C1300T (P434S) | ND | WT | D‡ | V1-46 | 99.3 | 46XY | C | C |
| D3-10 | ||||||||||
| B-CLL39 | N | A6295C(H2099Y) | ND | C1004T | D‡ | V1-69 | 100 | 46XY,del13q14 | B | C |
| (silent) | D3-10 | |||||||||
| B-CLL40 | R | G3556T(E1186stop) | ND | WT | D‡ | V4-59 | 98.9 | ND | C | C |
| D5-12 | ||||||||||
| B-CLL6† | R | 3994ins190 | Acq | WT | N | V3-73 | 100 | ND | A | No |
| B-CLL41 | R | 4829delG | ND | WT | N | V1-46 | 99.1 | ND | B | No |
| Dns | ||||||||||
| B-CLL42 | R | 7511del62 | Acq | WT | N | V3-33 | 99.7 | ND | A | No |
| D3-10 | ||||||||||
| BCLL5 | R | 3910del7 | Acq | WT | N | V3-23 | 99.6 | 46XYdel13q(q12;q32) or | A | No |
| Dns | (q14;q34) |
B-CLL 38 also has sequence change A1704G (silent).
B-CLL 6 also has sequence change C3161G (P1054R), a rare polymorphism, observed in breast tumors.
B-CLLs 38, 39, and 40 showed a normal induction of p53, but defective induction of MDM2 and p21 following IR. WT indicates wild type; ND, not done; A, absent; R, reduced; N, normal; D, defective; Acq, acquired; G, germline; Pre-l, prelymphoid; ns, not specified; Th, previous therapy; C, chlorambucil; P, prednisolone; CHOP, cyclophosphomide, doxorubicin, vincristine, and prednisolone; F, fludarabine; E, etoposide; and No, no previous therapy given; LOH, loss of heterozygosity.
Genotypic and phenotypic features of B-CLL tumors withTP53 mutations
| Tumor . | ATM protein . | ATM mutation . | TP53mutation (amino acid change) . | Germline/ somatic . | Allelic status . | p53/MDM2/p21 induction . | VH/D/J family . | % Homology . | Stage . | Th . |
|---|---|---|---|---|---|---|---|---|---|---|
| B-CLL9 | N | WT | C844T | Acq | Hemizygous | D | V3-30.3 | 100 | B | C/P |
| (R282W) | D3-22/J6 | |||||||||
| B-CLL43* | N | WT | C472G | Acq | Hemizygous | D | VH5-51 | 100 | C | CPFCy |
| (R158G) | D3-10 | |||||||||
| B-CLL44* | N | WT | G412C | Acq | Hemizygous | ND | V1-69 | 99.6 | C | NA |
| (A138P) | Dns/J5 | |||||||||
| B-CLL45 | N | WT | 1207ins133bp | Acq | Heterozygous | D | V3-30.3 | 99.4 | C | C |
| D1-7 | ||||||||||
| B-CLL66 | N | WT | 210ins2bp | Acq | Heterozygous | D | V3-11 | 100 | C | E/C |
| D3-9 | ||||||||||
| B-CLL46† | N | WT | A701G | G | Heterozygous | D | V3-21 | 94.6 | C | F |
| (Li-Fraumeni) | (Y234C) | D2-9 |
| Tumor . | ATM protein . | ATM mutation . | TP53mutation (amino acid change) . | Germline/ somatic . | Allelic status . | p53/MDM2/p21 induction . | VH/D/J family . | % Homology . | Stage . | Th . |
|---|---|---|---|---|---|---|---|---|---|---|
| B-CLL9 | N | WT | C844T | Acq | Hemizygous | D | V3-30.3 | 100 | B | C/P |
| (R282W) | D3-22/J6 | |||||||||
| B-CLL43* | N | WT | C472G | Acq | Hemizygous | D | VH5-51 | 100 | C | CPFCy |
| (R158G) | D3-10 | |||||||||
| B-CLL44* | N | WT | G412C | Acq | Hemizygous | ND | V1-69 | 99.6 | C | NA |
| (A138P) | Dns/J5 | |||||||||
| B-CLL45 | N | WT | 1207ins133bp | Acq | Heterozygous | D | V3-30.3 | 99.4 | C | C |
| D1-7 | ||||||||||
| B-CLL66 | N | WT | 210ins2bp | Acq | Heterozygous | D | V3-11 | 100 | C | E/C |
| D3-9 | ||||||||||
| B-CLL46† | N | WT | A701G | G | Heterozygous | D | V3-21 | 94.6 | C | F |
| (Li-Fraumeni) | (Y234C) | D2-9 |
For abbreviations, see Table 1.
Patients with tumors B-CLL 43 and 44 were identical twins.
Patient with B-CLL 46 had Li Fraumeni syndrome.
Cy indicates cyclophosphamide.
Genotypic and phenotypic features of B-CLL tumors with wild-type ATM/TP53
| Patient . | ATM gene . | TP53gene . | p53/MDM2/p21 induction . | Nearest VH/D/J gene . | % Homology . | Stage . | Th . |
|---|---|---|---|---|---|---|---|
| B-CLL47 | WT | WT | Normal | V4-34 | 95.5 | A | No |
| D5-5 | |||||||
| B-CLL48 | WT | WT | Normal | V1-03 | 81.9 | A | No |
| D2-2 | |||||||
| B-CLL13 | WT | WT | Normal | V3-30.3 | 99.6 | C | No |
| D3-3/J4 | |||||||
| B-CLL15 | WT | WT | Normal | V1-58 | 92.7 | B | No |
| D6-6/J4 | |||||||
| B-CLL19 | WT | WT | Normal | V3-30.3 | 89.9 | A | No |
| D3-3 | |||||||
| B-CLL49 | WT | WT | Normal | V5-11 | 95 | A | No |
| D21-7/J4 | |||||||
| B-CLL20 | WT | WT | Normal | V1-02 | 99.3 | C | C |
| D3-22/J4 | |||||||
| B-CLL50 | WT | WT | Normal | V1-12 | 95 | C | C |
| B-CLL51 | WT | WT | Normal | V3-23 | 98.6 | A | No |
| D4-17 | |||||||
| B-CLL52 | WT | WT | Normal | V1-69 | 94.8 | A | No |
| D3-16 | |||||||
| B-CLL53 | WT | WT | Normal | V3-21 | 97.3 | B | No |
| D3-9 | |||||||
| B-CLL54 | WT | WT | Normal | V3-74 | 93.5 | C | F |
| D5-12 | |||||||
| B-CLL553-150 | WT | WT | Normal | V3-74 | 100 | C | E/C/F |
| LOH | D3-10 | ||||||
| B-CLL56 | WT | WT | Normal | V4-41 | 92.1 | A | No |
| D1-7 | |||||||
| B-CLL57 | WT | WT | Normal | V1-08 | 90 | A | No |
| D3-10 | |||||||
| B-CLL583-150 | WT | WT | Normal | V4-15 | 97.9 | C | E/C/F |
| LOH | D5-24 | ||||||
| B-CLL59 | WT | WT | Normal | V3-30.3 | 98.2 | B | C |
| D3-3 | |||||||
| B-CLL60 | WT | WT | Normal | V3-21 | 97.2 | C | C |
| D4-4 | |||||||
| B-CLL61 | WT | WT | Normal | V3-23 | 95 | A | No |
| B-CLL68 | WT | WT | ND | V3-43 | 90 | A | No |
| B-CLL62 | WT | WT | ND | V3-33 | 96.8 | A | No |
| B-CLL63 | WT | WT | ND | V3-48 | 89.6 | C | F |
| D3-9/J6 | |||||||
| B-CLL29 | WT | WT | ND | V3-23 | 94.1 | A | No |
| B-CLL11 | WT | WT | ND | VDJ ns | ns | C | C/CHOP |
| B-CLL64 | WT | WT | ND | V1-2 | 99.5 | A | No |
| D3-10 | |||||||
| B-CLL10 | WT | WT | ND | V1-02 | 99.6 | B | C/P |
| D3-3 | |||||||
| B-CLL67 | WT | WT | ND | V4-39 | 95.3 | B | E/C |
| D2-9 | |||||||
| B-CLL65 | WT | WT | ND | V3-72 | 91.9 | A | C |
| D3-9 |
| Patient . | ATM gene . | TP53gene . | p53/MDM2/p21 induction . | Nearest VH/D/J gene . | % Homology . | Stage . | Th . |
|---|---|---|---|---|---|---|---|
| B-CLL47 | WT | WT | Normal | V4-34 | 95.5 | A | No |
| D5-5 | |||||||
| B-CLL48 | WT | WT | Normal | V1-03 | 81.9 | A | No |
| D2-2 | |||||||
| B-CLL13 | WT | WT | Normal | V3-30.3 | 99.6 | C | No |
| D3-3/J4 | |||||||
| B-CLL15 | WT | WT | Normal | V1-58 | 92.7 | B | No |
| D6-6/J4 | |||||||
| B-CLL19 | WT | WT | Normal | V3-30.3 | 89.9 | A | No |
| D3-3 | |||||||
| B-CLL49 | WT | WT | Normal | V5-11 | 95 | A | No |
| D21-7/J4 | |||||||
| B-CLL20 | WT | WT | Normal | V1-02 | 99.3 | C | C |
| D3-22/J4 | |||||||
| B-CLL50 | WT | WT | Normal | V1-12 | 95 | C | C |
| B-CLL51 | WT | WT | Normal | V3-23 | 98.6 | A | No |
| D4-17 | |||||||
| B-CLL52 | WT | WT | Normal | V1-69 | 94.8 | A | No |
| D3-16 | |||||||
| B-CLL53 | WT | WT | Normal | V3-21 | 97.3 | B | No |
| D3-9 | |||||||
| B-CLL54 | WT | WT | Normal | V3-74 | 93.5 | C | F |
| D5-12 | |||||||
| B-CLL553-150 | WT | WT | Normal | V3-74 | 100 | C | E/C/F |
| LOH | D3-10 | ||||||
| B-CLL56 | WT | WT | Normal | V4-41 | 92.1 | A | No |
| D1-7 | |||||||
| B-CLL57 | WT | WT | Normal | V1-08 | 90 | A | No |
| D3-10 | |||||||
| B-CLL583-150 | WT | WT | Normal | V4-15 | 97.9 | C | E/C/F |
| LOH | D5-24 | ||||||
| B-CLL59 | WT | WT | Normal | V3-30.3 | 98.2 | B | C |
| D3-3 | |||||||
| B-CLL60 | WT | WT | Normal | V3-21 | 97.2 | C | C |
| D4-4 | |||||||
| B-CLL61 | WT | WT | Normal | V3-23 | 95 | A | No |
| B-CLL68 | WT | WT | ND | V3-43 | 90 | A | No |
| B-CLL62 | WT | WT | ND | V3-33 | 96.8 | A | No |
| B-CLL63 | WT | WT | ND | V3-48 | 89.6 | C | F |
| D3-9/J6 | |||||||
| B-CLL29 | WT | WT | ND | V3-23 | 94.1 | A | No |
| B-CLL11 | WT | WT | ND | VDJ ns | ns | C | C/CHOP |
| B-CLL64 | WT | WT | ND | V1-2 | 99.5 | A | No |
| D3-10 | |||||||
| B-CLL10 | WT | WT | ND | V1-02 | 99.6 | B | C/P |
| D3-3 | |||||||
| B-CLL67 | WT | WT | ND | V4-39 | 95.3 | B | E/C |
| D2-9 | |||||||
| B-CLL65 | WT | WT | ND | V3-72 | 91.9 | A | C |
| D3-9 |
B-CLLs 55 and 58 had 11q deletion and LOH acrossATM region.
For abbreviations, see Table 1.
Cell separation, DNA/RNA extraction
The mononuclear cell fraction, containing tumor cells, was separated on density gradient (Lymphoprep, Axis-Shield, PoCAs, Oslo, Norway) and frozen viable in 90% fetal calf serum (FCS)/10% dimethyl sulfoxide (DMSO). Cell samples were thawed at 37°C, washed twice in RPMI medium and cultured overnight in RPMI/10% FCS cells prior to use in damage response assays or RNA and DNA extraction.
For the separation of B and T lymphocytes from case B-CLL 33, fresh mononuclear cells were stained with either CD19PE or CD3PE antibody (Beckman-Coulter) prior to magnetic selection using antiphycoerythrin (PE) supermagnetic microbeads, and the Posseld selection program on an AutoMACS cell separator (Miltenyi-Biotec, Bergisch, Gladbach, Germany). Purity was assessed by flow cytometry using a Coulter EPICS XL. Monocytes were purified by forward and side scatter gating and collection of CD14+/− cells. In B-CLL 34 CD19+ tumor cells were separated by magnetic beads coated with CD19 antibody (Dynal).
Patients' germline DNA was extracted from buccal cavity epithelial cells. Lymphoblastoid cell lines from an A-T patient and 2 healthy individuals were also used for control purposes.
Mutation analysis
Mutation analysis of the ATM coding sequence was performed on the tumor cDNA by reverse transcriptase–polymerase chain reaction (RT-PCR) and restriction endonuclease fingerprinting (REF),15,16 or by complete sequencing. Most of the mutations were verified in tumor DNA. Mutation analysis of theTP53 gene was performed using either amplification and sequencing of the whole TP53 coding sequence from the tumor cDNA, or alternatively, by amplification and sequencing of exons 4 to 10 from tumor DNA using primers previously described.11The allelic status of both ATM and TP53 genes (homozygous/heterozygous/hemizygous) was determined by the presence or absence of the second allele on REF gels, sequence chromatograms, and when possible, by microsatellite analysis of the paired tumor and germline DNA.
Haplotype analysis
Multiallelic short tandem repeats from DNA samples were analyzed for 8 loci, D11S1787, 1819, 2179, 1778, 1294, 1818, 2180, and 2178, on chromosome 11q22-23 as described before.15
Variable diversity joining region amplification and analysis of VH somatic mutations
IgH variable density joining region (VDJ) rearrangements were amplified using primers from the consensus sequences of the framework 1 (FR1) and joining region (J1H) of the immunoglobulin heavy chain gene. Products were column-purified (Boehringer) and subjected to automated DNA sequencing using the JPS primer internal to J1H. The nucleotide sequence of each rearrangement was used to search the European Molecular Biology Laboratory (EMBL) and GenBank databases and the variable region database (VBASE) directory of all human germline variable region sequences.17,18 Sequences that had fewer than 2% differences compared with the germline sequence of their V gene family were taken to indicate tumors lacking somatic hypermutation.17
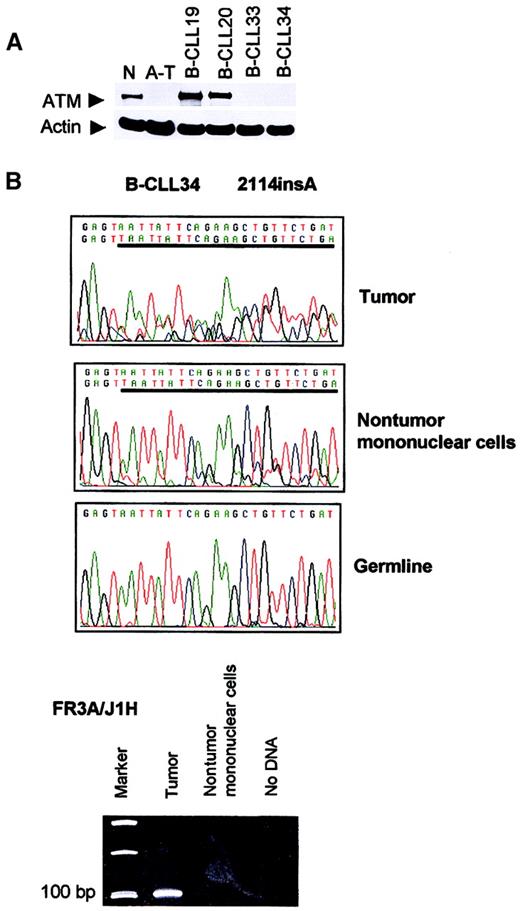
To identify the presence of tumor cells in the nonlymphoid compartment in B-CLL 34, VDJ rearrangements were amplified using a primer corresponding to the consensus sequence of the framework 3 IgH region (FR3A) together with J1H primer.
Protein analysis
Samples containing more than 95% viable cells, as assessed by trypan blue exclusion, were included in experiments. Cells were cultured overnight in 24-well plates with RPMI medium and 10% FCS, irradiated with 5 Gy gamma rays, kept at 37°C and harvested at defined time points.
The lysates were obtained by sonication of cells in 8 M urea, 150 mM β- mercaptoethanol, 50 mM Tris HCl, pH7.5, and centrifugation for 30 minutes at 9°C. Samples were subjected to electrophoresis on 6% to 10% sodium dodecyl sulfate (SDS)–polyacrylamide gels, blotted on nitrocellulose membranes, and screened for protein expression. A single nitrocellulose membrane was cut into adequate segments in order to perform simultaneous Western blotting using the ECL system (Amersham) and the following antibodies: ATM, rabbit FP8r15; p53, sheep Ab, donated by D. P. Lane (University of Dundee); MDM2, mAb 2A10, donated by R. Grand (University of Birmingham); and p21 rabbit Ab (Santa Cruz Biotechnology, Santa Cruz, CA). A separate gel was loaded for Western blotting with p53 serine 15P, rabbit Ab (New England Biolabs), and TRAIL-R2, goat Ab (Alexis). To verify that equivalent amounts of each sample were loaded, the filters were additionally probed with anti–actin-IgG (AC74, Sigma). Densitometry was performed routinely on each of the tested proteins as well as actin bands. A B-CLL tumor was assumed to have reduced ATM if expression of the protein was below the level of ATM expression in a group of 28 tumors with wild-type ATM/TP53.
Assessment of apoptosis
Apoptosis was measured by 2 independent methods: (1) propidium iodide (PI) incorporation and FACS analysis, and (2) staining with acridine orange and morphologic assessment. For PI staining and FACS analysis, cells were irradiated with gamma rays (5 Gy), harvested at defined time points, washed twice in phosphate-buffered saline (PBS), briefly fixed in 70% ethanol, and mixed with an equal volume of PI (10 mg/mL). Cell cycle analysis was performed using a Coulter XL flow cytometer. The apoptotic cell fraction was detected in the red fluorescent sub-G1 peak separate from cells with either G1 or S/G2M content. Ratios of the areas of the sub-G1 and G1 peaks were obtained for each time point after irradiation and expressed as surviving fraction.
For acridine orange staining, cells were mixed with an equal volume of acridine orange (10 mg/mL) and at each time point the proportion of cells showing fragmentation of nuclei was determined by microscopy.
Chromosome radiosensitivity and karyotype analysis
Peripheral blood from patients with B-CLL was cultured for 5, 6, or 7 days in RPMI medium containing 10% FCS, glutamine, penicillin, streptomycin, and 0.1 mg/mL phorbol myristate acetate (PMA) (Sigma). Cells were irradiated with 1 gray x-rays (1 Gy per minute) and harvested 4 hours later. Chromosome preparations were made, stained with Giemsa, and the G2 x-ray–induced damage was analyzed. Chromosome analysis of B-CLLs, for karyotyping, was carried out using standard G-banding techniques following culturing of the B-CLL tumor cells in PMA for 5, 6, or 7 days.
Results
ATM mutations are independent ofTP53 mutations in B-CLL
In a group of 50 B-CLL tumors (different from that in Pettitt et al11), where sufficient cells were available for a range of assays, we analyzed the status of TP53 and ATMusing the REF method in combination with the sequencing of the entireATM coding region. ATM mutations were identified in 16 B-CLLs, none of which also had a TP53 mutation (Table1). In 2 of these tumors (B-CLL 33 and B-CLL 36) there was evidence for the loss of one ATM allele and the presence of mutation in the other. In an additional 5 tumors (B-CLLs 1, 34, 2, 4, and 7), 2ATM mutations were identified. Five tumors (B-CLLs 35, 37, 38, 39, and 40) each had a single point mutation, 2 of which (in B-CLL 35 and B-CLL 40) were predicted to lead to the premature termination of the protein. The missense mutations identified were in regions conserved between mouse and man and not previously reported as polymorphisms. Finally, 4 tumors were detected carrying a single small deletion or insertion predicted also to lead to protein truncation (B-CLLs 6, 41, 42, and 5).
A separate group of 6 B-CLLs (B-CLLs 9, 43, 44, 45, 46, and 66), each with a single TP53 mutation, was identified. Three tumors were hemizygous and the other 3 heterozygous for a TP53mutation. The patient with B-CLL 46 had previously been diagnosed as having Li-Fraumeni syndrome and, as expected, the TP53mutation was present in the germline (Table 2). The remaining 28 B-CLL tumors showed absence of both ATM and TP53mutations (Table 3).
Compared with the ATM/TP53 wild-type B-CLLs, theATM mutant subset showed a higher percentage of stage B and C tumors (75% vs 50%). This difference, however, was not statistically significant (P = .125, Fisher exact test).
Although based on relatively small numbers, our results raise the possibility that the simultaneous occurrence of ATM andTP53 mutations is either a mutually exclusive event, or certainly rare, in the development of B-CLL.
ATM mutant tumors are of pregerminal center B-cell origin lacking VH somatic hypermutation
In order to determine whether all ATM-mutated B-CLLs were derived from the same stage of B-cell differentiation, we analyzed the VH status of the ATM mutant tumors and compared this with ATM/TP53 wild-type tumors.
Clonal IgH VDJ rearrangements were amplified and sequenced from 50 B-CLL tumors previously characterized for the presence of either theATM or TP53 mutation. Sequencing revealed that all 16 tumors with ATM inactivation showed absence of somatic hypermutation (Table 1) suggesting that these were derived from a progenitor bearing characteristics of pregerminal center B cells.
Absence of VH somatic mutations was also noted in all 5 tumors with acquired TP53 mutations, whereas the sixth TP53mutant B-CLL from a patient with Li-Fraumeni syndrome showed the presence of VH somatic hypermutations (Table 2). In contrast, the majority of tumors with wild-type TP53/ATM (20 of 28) showed the presence of multiple somatic mutations in their VH regions (Table3).
Our finding is consistent with a common mechanism of tumorigenesis in B-CLL tumors with ATM inactivation.
ATM mutations can be acquired early during tumorigenesis in B-CLL
If ATM inactivation is involved in B-CLL transformation at the pregerminal B-cell stage, mutations should be acquired prior to this point. We tested this possibility by searching for ATMmutations at 2 points: in the patients' germlines and in non–B cells.
First, ATM germline mutations were identified in 3 patients with B-CLL (B-CLLs 1, 4, and 35). The previously reported mutation 1058delGT10 in B-CLL 1 and mutation A8266T detected in B-CLL 35, were truncating and could not be polymorphisms. In addition, we identified mutation A8266AT in an A-T family15 (Table1). Microsatellite marker analysis (data not shown) provided convincing evidence that B-CLL 35 carried the same haplotype in the region of theATM gene as the affected individual from family 63, giving provenance to the pathogenic nature of this mutation. G6820A observed in the germline of B-CLL 4 was likely to be a real mutation as we were not able to identify this change in DNA from 140 normal meioses.
Second, we were able to obtain separated tumor and nontumor mononuclear cells in 2 additional patients with B-CLL (B-CLLs 33 and 34). Truncating mutation 2114insA in B-CLL 34 was present in both tumor cells and the nontumor mononuclear compartment containing T cells, NK cells, and monocytes, but not in the buccal smear DNA (Figure1). A clonal IgH rearrangement could be detected in the tumor population, but not in nontumor cells (Figure 1), suggesting that cross contamination of the nontumor cells with tumor cells was unlikely to be the cause of the presence of theATM mutations in this compartment. Rather, the results suggested that the mutation 2114insA arose in a progenitor at a stage prior to B-lymphoid commitment.
Absence of ATM in B-CLLs 33 and 34 and presence of
ATM mutation 2114insA in B-CLL 34 nontumor mononuclear cells. (A) Western blot showing complete loss of ATM in B-CLL tumors 33 and 34. N indicates normal lymphoblastoid cell line (LCL); A-T, LCL derived from an ataxia telangiectasia patient with 2 nullATM alleles; B-CLL 19 and 20, 2 tumors withoutATM mutations; B-CLL 33 and 34, tumors with 2 ATMmutations (or LOH). (B) Truncating mutation 2114insA (given as reverse sequence) in patient B-CLL 34 was present in DNA from both tumor (CD19+) and nontumor (CD19−) compartment, but not present in the buccal smear DNA. An immune system gene rearrangement could be detected only from the tumor population of patient B-CLL 34, but not from the nontumor cells, suggesting the absence of cross contamination between nontumor and tumor cells.
Absence of ATM in B-CLLs 33 and 34 and presence of
ATM mutation 2114insA in B-CLL 34 nontumor mononuclear cells. (A) Western blot showing complete loss of ATM in B-CLL tumors 33 and 34. N indicates normal lymphoblastoid cell line (LCL); A-T, LCL derived from an ataxia telangiectasia patient with 2 nullATM alleles; B-CLL 19 and 20, 2 tumors withoutATM mutations; B-CLL 33 and 34, tumors with 2 ATMmutations (or LOH). (B) Truncating mutation 2114insA (given as reverse sequence) in patient B-CLL 34 was present in DNA from both tumor (CD19+) and nontumor (CD19−) compartment, but not present in the buccal smear DNA. An immune system gene rearrangement could be detected only from the tumor population of patient B-CLL 34, but not from the nontumor cells, suggesting the absence of cross contamination between nontumor and tumor cells.
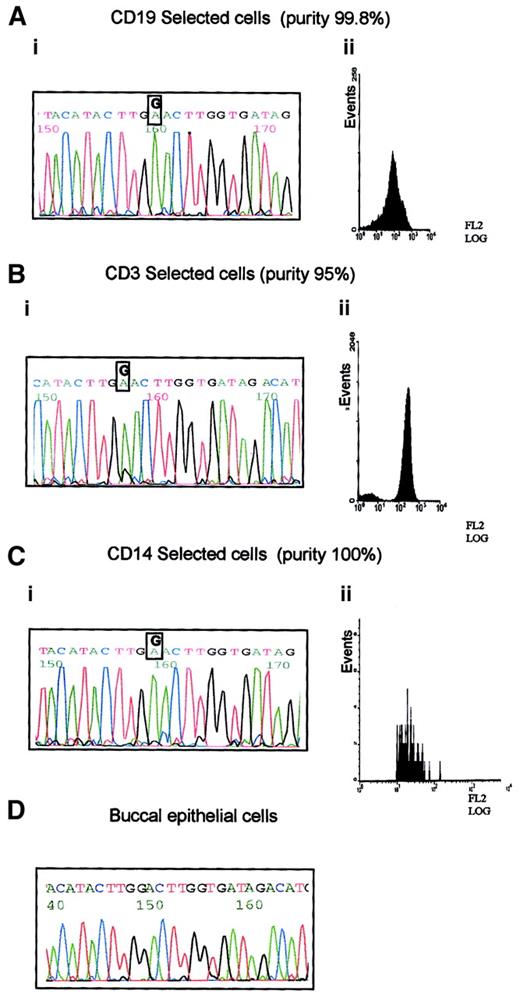
B-CLL33 carried the G8600A mutation, which is predicted to cause a significant amino-acid change in the ATM PI-3 kinase domain. This case was used to define the hematopoietic progenitor targeted by this mutation. The following fractions of high purity were obtained: CD19+ separated cells containing mostly B-CLL tumor cells; CD14+ sorted cells containing monocytes, and CD3+ sorted cells containing T lymphocytes (Figure2). Both the tumor B cells and the monocyte fraction showed the presence of the G8600A mutant allele alone, whereas the pure T cells, in addition to the mutant allele, revealed a small peak of the normal sequence (Figure 2). The prominence of the mutant allele in the DNA sequences of all analyzed subpopulations and the absence of the mutation in the germline from the buccal smear, suggested that all 3 blood lineages (T, B, and monocyte) were derived from a single hematopoietic precursor carrying the G8600A mutation together with the loss of a normal ATM allele. The presence of a minor germline peak in the T-cell population is consistent with a subpopulation of long-living T cells existing before the evolution of a lineage noncommitted progenitor carrying the G8600A mutation. We concluded that in B-CLL 33 not only ATMmutation, but complete ATM inactivation, was acquired early during hematopoiesis, well before the stage of tumor development, as a result of affecting more than one lineage.
Prelymphoid origin of G8600A mutation in B-CLL 33.
(A) G8600A mutation in DNA from tumor cells of patient with B-CLL 33 (i), separated to 99.8% purity using CD19 PE-coated magnetic beads (ii). (B) G8600A mutation together with a small peak of a normal sequence in DNA from CD3+ cells of patient with B-CLL 33 (i), separated to 95% purity using CD3 PE-coated magnetic beads (ii). (C) G8600A mutation in DNA from CD14+ cells of patient with B-CLL 33 (i) separated to 100% purity using CD14 labeling and FACS sorting (ii). (D) Absence of G8600A mutation in patient's buccal epithelial cells.
Prelymphoid origin of G8600A mutation in B-CLL 33.
(A) G8600A mutation in DNA from tumor cells of patient with B-CLL 33 (i), separated to 99.8% purity using CD19 PE-coated magnetic beads (ii). (B) G8600A mutation together with a small peak of a normal sequence in DNA from CD3+ cells of patient with B-CLL 33 (i), separated to 95% purity using CD3 PE-coated magnetic beads (ii). (C) G8600A mutation in DNA from CD14+ cells of patient with B-CLL 33 (i) separated to 100% purity using CD14 labeling and FACS sorting (ii). (D) Absence of G8600A mutation in patient's buccal epithelial cells.
Cellular features of ATM mutant B-CLLs
We first asked whether all ATM mutations in the B-CLLs were pathogenic. We took as possible indicators of a pathogenic effect (1) a decrease in ATM protein expression, and (2) a functional assay showing defective p53 response following exposure of the tumor cells to ionizing radiation (IR). In tumors with pathogenicATM mutations, we then focused on 2 aspects of the cellular phenotype: identification of a common defect in the downstream target which provides a link between aberrant IR induction of p53 and resistance to apoptosis in these cells and the ability ofATM mutant tumor cells to repair chromosome damage following gamma irradiation.
ATM mutations and protein expression.
Four tumors (B-CLLs 1, 2, 33, and 34) with apparent alterations of bothATM alleles showed a complete absence of ATM expression (Table 1). In the remaining 12 tumors with ATM mutations, protein expression was variable: 8 tumors showed a clearly reduced expression of ATM compared with the level of ATM protein expression in 28 ATM wild-type tumors, and 4 B-CLLs showed normal ATM expression. It is noteworthy that in each of the 4 tumors with normal ATM expression a single missense mutation was identified. In one of these, B-CLL 36, the mutant ATM allele was the only allele present as there was an 11q deletion and LOH across theATM gene region, presumably reflecting loss of the normal allele.
ATM mutations and p53 cellular response to IR.
Our previous study with a different group of B-CLL tumors demonstrated that ATM mutations can cause a defect in p53 response to DNA damage induced by ionizing radiation.11 We used this observation to analyze the pathogenicity of ATMmutations in the present study. Of 15 ATM mutant tumors that could be tested, 8 showed a clear defect in both p53 accumulation and its transcriptional activity as measured by p21/MDM2 up-regulation. This included 2 tumors with 11q deletion and mutation in the otherATM allele (B-CLLs 33 and 36).
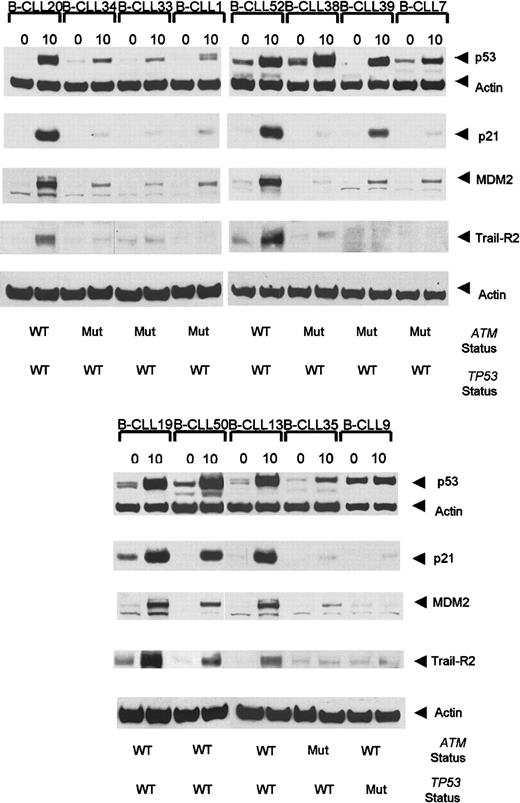
Interestingly, 3 ATM mutant tumors showed defective p21/MDM2 up-regulation despite the high accumulation of p53 (Table 1, Figure3). In addition, 4 B-CLL tumors, each with a single truncating ATM mutation, exhibited high p53/p21/MDM2 up-regulation (data not shown). As expected, p53/p21/MDM2 up-regulation was also high in 19 analyzed B-CLL tumors withATM/TP53 wild type (Table 3, Figure 3).
Ionizing radiation-induced accumulation of p53, p21, MDM2, and TRAIL-R2 at 0 hours and 10 hours following IR in
ATM mutant B-CLLs. B-CLLs 34, 33, 7, 1, and 35 are representative of 8 tumors with ATM mutations showing defective p53, p21, and MDM2 responses following exposure to IR. B-CLLs 38 and 39 are ATM mutant tumors with normal p53 accumulation but defective p21 and MDM2 up-regulation. A B-CLL tumor (B-CLL 9) withTP53 mutation showed an elevated basal p53 protein level but a defect in the ability to accumulate p53 and up-regulate p21 and MDM2. B-CLLs 20, 52, 19, 50, and 13 are representative of 19 analyzed tumors with wild-type ATM and p53 which showed normal p53, p21, and MDM2 responses. The TRAIL-R2 response 10 hours after irradiation was defective in ATM-deficient tumors (B-CLLs 34, 33, 1, 38, 39, 7, and 35) as well as in TP53 mutant (Mut) B-CLL 9, but it was normal in B-CLLs 20, 52, 19, 50, and 13 representative of 12 B-CLLs withATM/TP53 wild type (WT) analyzed for TRAIL-R2 response. The actin expression shows equal loading.
Ionizing radiation-induced accumulation of p53, p21, MDM2, and TRAIL-R2 at 0 hours and 10 hours following IR in
ATM mutant B-CLLs. B-CLLs 34, 33, 7, 1, and 35 are representative of 8 tumors with ATM mutations showing defective p53, p21, and MDM2 responses following exposure to IR. B-CLLs 38 and 39 are ATM mutant tumors with normal p53 accumulation but defective p21 and MDM2 up-regulation. A B-CLL tumor (B-CLL 9) withTP53 mutation showed an elevated basal p53 protein level but a defect in the ability to accumulate p53 and up-regulate p21 and MDM2. B-CLLs 20, 52, 19, 50, and 13 are representative of 19 analyzed tumors with wild-type ATM and p53 which showed normal p53, p21, and MDM2 responses. The TRAIL-R2 response 10 hours after irradiation was defective in ATM-deficient tumors (B-CLLs 34, 33, 1, 38, 39, 7, and 35) as well as in TP53 mutant (Mut) B-CLL 9, but it was normal in B-CLLs 20, 52, 19, 50, and 13 representative of 12 B-CLLs withATM/TP53 wild type (WT) analyzed for TRAIL-R2 response. The actin expression shows equal loading.
We conclude that ATM mutations in the majority of B-CLL tumors (11/15), led to a defective p53 response to IR-induced DNA damage, regardless of the level of ATM expression.
Resistance to IR-induced apoptosis in ATM-deficient B-CLL tumors is associated with defect in TRAIL-R2 up-regulation.
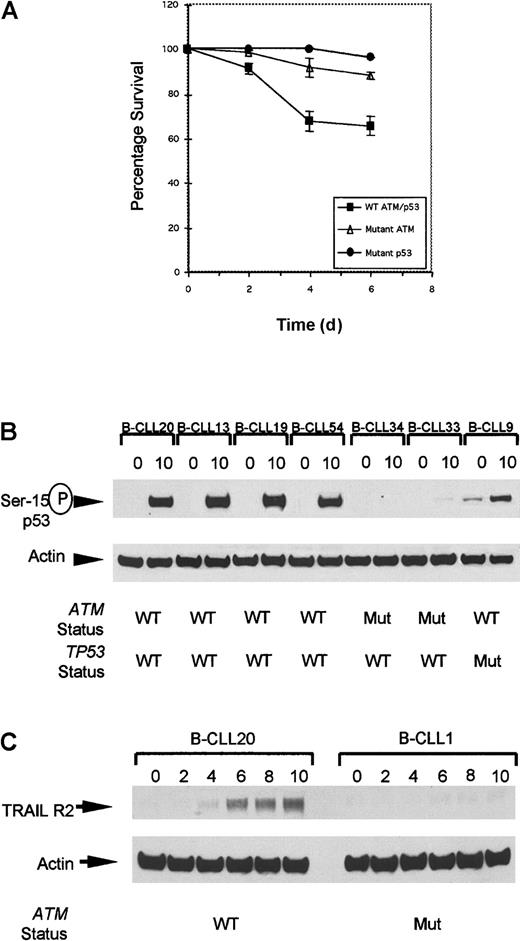
All 4 ATM mutant tumors completely lacking ATM expression in this series showed severely reduced apoptosis following IR (Figure4A). Furthermore, in 2 representatives of these tumors, despite some IR-induced p53 accumulation, there was a clear absence of p53 serine-15 phosphorylation (Figure 4B) at the time point used, consistent with the well-documented delay in p53 serine-15 phosphorylation in classical A-T cells. This modification is dependent on ATM and crucial for apoptotic activity of p53.19
Defect in cellular responses in
ATM mutant B-CLLs linking IR-induced p53 defect with resistance to apoptosis. (A) The rate of apoptosis after exposure to 5 Gy gamma irradiation was compared in B-CLL tumors either withATM mutations, TP53 mutations, or with wild-typeTP53/ATM sequence. Mean levels of apoptosis induced by ionizing radiation in B-CLLs with wild-type ATM (B-CLLs 13, 19, and 54), mutant ATM (B-CLLs 1, 2, 33, and 34), and also mutant p53 (B-CLLs 9 and 43) up to 6 days after irradiation. In unirradiated B-CLL cells the level of apoptosis was indistinguishable between the different genotypes at all 4 time points (data not shown). (B) Phosphorylation at serine-15 of p53 in B-CLLs 20, 13, 19, and 54 with wild-type ATM but not in B-CLLs 33 and 34 with mutantATM, or in TP53 mutant B-CLL 9 at 0 hours and 10 hours following irradiation. (C) Expression of TRAIL-R2 at 0, 2, 4, 6, 8, and 10 hours following IR with 5 Gy. Up-regulation reaches a peak 10 hours after irradiation in B-CLL20 with wild-type ATM. ATM mutant B-CLL 1 failed to up-regulate TRAIL-R2 following irradiation.
Defect in cellular responses in
ATM mutant B-CLLs linking IR-induced p53 defect with resistance to apoptosis. (A) The rate of apoptosis after exposure to 5 Gy gamma irradiation was compared in B-CLL tumors either withATM mutations, TP53 mutations, or with wild-typeTP53/ATM sequence. Mean levels of apoptosis induced by ionizing radiation in B-CLLs with wild-type ATM (B-CLLs 13, 19, and 54), mutant ATM (B-CLLs 1, 2, 33, and 34), and also mutant p53 (B-CLLs 9 and 43) up to 6 days after irradiation. In unirradiated B-CLL cells the level of apoptosis was indistinguishable between the different genotypes at all 4 time points (data not shown). (B) Phosphorylation at serine-15 of p53 in B-CLLs 20, 13, 19, and 54 with wild-type ATM but not in B-CLLs 33 and 34 with mutantATM, or in TP53 mutant B-CLL 9 at 0 hours and 10 hours following irradiation. (C) Expression of TRAIL-R2 at 0, 2, 4, 6, 8, and 10 hours following IR with 5 Gy. Up-regulation reaches a peak 10 hours after irradiation in B-CLL20 with wild-type ATM. ATM mutant B-CLL 1 failed to up-regulate TRAIL-R2 following irradiation.
We investigated the link between an aberrant IR-induced p53 response and decreased IR-induced apoptosis in B-CLL tumors. Many downstream targets have been proposed to mediate the apoptotic functions of the p53 protein, including the death effector Bax. The lack of ionizing radiation-induced up-regulation of Bax in any of our group of B-CLL tumors tested (data not shown) prompted us to investigate whether a defective induction of other p53-dependent downstream proapopototic targets could account for the apparent resistance to ionizing radiation–induced apoptosis in B-CLLs with ATM gene abnormalities.
Recently, transcriptional up-regulation of TRAIL Receptor 2 (TRAIL-R2) was reported to be a p53-dependent response specifically restricted to cells undergoing apoptosis and was shown to be defective in LCLs derived from classical A-T patients following exposure to IR.20 We determined first whether TRAIL-R2 was up-regulated in B-CLL cells without ATM mutations and then whether this was lost in ATM mutant B-CLLs. Western blot analysis revealed a peak of TRAIL-R2 up-regulation in aATM/TP53 wild-type B-CLL 8 hours to 10 hours following gamma irradiation (Figure 4C).
We next investigated whether ATM mutant tumors either with a complete p53/p21/MDM2 defect or with a defect in p21/MDM2 up-regulation, but with p53 accumulation, had the same impact on TRAIL-R2 up-regulation. Twenty B-CLL tumors from which material was available were analyzed at 0 hours and 10 hours after irradiation (Figure 3). Five B-CLLs (B-CLLs 1, 7, 33, 34, and 35) with a complete defect in p53 response, 2 B-CLLs (B-CLLs 38 and 39) with normal p53 response but defective p21/MDM2 up-regulation, and one TP53mutant (B-CLL9), clearly failed to up-regulate TRAIL-R2 10 hours after irradiation (Figure 3). In comparison, the 12 B-CLL tumors withATM/TP53 wild type (B-CLLs 13, 19, 20, 50, 52-55, 57, 59-61) showed strong up-regulation of TRAIL-R2. Five cases representative of the 12 analyzed ATM/TP53 wild-type tumors (B-CLLs 20, 52, 13, 19, and 50) are shown in Figure 3.
We conclude that TRAIL-R2 is a p53 downstream target linking IR-induced p53 response to apoptosis, and that this response is defective in bothATM and TP53 mutant B-CLL tumors.
ATM-deficient B-CLL tumors show reduced ability to repair chromosome damage.
Typical B-CLL is not characterized by the presence of translocations involving the immune system genes. Indeed, none of the 9 ATMmutant tumors for which karyotype was available showed the presence of translocations involving the immunoglobulin genes (Table 1), suggesting that errors during VDJ recombination did not play a role in their pathogenesis.
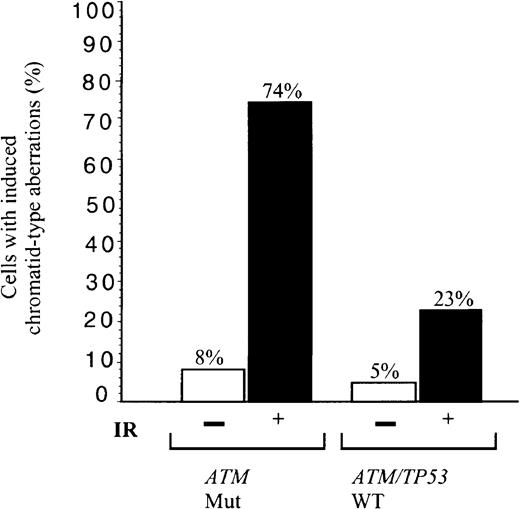
We asked, therefore, how effective tumor cells were in the repair of general non site-specific DNA damage induced by IR. B-CLLs either with total loss of ATM and defective induction of p53, p21/MDM2/TRAIL-R2 (B-CLLs 1 and 33) or mutant ATM with defective activation of p21/MDM2 (B-CLL 39) were irradiated. Of these irradiated ATM mutated B-CLL tumor cells, 74% showed unrepaired chromosome damage compared with 23% cells from 2 B-CLLs (51 and 56) with ATM/TP53 wild-type and normal p53 responses (Figure 5). In comparison, 16% of phorbol myristate acetate (PMA)–stimulated peripheral blood lymphocytes from 4 healthy individuals showed chromatid damage following exposure to the same dose of IR (not shown).ATM mutant B-CLLs, therefore, in addition to a defective p53 response to IR, have a defect in repair of some forms of chromosome damage.
Defect in repair of IR-induced chromosome damage in
ATM mutant B-CLLs. Percentage of cells showing induced chromatid type aberrations (gaps, breaks, and interchanges) in G2 at 4 hours following exposure to 1 Gy x-rays in B-CLL tumors 1, 33, and 39 (n = 100), with mutant ATM, and in B-CLLs 51 and 56 with wild-type ATM/TP53 (n = 60). The proportion of unirradiated cells with damage is also indicated with n = 65 and 55, respectively.
Defect in repair of IR-induced chromosome damage in
ATM mutant B-CLLs. Percentage of cells showing induced chromatid type aberrations (gaps, breaks, and interchanges) in G2 at 4 hours following exposure to 1 Gy x-rays in B-CLL tumors 1, 33, and 39 (n = 100), with mutant ATM, and in B-CLLs 51 and 56 with wild-type ATM/TP53 (n = 60). The proportion of unirradiated cells with damage is also indicated with n = 65 and 55, respectively.
Discussion
In this study we have shown that inactivation of theATM gene results in a combination of characteristics that defines a distinct subset of B-CLL tumors. The restriction to the same stage of differentiation, the timing of ATM mutations and their occurrence independently of TP53 mutations, all suggest that an ATM defect plays a role in the pathogenesis of B-CLL. Our findings, however, do not exclude the possibility of other pathways initiating the tumor process in B-CLL as a part of the heterogeneity of this malignancy.
The most consistent feature of ATM mutant tumors in our study was the absence of VH somatic hypermutation. VDJ sequences of all 16 ATM mutant tumors showed more than 98% sequence homology compared with the appropriate VH germline (9 of these tumors showed 100% homology with the germline sequence of the corresponding VH family). Apart from the V1-69 family, previously associated with VH unmutated B-CLL tumors2 and detected here in 3ATM mutant tumors, we have not observed bias toward any particular family of V genes. Two subsets of CLL that transform at distinct stages of differentiation, one with hypermutated IgH VDJ recombinations and another with unmutated recombinations, show distinct biologic behaviors with tumors derived from the nonmutated being more aggressive.2,3 ATM mutant B-CLLs revealed a tendency, although not significant in this study, to be associated with a later disease stage: 75% of tumors were at either stage B or C. It remains possible, therefore, that ATM deficiency accounts for a subset of VH unmutated tumors with an aggressive biologic behavior. The aggressive behavior of ATM-deficient B-CLLs was suggested in a previous report.21
The purpose of this study was to identify the common cellular features of ATM mutant B-CLL tumors. In order to attain this goal we used simultaneous analysis of mutations by RT-PCR, ATM protein expression, and cellular DNA damage response. All patients involved in this study had high lymphocyte counts, caused by the predominance of tumor cells. Consistent with this was the single clonal VDJH rearrangement readily detectable in peripheral blood mononuclear cells from all B-CLLs, apart from B-CLL 11.
The classic concept of tumor suppressor gene inactivation with mutation at one allele and loss of the wild-type allele does not appear to apply to the majority of ATM mutant B-CLLs. LOH across theATM gene region associated with 11q deletion was detected in only 2 of 16 B-CLL tumors with ATM mutations. The remainingATM mutant tumors revealed a spectrum of allelic combinations. In addition, in some tumors only a single missense or truncating mutation was detected.
It is possible that the number of tumors with inactivation of one or both ATM alleles is higher than we were able to demonstrate, since only about 70% of all ATM mutations are readily detectable14,22 irrespective of the method of detection. Experience shows that, overall, the missed mutations are likely to be missense, rather than nonsense. Although nonsense mutations can cause instability of RNA through the mechanism of nonsense-mediated mRNA decay (NMD), there is no evidence for such a mechanism operating in theATM gene23 and this is unlikely to contribute to missed mutations. There was also no evidence that ATM-dependent cellular functions in 19 tested tumors with ATM/TP53 wild type were anything other than normal. Thus, even if present, anyATM sequence changes in these tumors were unlikely to be pathogenic mutations.
We measured the functional consequences of ATM mutations on ATM expression and the ability of ATM mutant B-CLL tumor cells to stabilize p53 and transactivate p53 downstream genes following gamma irradiation. Twelve tumors with ATM mutations showed either absent or significantly reduced ATM expression. Four tumors, B-CLLs 36, 37, 38, and 39, however, showed normal ATM protein levels despite the presence of ATM mutations. We considered various explanations for this observation. In these 4 tumors p53 up-regulation was clearly defective, indicating that the ATM sequence changes were pathogenic rather than polymorphic. ATMpolymorphisms are not associated with a defect in ATM-dependent cellular responses. In all 4 tumors VDJH amplification revealed a single clonal rearrangement and in B-CLL 36, with a single missense mutation in one ATM allele and loss of the second ATMallele, there was no evidence of a normal sequence on the sequence chromatograms. We concluded that a substantial presence of a nontumor population was an unlikely cause of normal ATM expression in these tumors. Our observations favored the possibility that some mutant ATM proteins can be expressed at a high level in B-CLLs, as we previously noted in A-T individuals.15 24 In contrast, 4 tumors, with reduced level of ATM expression and each with a single identified truncating mutation, showed an apparently normal p53 damage response. The simplest explanation is that there was incomplete ATMinactivation in these tumors.
Unlike ATM and TP53 mutant tumors, all 19 analyzed tumors without apparent ATM/TP53 mutations showed normal p53 response to irradiation. Despite this, we considered the possibility that these tumors possessed some downstream defects in the p53/MDM2/p21 pathway. We examined TRAIL-R2 up-regulation as a link between IR-induced p53 response and apoptosis. All 12 analyzedATM/TP53 wild-type tumors showed normal up-regulation of TRAIL-R2, suggesting normal induction of the IR-induced apoptotic pathway. Furthermore, in a separate study11 we demonstrated that 30 B-cell tumors with normal IR-induced p53 response had comparable levels of IR-induced apoptosis. Therefore, if downstream defects in the p53 pathway exist in these B-CLLs, their overall contribution to the defect in IR-induced apoptosis is likely to be small.
It is difficult to determine the precise timing of ATMmutations detected only in B-CLL tumor cells. A targeted progenitor can be more precisely defined if a mutation is present not only in a B-CLL tumor but also in the nontumor hematopoietic cells. We showed that in the patient with B-CLL 33, mutation G8600A was present in tumor cells and T cells as well as in monocytes. In both tumor cells and monocytes a complete absence of the normal sequence at the coding position 8600 suggested a loss of the second ATM allele. Therefore, in this patient, complete ATM inactivation occurred in a progenitor at the stage prior to lymphoid commitment. Mutation 2114insA was present in both CD19+ and CD19−mononuclear cells of the patient with B-CLL 34, suggesting that this mutation probably also occurred in a cell at a stage prior to lymphoid commitment.
In each of 3 B-CLL patients (B-CLLs 1, 4, and 35), we identified a single ATM germline mutation. B-CLL is currently the only sporadic lymphoid tumor in which germline ATM mutations have been observed. If carrying ATM mutations can confer an increased risk of developing B-CLL with low penetrance, in much the same way that an increased risk of breast cancer in female A-T carriers has been suggested,25 it is unlikely that families withATM mutation carriers will show multiple cases of B-CLL.26,27 Indeed, a slightly increased number of patients with CLL among relatives of A-T individuals was noted previously by some28 but not by others.29 30Thus, the role of A-T heterozygosity in predisposition to B-CLL might be best clarified by comparing the frequency of A-T heterozygotes in a large group of B-CLL cases with that in matched controls.
The origin, nature, and distribution of ATM mutations in sporadic B-CLL10-13 is different than sporadic T-PLL.7-9ATM mutations in B-CLL represent a mixture of missense and truncating errors evenly distributed across theATM coding sequence, mirroring the picture observed in a wide range of A-T individuals.15 In contrast, mutations in sporadic T-PLL are predominantly missense, tend to cluster in the catalytic domain of the protein, and are almost invariably associated with loss of the second ATM allele.7-9 Also, there is no evidence of ATM germline mutations orATM mutations in non–T lineage in sporadic T-PLL.31 The reason for this difference is unclear, but the difference itself supports the notion that the pathogenesis of B and T cell tumors on an ATM-deficient background is different.
Inactivation of ATM and p53 appears to be mutually exclusive in B-CLL and despite the similarity in their cellular phenotype, the pathogenesis of these 2 types of tumor may be different. In human lymphomas, TP53 mutations are infrequent and are primarily associated with aggressive subtypes or with relapse, suggesting thatTP53 mutation may occur later during tumor proliferation.32-35 Moreover, lymphoid malignancies are not prominent among tumors in patients with Li-Fraumeni syndrome, who carry germline TP53 mutations.36 Another genetic event (not involving ATM) may be required in theTP53 mutant B-CLLs for the development of the tumor. This notion is consistent with our observation that differentTP53 mutations occurred in identical twins with B-CLL 43 and 44.
In contrast to ATM mutant B-CLLs and B-CLLs with acquiredTP53 mutations, a tumor with a germline TP53mutation (Li-Fraumeni patient) showed VH somatic hypermutation. IfTP53 mutations are, as proposed, a secondary event in B-CLL34 then it is not inconceivable that the pathogenesis, and therefore the stage of differentiation at which malignant transformation occurs, may be different when theTP53 mutation is present in the germline.
It is significant that VH unmutated B-CLL is not the only tumor of pregerminal center origin with frequent ATM mutations. It has been reported that a high proportion of mantle-cell lymphomas (MCL), a tumor characterized by the presence of t(11;14), and absence of VH somatic hypermutation also show inactivation of theATM gene.37 Consequently, there is a possibility that B cells at the pregerminal center stage are particularly sensitive to ATM loss and that the pathogenesis of ATM-deficient MCL and ATM-mutated B-CLL is similar.37-39 Interestingly, and in support of this notion, we observed a high expression of ATM in the interfollicular and mantle zones of the germinal center in normal tonsil and low ATM expression in the center of the follicle (J. Oates, unpublished data, March 2001).
In this study we have addressed the question of which cellular defects caused by ATM loss might be important for transformation at the pregerminal stage of B-cell differentiation. We show thatATM mutant tumors are defective in TRAIL-R2 up-regulation following irradiation which provides an explanation for why neitherATM nor TP53 mutant B-CLLs can undergo apoptosis after exposure to gamma rays. One can envisage that, if acquired early during B-cell ontogeny, ATM inactivation can produce an inability of mutant B cells to respond normally to DNA DSBs and consequently cause genomic instability. Indeed, our preliminary observations suggest that, compared with normal B lymphocytes and B-CLL tumors without ATM mutations, ATM mutant tumors retain a high level of unrepaired chromosome damage following exposure to IR.
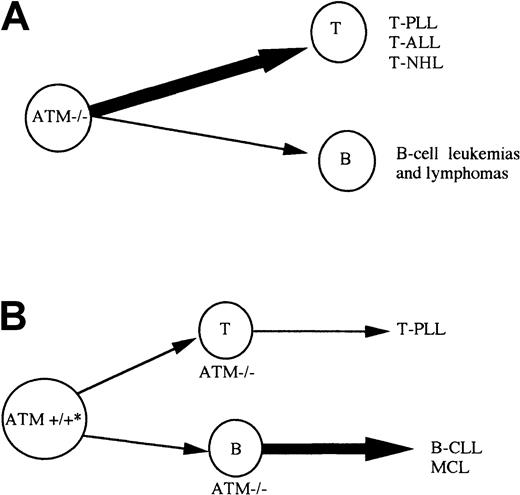
ATM mutation can give rise to lymphoid tumors sporadically as well as in A-T patients. It is interesting that, although the same tumor types can be observed in both A-T and non–A-T individuals (eg, T-PLL), the predominant cell of origin for tumors is different in each group. In the case of inherited ATM inactivation of both alleles, in A-T and in the Atm−/− mouse model, T-cell tumors are the most frequent (Figure 6A) and translocations involving TCR genes play an important role in their development.5 40 In contrast, when acquired in hematopoietic tissue, ATM inactivation leads predominantly to the development of B-cell tumors (Figure 6B), where translocations involving IgH genes may or may not play a role. This may be related to the different role of ATM during B-cell and T-cell development. It is possible that the role of ATM in T-cell development is particularly important in the process of VDJ recombination. Although ATM inactivation does not cause a gross defect in VDJ recombination in T cells, it does not result in TCR interlocus translocations. The absence of ATM in T cells could cause tumorigenic VDJ errors. In contrast, during B-cell development, the role of ATM may not be restricted to the stage of VDJ recombination but rather is evenly distributed throughout B-cell ontogeny. This would allow ATM inactivation to initiate tumorigenesis at any point during early B-cell development. In contrast to lymphoid malignancies initiated by VDJ errors, the pathogenesis of B-CLL may involve more indolent events requiring a longer period of time to promote complete malignant transformation.ATM inactivation that drives B-CLL tumorigenesis independently of VDJ translocations would certainly fit with this concept.
Model for development of lymphoid malignancies on the background of inherited or acquired
ATM inactivation. (A) A-T patients with inherited inactivation of both ATM alleles develop both B (B-cell lymphoma) and T-cell tumors, but T-cell tumors are largely predominant. Atm−/− mice develop only T-cell tumors. (B) Individuals with both germline ATM alleles wild type, can acquire complete ATM inactivation during lymphoid development and develop either B-cell malignancies of pregerminal center origin (B-CLL and MCL) or T-cell tumors (T-PLL). * Patients with sporadic B-CLL can be ATM mutation carriers. In contrast to A-T patients with inherited inactivation of both ATMalleles, B-cell tumors are more common than T-cell malignancies in non–A-T individuals.
Model for development of lymphoid malignancies on the background of inherited or acquired
ATM inactivation. (A) A-T patients with inherited inactivation of both ATM alleles develop both B (B-cell lymphoma) and T-cell tumors, but T-cell tumors are largely predominant. Atm−/− mice develop only T-cell tumors. (B) Individuals with both germline ATM alleles wild type, can acquire complete ATM inactivation during lymphoid development and develop either B-cell malignancies of pregerminal center origin (B-CLL and MCL) or T-cell tumors (T-PLL). * Patients with sporadic B-CLL can be ATM mutation carriers. In contrast to A-T patients with inherited inactivation of both ATMalleles, B-cell tumors are more common than T-cell malignancies in non–A-T individuals.
We thank the Leukaemia Research Fund, the Cancer Research Campaign, the Kay Kendall Leukaemia Fund, and the Ataxia Telangiectasia Society of the United Kingdom for their continued support; Dr D. W. Milligan, and Dr R. J. Johnson (Birmingham Heartlands Hospital), Dr T. Tighe (Aberdeen Royal Infirmary), and Dr J. Rees (Addenbrooks Hospital, Cambridge) for providing tumor samples from their patients; Sarah Reid and Tina Bedenham for technical assistance; Dr Judith Powell for statistical analysis; Mr Kim Ward for his help with flow cytometry; and Mike Griffiths (Birmingham Women's Hospital) for help with cytogenetics.
Supported by grants from the Leukaemia Research Fund, United Kingdom, the Cancer Research Campaign, the Kay Kendall Leukaemia Fund, and the Ataxia Telangiectasia Society of the United Kingdom.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
A. M. R. Taylor, CRC Institute for Cancer Studies, University of Birmingham, Vincent Drive, Edgbaston, Birmingham B15 2TT, United Kingdom; e-mail: a.m.r.taylor@bham.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal