The tumor necrosis factor receptor family molecule CD30 is expressed by activated and memory T cells, depending on IL-4 stimulation preferentially in association with Th0- and Th2-type responses. It mediates pleiotropic effects primarily of the inhibitory type. Arguing that CD30+ cells have a peculiar redistribution in disease, it is demonstrated here, in the Hodgkin-derived L540 cell line (an established model for studying CD30 signaling), that CD30 regulates the prototypic lymphoid chemokine receptor CXCR4 (CD184), which plays an important role in many organ systems and is a coreceptor for human immunodeficiency virus-1 entry. CD30 stimulation with agonistic antibodies in L540 cells led to the accumulation of CXCR4 mRNA, which reached a plateau after 4 hours and did not require protein synthesis. It has been reported recently that CD30 up-regulates the transcription of CCR7 mRNA in YT lymphoma cells. After mRNA transcription, membrane expression of CXCR4 in L540 cells increased as early as 12 hours, reached a plateau after 24 hours (MFI ± SD, 839 ± 122 vs basal 168 ± 28;P < .01) and was still increased after 5 days, permitting enhanced sensitivity to the chemotactic activity of CXCR4-ligand CXCL12 (CI ± SD, 10 ± 1 vs basal 5 ± 2;P < .01). CD30 cross-linking also induced the release of CCL5 and CCL3 and the up-regulation of membrane binding capacity for CCL3 and CCL4 and decreased proliferative activity. This new regulatory role of CD30 may be relevant for T-cell maturation and effector responses and for promoting cancer biology.
Introduction
The tumor necrosis factor receptor (TNFR) family includes molecules such as TNFR1, TNFR2, CD27, CD30, CD40, CD95, and OX40, which interact with a corresponding family of TNF-like cytokines1,2 integrating signals of activation, proliferation, apoptosis, or differentiation, depending on cell type, specific receptor expression, coactivation signals, and mobilization of transduction molecules.1-4 CD30, CD40, OX40, and CD95 at least have been involved in lymphoid differentiation and effector functions.5 CD30, originally described as a marker of a number of lymphoma cells and reactive lymphoblasts,6 is expressed by activated and memory T cells,6-9 medullary thymocytes,10,11 B cells, natural killer cells, and some nonhematologic cells.12 Its expression by T cells is essentially dependent on activation involving IL-4 and CD28 signaling.9,13 The IL-4 requirement is probably the reason for the association of CD30 expression with preferential Th0- and Th2-type immune responses in vitro8 and in vivo.14-16 Stimulation of CD30 by CD153 (CD30 ligand) leads to nuclear mobilization of NF-κB complexes17 while inducing a signal-coupled depletion of TRAF2, which increases the cell sensitivity to TNFR1-dependent apoptosis.3 This dual signaling may explain the pleiotropic effects after CD30 stimulation in CD30+ lymphoma cell lines18 and T cells, the latter acquiring a Th2-type function,19 associated with inhibitory functions,20 based on impaired clonal expansion of T cells.10,20,21 High levels of soluble CD30 or TNF-α in the sera of patients infected with human immunodeficiency virus (HIV-1) at diagnosis have been shown to be an independent prognostic indicator of disease progression,22,23 and the suggested role of CD30 in HIV-1 infection15 has been linked to its ability to promote HIV-1 shedding through NF-κB activation in infected cells.24
The role of CD30 in HIV-1 infection,15,22 the preferential association of CD30 with compartmentalized lymphoid subsets during differentiation11 and effector responses,16and the evidence that Th1 versus Th2 or naive versus primed T cells are distinguished by specific patterns of CD308 and chemokine receptor expression25,26 suggest some kind of relationship between CD30 signals and chemokine function. Recently, Muta et al27 reported that CD30 signals increased amounts of CCR7 mRNA in the YT lymphoma cell line. Actually, a number of CD30-related activities seem to integrate cellular functions driven by lymphoid chemokines, namely their prototypic member CXCL12 (SDF-1) and its receptor CXCR4 (CD184).28 CXCL12 is expressed constitutively in various cell types28-30 and is an efficient cell chemoattractant to specific sites.30-32CXCR4 is expressed by neutrophils, monocytes, naive T lymphocytes, thymocytes, mature and immature B cells, CD34+cells,31,32 vascular endothelial cells,33 and neurologic cells.32 It has been implicated in platelet formation34 and is a coreceptor for HIV-1 entry.35 CD30 and CD153 are reasonable candidates for regulating lymphoid chemokines, as are other TNF family–related molecules, namely lymphotoxin α/β36 and CD40.37 Both CXCR4 and CD30 are up-regulated at the promoter level at least by a combination of CD3, CD28, and IL-2 or IL-4 stimulation in T lymphocytes9,13,35 and are remarkably coexpressed in a number of tissues,6,10-12,38 including the thymus. CXCL12 and CXCR4 are relevant to prothymocyte homing,28,38 and CD30 has been found to be involved in negative selection.10 11
Using the Hodgkin-derived CD4+CD30+ cell line L540, which is an established in vitro model for studying CD30 signaling,17,18 we found that CD30 cross-linking modifies CXCR4 expression and function. This activity is potentially relevant to differentiation, immunity, and cancer because of the extensive roles of the CXCR4–CXCL12 signaling system from embryogenesis28,38to lymphoid maturation and function32 to promoting tumor biology.30
Materials and methods
Culture of the cell line L540
L540 cells (kindly provided by Dr Volker Diehl, Cologne, Germany) are human Hodgkin disease-derived (T-cell–like, Epstein-Barr virus−, CD30+) and have been thoroughly described.39 Cells were cultured in 24-well tissue culture plates (Greiner, Nurtingen, Germany) at 0.15 to 10 × 105cells/mL (see below) in RPMI 1640 medium (Life Technologies, Paisley, United Kingdom) supplemented with 10% low endotoxin (1.6 pg/mL) fetal calf serum (Life Technologies), 50 U/mL penicillin, and 50 μg/mL streptomycin, under 5% CO2 humidified air at 37°C. After culturing for appropriate times with appropriate factors (see below), cells were collected for flow cytometry, total RNA extraction, electrophoretic mobility shift assay, and proliferation or specific functional assay. Cell-free supernatants (SN) were harvested, centrifuged at 2000g for 10 minutes, and stored frozen at −70°C until use.
Stimulation of the L540 cell line and CD30 ligation
Soluble factors.
L540 cells were treated with one of the following factors: 20 ng/mL interleukin-2 (IL-2), 40 ng/mL IL-4, 10 ng/mL IL-6, 10 ng/mL IL-10, 10 ng/mL interferon (IFN)–γ (PeproTech, Rocky Hill, NJ), 20 ng/mL granulocyte macrophage–colony-stimulating factor (GM-CSF; Genzyme, Cambridge, MA), 200 ng/mL CXCL12 (SDF-1α; BD PharMingen, San Diego, CA), or 15 ng/mL phorbol 12-myristate 13-acetate (PMA) + 500 ng/mL ionomycin (PMA–ion; Sigma, St Louis, MO).
Plastic-anchored agonistic anti-CD30 monoclonal antibodies.
L540 cells were stimulated in 24-well plates containing immobilized anti-CD30 agonistic M44 or M67 monoclonal antibodies (mAbs) (kindly provided by Immunex Research and Development, Seattle, WA) according to the method described by McDonald et al17 with minor modifications. Briefly, 24-well polystyrene plates were coated overnight at 4°C with 10 μg/mL M44, M67, or isotype-matched immunoglobulin (Ig)G1 anti-CD34 mAb (Becton Dickinson, San Jose, CA) and an isotype-matched nonagonistic anti-CD30 mAb (BerH2; DAKO, Glostrup, Denmark) in 100 mM carbonate buffer at pH 9.6. Wells were washed twice with phosphate-buffered saline (PBS) and were blocked with PBS containing 5 mg/mL bovine serum albumin (BSA) for 90 minutes at 37°C. After further washes, cells were dispensed in wells (0.15 × 105 cells/well) containing immobilized antibodies and were cultured up to 72 hours. The optimal stimulation time (4, 24, 48, or 72 hours) was assessed in preliminary experiments. There were no substantial differences between M44 and M67 anti-CD30 agonistic mAbs as far as the effects in L540 cells were concerned. Aliquots of cells were also pretreated with 20 μg/mL cycloheximide (CHX) before stimulation with PMA–ion or M67 mAb for 4 hours.
Northern blot and reverse transcription–polymerase chain reaction
After 4 and 24 hours of culture in the presence or absence of M44, M67, BerH2 or PMA–ion with or without CHX pretreatment as reported above, total RNA was extracted from L540 cells.
Northern blot analysis.
Northern blot analysis (10 μg/lane) was performed as previously described.40 Filters were hybridized with32P-labeled complementary DNA (cDNA) probes to CCR5 and CXCR4 obtained as specified above and with a 32P-labeled plasmid containing a cDNA probe to β-actin.
Reverse transcription–polymerase chain reaction.
For RT-PCR, 4 μg RNA was reverse-transcribed as previously described41 42 and was cDNA amplified using the following primers: (1) CXCR4 sense 5′-AGAACCAGCGGTTACCATGGA-3′ and antisense 5′-GAGTGTGACAGCTTGGAGATG-3′ (702 bp); (2) CCR1 sense 5′-GAAACTCCAAACACCACAGAG-3′ and antisense 5′-CAGCTTCCACTCTCGTAGGCTT-3′ (588 bp); (3) CCR3 sense 5′-TCCTT CTCTCTTCCTATCAATC-3′ and antisense 5′-GGCAATTTTCTGCATCTG-3′ (313 bp); (4) CCR5 sense 5′-CGCATCAAGTGTCAAGTCCAATC-3′ and antisense 5′-TG TAAACTGAGCTTGCTCGCT-3′ (1014 bp); (5) Vimentin sense 5′-GCTCAGATTCAGGAACAGCAT-3′ and antisense 5′-TAAGGGCATCCACTTCACAGG-3′ (266 bp). cDNAs were denatured for 5 minutes at 94°C before 35 runs at 58, 52, 57, and 60°C for 40, 30, 40, and 40 seconds, respectively (annealing) and at 72°C for 50 seconds (elongation) followed by 5 minutes at 72°C in a thermal cycler (GeneAmp PCR System 2400; PE Biosystems, Foster City, CA) using 1.25 U Taq polymerase (PE Biosystems) in 50 μL. PCR products were separated by electrophoresis on 1.5% agarose gel. CXCR4 and CCR5 cDNAs amplified from the L540 cell line using the primers specified above were analyzed for theBamHI and EcoRI (Life Technologies) restriction sites, respectively (which gave the expected CXCR4 fragments of 498 and 204 bp and the expected CCR5 fragments of 780 and 234 bp). They were sequenced (Sequenase 2.0 sequencing kit; USB, Cleveland, OH) as plasmid inserts (TA cloning kit; Invitrogen, San Diego, CA) from which the CXCR4 and CCR5 probes were generated for Northern blot analysis.
Electrophoretic mobility shift assay
After 20 to 60 minutes of culture in the presence or absence of M44, M67, or PMA–ion as reported above, nuclear extracts were prepared as previously described.17 Briefly, cells were harvested and centrifuged at 2000g for 2 minutes. The pellets were washed and resuspended in 100 μL ice-cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 1.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA) containing 0.15% NP40 and antiproteases (1 mM phenylmethylsulfonyl fluoride and 10 μg/mL each of aprotinin, leupeptin, and pepstatin A). Cells were incubated on ice for 10 minutes, vortexed briefly, and centrifuged at 1000g for 10 minutes at 4°C. Nuclear pellets were washed twice in lysis buffer containing the antiproteases and were incubated for 15 minutes on ice in nuclear extraction buffer (20 mM HEPES, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 20% vol/vol glycerol) containing the antiprotease cocktail. Extracted nuclear fractions were then centrifuged at 16 000g for 15 minutes at 4°C, and the resultant supernatants were made into aliquots and were stored at −70°C before use. Binding reactions were performed in 20 μL binding buffer (20 mM Tris-HCl, pH 7.5, 80 mM KCl, 1 mM dithiothreitol, 0.1% NP40, 6% glycerol) supplemented with 20 μg acetylated BSA and 1 μg poly(dI).poly(dC). The following (γ-32P) adenosine triphosphate (Amersham International, Little Chalfont, United Kingdom) end-labeled, double-strand oligonucleotides were used as probes: (1) 5′-GATCAGGGACTTTCCGCTGGGGACTTTCC-3′, containing tandem repeats of the consensus NF-κB binding site identical to those found in the HIV promoter; (2) 5′-GATCGAT CGGGGCGGGGCGATC-3′, containing the consensus Sp1-binding site; (3) 5′-CAGCGGCGCATGCGCCGCGCTC-3′, containing the binding sequence for NRF-1; (4) 5′-CGCTTGATGACTCAGCCGGAA-3′, containing the DNA binding site for the AP-1/c-jun homodimer complex. Samples were separated on 5% native polyacrylamide gels and were visualized by autoradiography.
RNase protection assay
Detection of chemokine message expression was performed with an RNase protection analysis system (RiboQuant; BD PharMingen) on cells in basal conditions or treated as reported above. A multiprobe template set (hCK-5) was used for the in vitro transcription reaction using T7 polymerase to direct the synthesis of high-specific–activity32P-labeled antisense RNAs that hybridize with human RNAs encoding XCL1 (lymphotactin α), CCL5 (RANTES), CXCL10 (IP-10), CCL4 (MIP-1β), CCL3 (MIP-1α), CCL2 (MCP-1), CXCL8 (IL-8), CCL1 (I-309), and 2 housekeeping control gene products, L32 and GAPDH. After hybridization, samples were treated with RNase A and proteinase K, and the protected probes were resolved on a 5% acrylamide–urea gel and were quantified by autoradiography.
Flow cytometry analysis
Cells in basal conditions or stimulated for 72 hours as reported above were harvested, washed twice with PBS, and stained (1 × 106) for 1 hour at 4°C with anti–CD30-fluorescein isothiocyanate (FITC; DAKO), anti–CXCR4-phycoerythrin (PE) (12G5; BD PharMingen), anti–CCR5-PE (BD PharMingen) mAbs, and isotype–fluorochrome-matched controls, as previously described.43 In selected experiments, CXCR4 internalization in L540 cells was evaluated according to the method described by Signoret et al44 adapted to flow cytometry. Cells were also tested for the binding of biotinylated CCL3 and CCL4 using commercially available kits (R&D Systems, Minneapolis, MN). The manufacturer's protocols were followed with minor modifications. Briefly, 1 × 106 cells were washed twice in PBS, incubated for 90 minutes at 4°C with biotinylated cytokine, added to avidin–FITC reagent for 30 minutes, and washed twice with the buffer supplied. Controls for specificity of binding were performed using neutralizing concentrations of nonbiotinylated CCL3 and CCL4. Flow cytometry analysis was performed on a FACScan (Becton Dickinson, Mountain View, CA) using the CELLQuest analysis program.
Detection of soluble molecules in culture supernatants
Soluble CD30, CCL3, CCL4, and CCL5 proteins in culture cell-free SN derived from cells in basal conditions or stimulated for 72 hours with mAb M67 or M44 or with PMA–ion as reported above were measured using commercially available ELISA kits (DAKO and R&D Systems, respectively). Briefly, 100 to 200 μL SN alone (CCL3, CCL4, CCL5) or SN added to enzyme-conjugated antibody (CD30) were incubated in duplicate for 2 hours at room temperature in 96-well microplates coated with specific antibodies. After washing, the samples were directly treated with substrate solution (CD30) or were added to enzyme-conjugated antibodies for 2 hours, washed, and treated with substrate solution (CCL3, CCL4, CCL5). The enzymatic reactions were stopped and read within 30 minutes at 450 nm on an AutoReader III (Ortho Diagnostic Systems, Raritan, NJ).
Chemotaxis assay
Chemotaxis assay was performed following a modification of a published procedure.45 Cells in basal conditions or stimulated for 72 hours with mAb M67 or M44 or PMA–ion with, as reported above, were washed twice in chemotaxis buffer (RPMI 1640, 20 mM HEPES, 0.4% BSA), resuspended at a density of 5 × 106/mL, and incubated for 1 hour in the presence or absence of 50 μg/mL of neutralizing anti-CXCR4 (12G5) or isotype-matched control mAbs (BD PharMingen). Aliquots of 100 μL cell suspensions were seeded in the upper chamber of a 24-well, 8-μm pore polycarbonate Transwell culture insert (Costar, Cambridge, MA) and were incubated for 90 minutes at 37°C. The lower chamber was filled with 600 μL chemotaxis buffer alone or containing 200 ng/mL CXCL12 (SDF-1α; BD PharMingen). In selected experiments, the inhibitory effect of CXCL12 in the upper chamber on migration of L540 cells was also evaluated according to Wang et al.34 Cells that migrated into the lower chamber through the membrane pores were resuspended and counted with a FACScan (Becton Dickinson) for 20 seconds at a 60 μL/min flow rate. Results were expressed as percentage of migrated cells or chemotactic index (CI = cells migrating to CXCL12/cells migrating to medium).
Proliferation of L540 cells
The L540 cell proliferation in basal conditions and after stimulation with IL-2, M67 or M44 mAb, or PMA–ion, as reported above, was measured after 72-hour culture as follows.
Incorporation of 5-bromo-2′-deoxyuridine.
Cells were added with 10 μM BrdU (Sigma), incubated for 1 hour at 37°C, washed twice with PBS, and fixed in 70% ethanol. Cell suspensions were centrifuged and incubated with 3 N HCl for 20 minutes. After neutralizing with 0.1 M Na2B4O7, cells were resuspended in 100 μL PBS containing 0.5% BSA and were stained with 10 μL anti-BrdU–FITC or isotype-matched control mAbs (Becton Dickinson) for 30 minutes at room temperature, washed twice in PBS, and analyzed with a FACScan (Becton Dickinson). Results were expressed as a percentage of BrdU-positive cells of the total number of acquired cells.
3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay.
MTT assay was performed as previously described.46 Cells were added with 10 μL 5 mg/mL MTT solution, incubated at 37°C for 4 hours, lysed with 0.04 N HCl in isopropanol, and read (wavelengths: test 570, reference 630 nm) on an AutoReader III (Ortho Diagnostic Systems). Results were expressed as percentage proliferation of stimulated versus unstimulated cells.
Statistical analysis
Student t test, Mann-Whitney U test, and Kruskall-Wallis analysis of variance by ranks were used. Differences were considered statistically significant whenP < .05.
Results
Expression pattern in resting L540 cells
The pattern of relevant molecules in resting L540 cells is listed in Table 1. L540 cells were CD3−CD4+ (MFI ± SD, 900 ± 30) and strongly CD30+ (1370 ± 252). When we examined the presence of a number of CC and CCR mRNAs or molecules using RPA, RT-PCR, flow cytometry, and ELISA, we found that L540 cells transcribed CCR1, CCR2, CCR3, and CXCR4 mRNAs, released CCL3 CCL4, and CCL5, and had clear membrane expression of CXCR4 at flow cytometry (168 ± 28). Specific binding capacities for fluorochrome-labeled CCL3 and CCL4 were also expressed on resting L540 cell membrane (70 ± 15 and 439 ± 47, respectively).
Expression of some relevant molecules in resting L540 cells
| . | mRNA . | Protein . | Receptor* . |
|---|---|---|---|
| CD3 | ND | −* | ND |
| CD4 | +† | ++* | ND |
| CD30 | +† | ++* | ND |
| CXCR4 | +†,‡ | +* | ND |
| CCR1 | +‡ | ND | ND |
| CCR2 | +‡ | ND | ND |
| CCR3 | +‡ | ND | ND |
| CCR5 | −†,‡ | −* | ND |
| CXCL8 | +1-153 | ND | ND |
| CXCL10 | +1-153 | ND | ND |
| CCL1 | +1-153 | ND | ND |
| CCL2 | −1-153 | ND | ND |
| CCL3 | +1-153 | +1-160 | + |
| CCL4 | +1-153 | +1-160 | + |
| CCL5 | +1-153 | +1-160 | + |
| XCL1 | +1-153 | ND | ND |
| . | mRNA . | Protein . | Receptor* . |
|---|---|---|---|
| CD3 | ND | −* | ND |
| CD4 | +† | ++* | ND |
| CD30 | +† | ++* | ND |
| CXCR4 | +†,‡ | +* | ND |
| CCR1 | +‡ | ND | ND |
| CCR2 | +‡ | ND | ND |
| CCR3 | +‡ | ND | ND |
| CCR5 | −†,‡ | −* | ND |
| CXCL8 | +1-153 | ND | ND |
| CXCL10 | +1-153 | ND | ND |
| CCL1 | +1-153 | ND | ND |
| CCL2 | −1-153 | ND | ND |
| CCL3 | +1-153 | +1-160 | + |
| CCL4 | +1-153 | +1-160 | + |
| CCL5 | +1-153 | +1-160 | + |
| XCL1 | +1-153 | ND | ND |
Evaluated using flow cytometry (see “Materials and methods”).
Northern blot analysis.
RT-PCR.
RPA.
ELISA on culture supernatants.
ND indicates not determined.
CD30-triggering–induced increase in CXCR4 mRNA
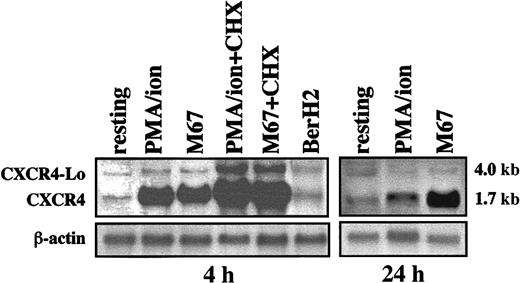
To investigate whether CXCR4 mRNA was transcribed, L540 cells were initially cultured for 4 hours in the presence or absence of PMA–ion, 10 μg/mL plastic-bound anti-CD30 agonistic mAb M67 or M44, and isotype-matched anti-CD34 mAb and anti-CD30 nonagonistic, indifferent BerH2 mAb as controls. Total RNA was then processed for Northern blot analysis. Figure 1 shows that resting L540 cells produced small amounts of CXCR4 mRNA. By contrast, PMA–ion- and CD30-stimulated (M67) L540 cells showed a considerable accumulation of CXCR4 transcripts of the expected 1.7-kb size, with a pattern matching that observed in other cell types.33 In addition, transcripts corresponding to a larger 4.0-kb mRNA present in all conditions were not subject to regulation by anti-CD30 agonistic mAbs or PMA–ion. These transcripts are likely to correspond to the unspliced isoform of CXCR4, termed CXCR4-Lo.47 Consistent with the genuine ability of L540 cells to accumulate CXCR4 mRNA in response to CD30 stimulation were additional experiments in which, by contrast, a wide panel of cytokines (Table2) did not influence the levels of CXCR4 mRNA.
Production of CXCR4 mRNA by L540 cells.
2 × 106 cells/mL were incubated alone and with PMA–ion (15 ng/mL PMA + 500 ng/mL ionomycin), plastic-bound anti-CD30 agonistic mAb M67 (10 μg/mL), or BerH2 or anti-CD34 isotype-matched mAb as a control. At the time-points indicated, total mRNA was extracted and analyzed for CXCR4 and actin mRNA expression. Aliquots of cells were also pretreated with 20 μg/mL CHX before stimulation with PMA–ion and M67 mAb for 4 hours. Resting cells produced small amounts of CXCR4 (1.7 kb) and CXCR4-Lo mRNA (4 kb). At 4 hours, both PMA–ion and M67 strongly up-regulated CXCR4 mRNA. At 24 hours, PMA–ion–dependent CXCR4 mRNA transcription had reverted to basal levels, whereas the M67-dependent up-regulation persisted. CXCR4-Lo mRNA was not regulated, except in the case of pretreatment with CHX, which superinduced both CXCR4 and CXCR4-Lo mRNA in M67 and PMA–ion-stimulated cells. The experiment depicted is representative of 5.
Production of CXCR4 mRNA by L540 cells.
2 × 106 cells/mL were incubated alone and with PMA–ion (15 ng/mL PMA + 500 ng/mL ionomycin), plastic-bound anti-CD30 agonistic mAb M67 (10 μg/mL), or BerH2 or anti-CD34 isotype-matched mAb as a control. At the time-points indicated, total mRNA was extracted and analyzed for CXCR4 and actin mRNA expression. Aliquots of cells were also pretreated with 20 μg/mL CHX before stimulation with PMA–ion and M67 mAb for 4 hours. Resting cells produced small amounts of CXCR4 (1.7 kb) and CXCR4-Lo mRNA (4 kb). At 4 hours, both PMA–ion and M67 strongly up-regulated CXCR4 mRNA. At 24 hours, PMA–ion–dependent CXCR4 mRNA transcription had reverted to basal levels, whereas the M67-dependent up-regulation persisted. CXCR4-Lo mRNA was not regulated, except in the case of pretreatment with CHX, which superinduced both CXCR4 and CXCR4-Lo mRNA in M67 and PMA–ion-stimulated cells. The experiment depicted is representative of 5.
Effect of various agonists on CXCR4 expression in L540 cells
| No variation | |
| Medium | * |
| BerH2 | * |
| Anti-CD34 | * |
| GM-CSF | * |
| IL-2 | * |
| Cycloheximide | * |
| Down-regulation | |
| PMA + ionomycin | −− |
| IFN-γ | − |
| IL-4 | − |
| IL-6 | − |
| Up-regulation | |
| IL-10 | + |
| M44 | ++ |
| M67 | ++ |
| M44/M67 + cycloheximide | +++ |
| No variation | |
| Medium | * |
| BerH2 | * |
| Anti-CD34 | * |
| GM-CSF | * |
| IL-2 | * |
| Cycloheximide | * |
| Down-regulation | |
| PMA + ionomycin | −− |
| IFN-γ | − |
| IL-4 | − |
| IL-6 | − |
| Up-regulation | |
| IL-10 | + |
| M44 | ++ |
| M67 | ++ |
| M44/M67 + cycloheximide | +++ |
See “Materials and methods” for concentrations.
CXCR4 mRNA accumulation pattern
Kinetic experiments revealed that maximal expression of CXCR4 1.7-kb mRNA occurred after 4 hours of stimulation with anti-CD30 mAb (Figure 1) and that an increase of CXCR4 density on the membrane of the L540 cells followed (see below). A similar accumulation of CXCR4 1.7-kb mRNA was detectable after stimulation with PMA–ion, but was associated with a reduced membrane expression of the CXCR4 molecule, in line with previous reports.33 44 After 24 hours, increased levels of CXCR4 mRNA transcripts were still detectable in cells treated with PMA–ion and especially with anti-CD30 mAb (Figure 1). We also examined whether de novo protein synthesis was necessary for the CD30-driven induction of CXCR4 mRNA in L540 cells. As shown in Figure 1, the inhibitor of protein synthesis CHX not only did not inhibit, but actually superinduced, the anti–CD30-dependent accumulation of 1.7- and 4.0-kb mRNA for CXCR4.
NF-κB nuclear translocation after CD30 ligation
We have previously shown that L540 cells represent an adequate model to evaluate CD30-related transduction events. The CD30-dependent activation of NF-κB is substantially similar in L540 cells, peripheral T lymphocytes, and T-cell clones derived from peripheral T cells.17 After stimulation of CD30 on L540 cells through plastic-bound agonistic mAb M44 or M67, NF-κB nuclear translocation followed as expected (Figure 2), involving p50 and p65 NF-κB complexes, as previously shown.17 The effects on NF-κB complexes induced by stimulation with M67, M44, or PMA–ion were similar in the current experiments, as previously reported.17
Effect of CD30 ligation on NF-κB, SP1, AP-1, and NRF-1 binding activity in L540 cells.
2 × 106 cells/mL were cultured for 30 minutes in 24-well plates coated with 10 μg/mL plastic bound anti-CD30 agonistic mAb M67 or BerH2 as a control and in the absence or presence of PMA–ion (15 ng/mL PMA + 500 ng/mL ionomycin). M67 induced NF-κB but not Sp1, AP-1, or NRF-1 DNA binding activity in nuclear extracts from L540 cells. PMA–ion induced not only NF-κB but also AP-1 binding activity. The BerH2 control was not represented if stimulation with M67 mAb was ineffective. A section of gels is shown. The experiment depicted in this figure is representative of 5.
Effect of CD30 ligation on NF-κB, SP1, AP-1, and NRF-1 binding activity in L540 cells.
2 × 106 cells/mL were cultured for 30 minutes in 24-well plates coated with 10 μg/mL plastic bound anti-CD30 agonistic mAb M67 or BerH2 as a control and in the absence or presence of PMA–ion (15 ng/mL PMA + 500 ng/mL ionomycin). M67 induced NF-κB but not Sp1, AP-1, or NRF-1 DNA binding activity in nuclear extracts from L540 cells. PMA–ion induced not only NF-κB but also AP-1 binding activity. The BerH2 control was not represented if stimulation with M67 mAb was ineffective. A section of gels is shown. The experiment depicted in this figure is representative of 5.
Specific inducers for the CXCR4 promoter
Because NF-κB–related binding sites are not present on the CXCR4 promoter,35 we conducted shift experiments to study the following molecules known to be involved in CXCR4 gene regulation35,48,49: NRF-1, a specific transcription factor for the CXCR4 promoter, usually involved in the constitutive expression of the CXCR4 gene35,49; Sp1 complexes targeted to GC boxes on the CXCR4 promoter35,48; and AP-1 (c-jun) factors, which recognize a specific motif on the CXCR4 promoter35,49 and are putatively mobilized by CD30 signaling through modulation of TRAF-2.3 NRF-1, SP1, and AP-1 transcription factors were detectable in L540 cell nuclei in basal conditions (Figure 2). Their amounts, however, proved to be unaffected after CD30 stimulation. By contrast, PMA–ion induced nuclear mobilization of AP-1 (Figure 2), accounting for the PMA-dependent CXCR4 mRNA up-regulation.
CD30 triggering-induced increase in CXCR4 membrane expression
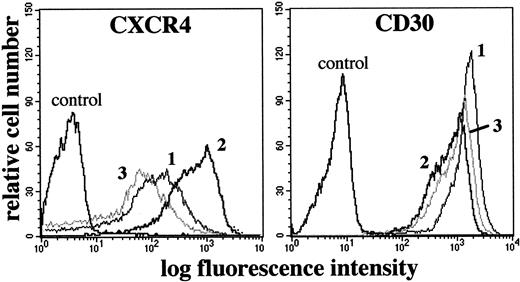
CD30 stimulation was associated with a clear-cut increase in CXCR4 membrane expression as early as 12 hours, which reached a plateau after 24 hours (MFI ± SD, 839 ± 122 versus basal condition 168 ± 28; P < .01) (Figure3) and was still present after 5 days. After PMA–ion stimulation, despite increased levels of CXCR4 mRNA similar to those induced after CD30 stimulation, a clear-cut reduction of the membrane density of CXCR4 was detectable33,44(Table 2, Figure 3). Consistent with a reduction from internalization33,44 were 3 additional experiments in which PMA–ion stimulation induced cytoplasmic translocation of fluorescent mAbs previously bound to membrane CXCR4. The cytoplasmic translocation of the CXCR4-fluorescent mAb complex was abolished by 1 M sucrose-containing medium, which inhibits clathrin-coated vesicle formation50 (data not shown). The membrane expression of CXCR4 was variously affected when L540 cells were stimulated with a panel of other factors known to regulate the surface expression of a number of receptors (Table 2), and we never observed any spontaneous increase in CXCR4 expression as a result of the culture conditions. Thus, the up-regulation of CXCR4 was a genuine, specific effect of CD30 ligation.
Flow cytometric analysis of CXCR4 and CD30 expression in the L540 cells.
1 × 106 cells resting (1) or stimulated for 72 hours with plastic-bound anti-CD30 agonistic mABs M67 (10 μg/mL) (2) or PMA/ion (15 ng/mL PMA + 500 ng/mL ionomycin) (3) were stained with anti-CXCR4-PE and anti-CD30-FITC mAbs and isotype/fluorochrome matched controls. Compared with resting cells (1), M67 (2), but not the nonagonistic anti-CD30 mAb BerH2, induced a clear-cut increase in CXCR4 and a decrease in CD30 MFI, whereas PMA/ion (3) induced a decrease in both CXCR4 and CD30 MFI. The experiment depicted is representative of 10.
Flow cytometric analysis of CXCR4 and CD30 expression in the L540 cells.
1 × 106 cells resting (1) or stimulated for 72 hours with plastic-bound anti-CD30 agonistic mABs M67 (10 μg/mL) (2) or PMA/ion (15 ng/mL PMA + 500 ng/mL ionomycin) (3) were stained with anti-CXCR4-PE and anti-CD30-FITC mAbs and isotype/fluorochrome matched controls. Compared with resting cells (1), M67 (2), but not the nonagonistic anti-CD30 mAb BerH2, induced a clear-cut increase in CXCR4 and a decrease in CD30 MFI, whereas PMA/ion (3) induced a decrease in both CXCR4 and CD30 MFI. The experiment depicted is representative of 10.
Variable regulation of CXCR4 membrane expression by interleukins
For comparative purposes, we examined the effect of IFN-γ, IL-4, IL-6, and IL-10 on the membrane expression of CXCR4 in L540 cells. The effect of pretreatment with CHX of L540 cells stimulated with anti-CD30 agonistic mAbs was also evaluated. According to the results reported in Table 2, the membrane expression of CXCR4 was unaffected after stimulation with GM-CSF, IL-2, and CHX alone. A decrease in CXCR4 expression was induced by stimulation with IFN-γ, IL-4, and IL-6. Stimulation with IL-10 led to up-regulation of the CXCR4 molecule on the membrane of L540 cells. IL-10 was a weaker inducer of CXCR4 than M44 or M67 alone, whereas M44 and M67+CHX were the strongest inducers of CXCR4 (Table 2).
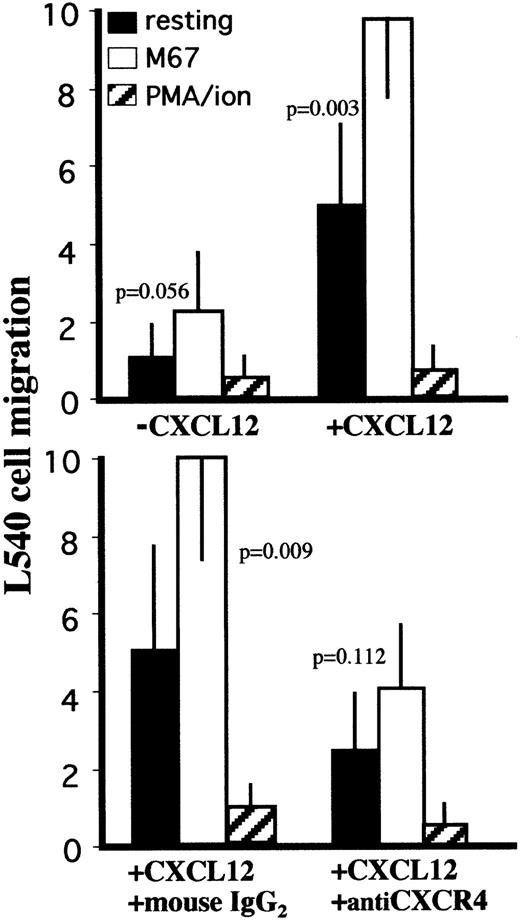
CXCL12-dependent chemotaxis after CD30-induced increase in CXCR4
Cells exposing CXCR4 on their membranes are usually sensitive to chemotactic activity induced by CXCL12. When tested in basal conditions, CXCR4+ L540 cells were responsive to CXCL12-dependent chemotactic signals with a CI of 5 ± 2, compared to a CI of 10 ± 1 when stimulated with anti-CD30 agonistic mAbs (P = .003; Figure 4). CXCL12 concentrations in the upper chamber inhibited the migration of L540 cells34 (data not shown). This migration pattern was abolished in experiments involving preincubation of L540 cells with neutralizing anti-CXCR4 mAbs, whereas indifferent isotype-matched mAbs were ineffective (Figure 4). L540 cells stimulated with PMA–ion were poorly sensitive to CXCL12-dependent chemotaxis (Figure 4).
Effect of CD30 ligation on CXCL12-induced chemotaxis in the L540 cells.
5 × 105 cells resting or stimulated with PMA–ion (15 ng/mL PMA + 500 ng/mL ionomycin), plastic-bound anti-CD30 agonistic mAb M67 (10 μg/mL), or BerH2 (or anti-CD34 isotype-matched mAb) as a control were subjected to chemotaxis through 8-μm pore Transwell filters to CXCL12 (200 ng/mL) in the lower chamber. Stimulation with M67 increased the sensitivity of L540 cells to the chemotactic effect of CXCL12. The increase in sensitivity was inhibited after 60 minutes of preincubation with neutralizing anti-CXCR4 mAb (12G5, 50 μg/mL). Data are expressed as mean ± SD of 5 experiments.
Effect of CD30 ligation on CXCL12-induced chemotaxis in the L540 cells.
5 × 105 cells resting or stimulated with PMA–ion (15 ng/mL PMA + 500 ng/mL ionomycin), plastic-bound anti-CD30 agonistic mAb M67 (10 μg/mL), or BerH2 (or anti-CD34 isotype-matched mAb) as a control were subjected to chemotaxis through 8-μm pore Transwell filters to CXCL12 (200 ng/mL) in the lower chamber. Stimulation with M67 increased the sensitivity of L540 cells to the chemotactic effect of CXCL12. The increase in sensitivity was inhibited after 60 minutes of preincubation with neutralizing anti-CXCR4 mAb (12G5, 50 μg/mL). Data are expressed as mean ± SD of 5 experiments.
CD30 shedding after CD30 ligation and PMA stimulation
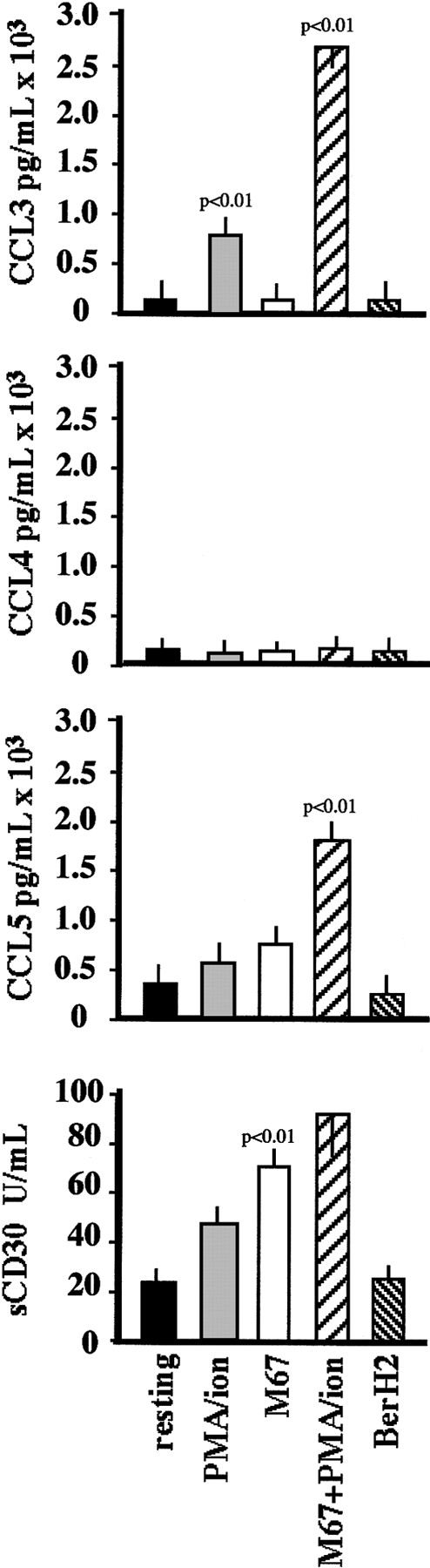
Stimulation of CD30 induced a decrease in the membrane density of the CD30 molecule itself (Figure 3), paralleled by an increase in its concentration in culture SN (Figure 5). Forty-eight–hour stimulation of L540 cells with M67 reduced the CD30 membrane MFI ± SD from 1370 ± 252 to 600 ± 138 (P < .01). A similar trend was observed using PMA–ion as the stimulator, which induced a decrease to 854 ± 119 (P < .01). These effects lasted up to 96 hours after stimulus delivery. The concentration of CD30 in 48-hour culture SN was 24 ± 9 U/mL for unstimulated L540 cells. It was 64 ± 10 (P < .01), 50 ± 8 (P < .01), and 95 ± 22 (P < .01) U/mL after stimulation with M67, PMA–ion, and M67+PMA–ion, respectively. By contrast, the nonagonistic anti-CD30 mAb BerH2 induced no statistically significant variations compared with controls, either in CD30 membrane MFI (1343 ± 230) or in culture SN concentration (20 ± 7 U/mL).
Effect of CD30 ligation on the release of CD30, CCL3, and CCL5 by L540 cells.
5 × 104 cells/mL were cultured for 72 hours in 24-well plates coated with 10 μg/mL M67 mAb or BerH2 as a control in the presence or absence of PMA–ion (15 ng/mL PMA + 500 ng/mL ionomycin). CD30, CCL3, and CCL5 concentrations in the culture medium were evaluated by commercially available ELISA. Anti-CD30 agonistic mAb M67 induced increased levels of soluble CD30, CCL3, and CCL5 in the L540 cell culture medium. Data are expressed as mean ± SD of 5 experiments.
Effect of CD30 ligation on the release of CD30, CCL3, and CCL5 by L540 cells.
5 × 104 cells/mL were cultured for 72 hours in 24-well plates coated with 10 μg/mL M67 mAb or BerH2 as a control in the presence or absence of PMA–ion (15 ng/mL PMA + 500 ng/mL ionomycin). CD30, CCL3, and CCL5 concentrations in the culture medium were evaluated by commercially available ELISA. Anti-CD30 agonistic mAb M67 induced increased levels of soluble CD30, CCL3, and CCL5 in the L540 cell culture medium. Data are expressed as mean ± SD of 5 experiments.
Release of CCL3, CCL4, and CCL5 after CD30 ligation
We investigated whether M67-treated L540 cells released CCL3, CCL4, and CCL5 proteins into the culture SN. To this end, we used specific commercial ELISAs with threshold sensitivities of 6, 4, and 8 pg/mL (pM) for CCL3, CCL4, and CCL5, respectively. As shown in Figure5, detectable yields of CCL3 and CCL5 were measured in SN harvested from resting L540 cells (6 ± 2 and 270 ± 38 pg/mL, respectively), which increased to 710 ± 50, 2805 ± 235, 600 ± 136, and 1775 ± 278 pg/mL after treatment with PMA–ion and M67+PMA–ion, respectively (P < .01). M67 alone had no effect on the release of CCL3, whereas it induced a CCL5 SN concentration of 780 ± 165 (P < .01). The basal release of CCL4 (10 ± 3 pg/mL) was not substantially affected by the stimuli reported above.
CCL3 and CCL4 binding to membrane receptors after CD30 ligation
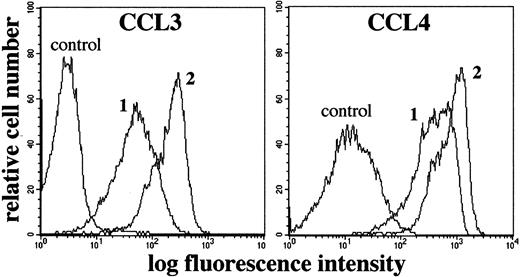
The ability of biotinylated CCL3 and CCL4 to specifically bind to receptors on the L540 cell membrane was also modified by CD30 triggering. The basal binding for these chemokines gave an MFI ± SD of 70 ± 15.3 and 439 ± 47 for CCL3 and CCL4, respectively. After stimulation with M67 alone, the binding of both chemokines was greatly increased (210 ± 12, P = .006, and 818 ± 89,P = .019, respectively). PMA–ion stimulation did not affect the binding capacity for CCL3 (74 ± 12), and it induced a modest increase in that for CCL4 (535 ± 56;P = .029).
L540 cell growth modulation after CD30 ligation and IL-2 stimulation
CD30 stimulation has pleiotropic effects on lymphoma cell lines. L540 cells have been reported to proliferate in response to stimulation by CD30.18 In our experiments, this was not the case. As shown in Table 3, agonistic anti-CD30 mAbs reduced the proliferative activity of L540 cells by at least 30%, as evaluated by BrdU incorporation analysis. IL-2, which we evaluated for comparative purposes, induced a similar down-regulation of L540 cell proliferative activity, as previously reported.51
Effects of CD30 triggering on L540 cell proliferation
| L540 cell treatment3-150 . | Cycling cells,3-151 % . | Living cells,3-151 % . |
|---|---|---|
| M67 | 63 ± 1.83-152 | 81 ± 10 |
| BerH2 | 96 ± 2.1 | 84 ± 6 |
| PMA + ionomycin | 87 ± 8.4 | 94 ± 5 |
| L540 cell treatment3-150 . | Cycling cells,3-151 % . | Living cells,3-151 % . |
|---|---|---|
| M67 | 63 ± 1.83-152 | 81 ± 10 |
| BerH2 | 96 ± 2.1 | 84 ± 6 |
| PMA + ionomycin | 87 ± 8.4 | 94 ± 5 |
Values are mean ± SD of 5 experiments.
10 μg/mL plastic-bound mAb M44, M67, or BerH2; 15 ng/mL PMA + 500 ng/mL ionomycin.
Normalized to the corresponding percentage for untreated L540 cells.
P = .001 compared with untreated L540 cells.
Discussion
The current study defines a new mechanism, the induction of CXCR4, whereby CD30 regulates the cellular functions involved in directing the specific homing of activated cells in specific microenvironments.
In the CD30+ cell line L540, previously exploited to dissect CD30-mediated activities,17,18 CD30 cross-linking with agonistic anti-CD30 mAb M67 or M44 led to 3 occurrences: up-regulation of the expression of CXCR4 (Figure 3), after accumulation of its mRNA (Figure 1), permitting increased cell sensitivity to the specific chemotactic activity of CXCL12 (Figure 4); release of CCL5 and CCL3 (Figure 5) associated with up-regulation of membrane-binding capacity for CCL3 and CCL4 (Figure 6); and decreased proliferative activity (Table 3). The up-regulation of CXCR4 mRNA and membrane molecules induced by CD30 triggering in L540 cells was genuine and specific, as demonstrated by the different effects induced by other factors and by the selective response to CXCL12. Among the other factors tested (Table 2), only IL-10 up-regulated CXCR4, though to a lesser extent than CD30. The classic stimulator, PMA–ion, induced an increase in CXCR4 mRNA (Figure 1), but the surface density of CXCR4 decreased (Figure 3) primarily because of internalization.33,44 Moreover, while this work was in progress, Muta et al27 reported the surprising, unexpected finding that CD30 signals led to a 5.8-fold increase in CCR7 mRNA in YT lymphoma cells, arguing that, through CCR7, CD30 signals integrate lymphocyte trafficking, proliferation, and apoptosis.
Flow cytometric analysis of specific binding of biotinylated CCL3 and CCL4 to L540 cells.
1 × 106 cells resting (1) or stimulated for 72 hours with plastic-bound anti-CD30 agonistic mAbs M67 (10 μg/mL) (2) were incubated with biotinylated CCL3 or CCL4 and stained with avidin-FITC reagent. Controls for specificity of binding were performed using non-biotinylated CCL3 and CCL4. Compared with resting cells (1), stimulation with M67 (2), but not with nonagonistic anti-CD30 mAb BerH2, induced a clear-cut increase in both CCL3 and CCL4 binding. Stimulation with 15 ng/mL PMA + 500 ng/mL ionomycin did not modify the binding of CCL3 and CCL4 (not shown), compared with basal conditions (1). The experiment depicted is representative of 5.
Flow cytometric analysis of specific binding of biotinylated CCL3 and CCL4 to L540 cells.
1 × 106 cells resting (1) or stimulated for 72 hours with plastic-bound anti-CD30 agonistic mAbs M67 (10 μg/mL) (2) were incubated with biotinylated CCL3 or CCL4 and stained with avidin-FITC reagent. Controls for specificity of binding were performed using non-biotinylated CCL3 and CCL4. Compared with resting cells (1), stimulation with M67 (2), but not with nonagonistic anti-CD30 mAb BerH2, induced a clear-cut increase in both CCL3 and CCL4 binding. Stimulation with 15 ng/mL PMA + 500 ng/mL ionomycin did not modify the binding of CCL3 and CCL4 (not shown), compared with basal conditions (1). The experiment depicted is representative of 5.
Thus, engagement of CD30 by CD153, which is associated preferentially with Th2-type effector responses,19regulates CXCR4 and CCR727 expression and delivers inhibitory signals,20,27 leading to impaired clonal expansion of peripheral T cells,21 sensitizing to TNFR1-dependent apoptosis,3 and counteracting IgG and IgA production.52 This cellular-level regulation seems to have a counterpart in a number of conditions, sharing the tendency to selective compartmentalization of low-proliferating CD30+cells, which deliver locally anti-inflammatory cytokines, such as IL-4 and IL-10, whenever tested for16 and which generate a molecular milieu characterized by features of immune suppression.16,20 These conditions include autoimmune diabetes in mouse models,21 rheumatoid arthritis,16 Omenn syndrome,14 a severe immunodeficiency characterized by features reminiscent of a Th2-type response in which large proportions of tissue-infiltrating CD30+ cells are present, measles53 in which CD30+ cell expansion is related to the impaired mitogenic capability of T lymphocytes, inadequate Th1-type responses, and cell recruitment to lymph nodes and skin. A faster progression to acquired immunodeficiency syndrome reported for asymptomatic HIV-1+patients with immune responses characterized by higher CD30 expression22,23 may be explained by the combined effect of NF-κB mobilization,24 up-regulation of viral coreceptors such as CXCR4,37 and diminishing ability to mount effective Th1-type responses, also related to the activity of anti-inflammatory CD30+ cells.16Pathology-based evidence supports a functional association of CD30 with chemokines or receptors, such as CXCR4 and CCR7, that cooperate in directing cell trafficking. This association is also suggested by cellular events during thymic maturation10,11 and by the natural history of CD30+ neoplasias.6,12 30
CXCR4–CXCL12 and CCR7–CCL21 systems are involved in the migration of hematopoietic and nonhematopoietic cells in vivo.28,29,31,38 CXCR4–CXCL12 mediates chemotaxis for monocytes, neutrophils, and T cells, but it is also involved in early activation events and in the production of IL-2, IFN-γ, IL-4, and IL-10 by anti-CD3–stimulated CD4+ T cells.29CXCR4- or CXCL12-deficient mice share perinatal lethality because of hematopoietic, cardiovascular, and nervous system development defects.29,38 The CCR7–CCL21 system plays an important role in naive lymphocyte homing to secondary lymphoid organs, which is impaired by disruption of the CCR7 gene54-56and is required for typical lymphoid tissue localization of Th1-type lymphocytes.56,57 Both CCR7 and CXCR4 participate in the recruitment of lymphocytes from blood,30,54 but CXCR4− lymphocytes develop and seed peripheral lymphoid tissues normally.54,58,59 The regulation of CXCR4 and CCR7 on CD30+ cells is likely to provide signals for the homing of activated T cells to specific sites28,29,31,60 to inhibit the delivery of B-cell help and the activity of cytolytic CD8+ cell trafficking.57
CD30+ cancers either of hematologic6 or nonhematologic lineage12 have distinctive clinical features and localize to CXCL12–CCL21-rich tissues, including the lymph nodes, spleen, liver, bone marrow, lung, and brain,4,30,61,62 with a diffusion pattern that may be conditioned by CD30-dependent regulation of CXCR4 and CCR7.30
The thymic microenvironment is rich in CXCL12+CD153+ epithelial cells and in CD30+CXCR4+ T cells. Overexpression of CD30 has been related to negative selection of double-positive cells in thymic medulla10 and the interaction of CD30 with CD153 to the constitution of Hassal corpuscles.11 Although there is no appreciable change in CXCL12 responsiveness from the cortex to the medulla,58 a less intense medullary expression of CXCR4 has been reported63 that may reflect the reduced density of lymphocytes there,58,63 as a result of apoptotic events favored by CD30 stimulation in the medullary context.10CCR7 is preferentially expressed in transitional cells57and medullary CD30+ cells.11,58 Thus, synergistic effects of CXCL12 and CCL2130 may be driven by CD30 stimulation during thymic differentiation.
One point of interest is whether the CD30-dependent increase in CXCR4 mRNA was transcriptionally regulated. It reached a plateau after 4 hours and was independent of protein synthesis. Elements regulating the transcription unit of the human CXCR4 gene include NRF-1, Sp1, and AP-1, but no NF-κB–related complexes.35,48,49 NRF-1 is a nuclear-encoded gene product important for the transcriptional regulation of mitochondrial cellular respiration genes64,65 and related to constitutive transcription of CXCR4 mRNA.35,49 Putative NRF-1 binding sites have been identified in several cell growth genes, including bcl-2 and murine GM-CSF.64,65 CD30 signaling leads to activation of at least NF-κB through a TRAF-dependent mechanism65 and AP-1 through jun N-terminal kinases. TRAF2 and related proteins are rapidly depleted by proteolysis after binding to the cytoplasmic domain of CD30,3,66,67 resulting in down-modulation of the proliferative potential and sensitization to TNFR1-mediated proapoptotic effects.3 NRF-1, Sp1, AP-1, and NF-κB were all translocated in unstimulated L540 cells. After stimulation with PMA–ion, both AP-1 and NF-κB further accumulated in the nuclei. This suggests that PMA–ion induces CXCR4 gene transcription through AP-1–mediated regulation. After CD30 cross-linking, NF-κB underwent further nuclear translocation, whereas NRF-1, Sp1, and AP-1 did not. AP-1, however, might be activated through phosphorylation mechanisms67 without evidence of further nuclear accumulation. Thus, CD30-mediated signals did not transcriptionally regulate the CXCR4 gene through Sp1 or NRF-1, which is probably responsible for the constitutive expression of CXCR4 in L540 cells.
In conclusion, our data demonstrate that CD30 participates in the regulation of molecules that permit selective migration to specific sites. This role of CD30 may be relevant during lymphoid differentiation or antigen-driven effector responses or in promoting cancer biology, and it may provide a unifying basis for explaining the significance of CD30 in physiology and pathology.
This article is dedicated to the memory of the hematologist Professor Giuseppe Perona, who passed away on April 30, 2001. He was a radical humanitarian, a source of wisdom, and a man of ideas, gifted with uncommon intelligence, culture, and scientific insight. The authors are indebted to him for their knowledge and sense of purpose.
Supported by grants from ISS-National AIDS Research Program, MURST (60% funds and cofinanziamento MURST–Università 40%), Associazione Italiana per la Ricerca sul Cancro (Milan), and Progetto Sanità, Fondazione Cassa di Risparmio VR-VI-BL-AN (Verona).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Fabrizio Vinante, Section of Hematology, Policlinico GB Rossi, 37134 Verona, Italy; e-mail:fabrizio.vinante@univr.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal