To investigate the role of viral expression in individuals infected with human T-cell lymphotropic virus type 1 (HTLV-1), a real-time quantitative reverse transcription-polymerase chain reaction (RT-PCR) of HTLV-1 tax messenger RNA (mRNA) using ABI Prism 7700 Sequence Detection System was developed. Using this system, the HTLV-1tax mRNA load was compared with HTLV-1 proviral DNA load, HTLV-1 Tax protein expression, HTLV-1 Tax-specific CD8+T-cell frequency, and disease severity of HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP). This approach was a sensitive and specific technique for the precise quantification of HTLV-1 tax mRNA. The total amount of HTLV-1 taxmRNA and mRNA expression level in HTLV-1–infected cells (mRNA/DNA ratio) were higher in HAM/TSP patients than in asymptomatic HTLV-1 carriers. The HTLV-1 tax mRNA load correlated with the HTLV-1 proviral DNA load ex vivo, the Tax protein expression in vitro, and the Tax-specific CD8+ T-cell frequency ex vivo. The HTLV-1 tax mRNA load also correlated with disease severity in HAM/TSP patients. These data suggest that increased HTLV-1 expression plays an important role in the pathogenesis of HAM/TSP, and the HTLV-1 tax mRNA level could be a useful predictor of disease progression in patients with HAM/TSP.
Introduction
Human T-cell lymphotropic virus type 1 (HTLV-1) is a human retrovirus that infects approximately 10 million people worldwide.1 The majority of infected individuals remain healthy lifelong asymptomatic carriers (ACs), approximately 0.25% to 3% develop an inflammatory disease of the central nervous system termed HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP),2-5 and another 2% to 3% develop an aggressive mature T-cell malignancy termed adult T-cell leukemia (ATL).6,7 Patients with HAM/TSP generally harbor a much larger population of HTLV-1–infected T cells in peripheral blood mononuclear cells (PBMCs)8-10 and a higher immune response against HTLV-1 antigens than ACs.11-13 These findings indicate that an abundant viral load plays an important role in the pathogenesis of HAM/TSP, suggesting that high viral expression may be associated with the high immune response against the virus.
Even though a high proviral DNA load is characteristic of patients with HAM/TSP, the expression of HTLV-1 in PBMCs appears to be very low,14-18 which suggests that HTLV-1 may be latent in peripheral blood. However, this suggestion is difficult to reconcile with the observation of high activated immune responses in patients with HAM/TSP.11-13 Moreover, it has been reported that a significant proportion of PBMCs infected with HTLV-1 are capable of expressing HTLV-1 proteins after short-term cultivation in vitro,17,19 supporting the hypothesis that HTLV-1 is not inherently latent. Recently, a correlation between the HTLV-1 proviral DNA load and the frequency of HTLV-1 Tax-specific CD8+ T cells20,21 has been described in patients with HAM/TSP. Collectively, these data suggest HTLV-1 antigens continuously restimulate the cellular immune system in vivo. However, no reports have characterized the HTLV-1 tax messenger RNA (mRNA) expression in the relationship between the HTLV-1 proviral DNA load and the virus-specific T-cell immune response in individuals infected with HTLV-1. In this study, to determine the importance of HTLV-1tax mRNA expression in that relationship and in the pathogenesis of HAM/TSP, a rapid, sensitive, and precise quantification of HTLV-1 tax mRNA expression was developed by a real-time reverse transcription-polymerase chain reaction (RT-PCR) method using the ABI Prism 7700 Sequence Detection System. Because the level of human immunodeficiency virus type 1 (HIV-1) RNA has been found to be a valid predictor of the clinical progression of HIV-1–related disease and efficacy of anti-HIV therapy,22 23 it was also of interest to determine if the HTLV-1 tax mRNA load as quantified by this novel method might be a marker of disease activity and of relative prognostic value in HAM/TSP.
In this study, the HTLV-1 tax mRNA load in PBMCs from patients with HAM/TSP and ACs was measured and compared with HTLV-1 proviral DNA load, HTLV-1 Tax protein expression, HTLV-1 Tax-specific CD8+ T-cell frequency, and disease severity in patients with HAM/TSP. The data presented demonstrate that increased HTLV-1 expression plays an important role in the pathogenesis of HAM/TSP, and HTLV-1 tax mRNA level could be a useful predictor of disease progression in HAM/TSP patients.
Patients, materials, and methods
Subjects and cells
The study population consisted of 16 patients with nontreated HAM/TSP (HAM-1 to HAM-16), 8 asymptomatic HTLV-1 carriers (AC-1 to AC-8), and 5 seronegative healthy donors (HDs). Infection of HTLV-1 was confirmed by enzyme-linked immunosorbent assay (Abbot Laboratories, North Chicago, IL) and Western blot analysis (Gene Labs, Singapore). The diagnosis of HAM/TSP was assessed according to the World Health Organization guidelines.24 The HAM-2 to -5, -11, -13, and -16 and AC-4, -6, and -7 individuals were positive for HLA-A*0201. All subjects were screened for HIV-1/2, hepatitis virus type B/C, HTLV-2, Epstein-Barr virus, Borrelia burgdorferi,and syphilis antibodies. HAM-8 and HAM-16 were coinfected with hepatitis virus type C. All samples were taken under informed consent. The clinical features of the patients included in this study are shown in Table 1. Disease severity was measured by using an expanded disability status scale (EDSS).25MT-2 cells and Hut 102 cells are lines of human CD4+ T cells infected with HTLV-1.26 27 Jurkat cells are human T-cell lines not infected with HTLV-1.
HTLV-1 tax mRNA load, HTLV-1 proviral DNA load, mRNA/DNA ratio, HTLV-1 Tax-specific CD8+ T-cell frequency, and clinical profiles of HAM/TSP patients and ACs
| Patients . | Age, y . | Sex . | Origin . | tax mRNA load . | Proviral DNA load . | mRNA/DNA ratio . | Tax-specific CD8+ T-cell frequency, % . | EDSS . | Duration of illness, y . |
|---|---|---|---|---|---|---|---|---|---|
| HAM-1 | 38 | F | USA | 0.06 | 1.62 | 3.70 | — | 5.5 | 6 |
| HAM-2 | 47 | F | USA | 1.61 | 11.92 | 13.51 | 0.52 | 6.0 | 10 |
| HAM-3 | 63 | M | USA | 2.32 | 25.16 | 9.22 | 5.70 | 1.5 | 18 |
| HAM-4 | 78 | F | USA | 5.00 | 23.20 | 21.55 | 3.53 | 2.0 | 12 |
| HAM-5 | 46 | F | USA | 5.95 | 12.38 | 48.06 | 4.21 | 3.5 | 12 |
| HAM-6 | 47 | F | Caribbean | 9.76 | 9.37 | 104.16 | — | 6.5 | 2 |
| HAM-7 | 57 | F | USA | 11.79 | 20.98 | 56.20 | — | 6.5 | 3 |
| HAM-8 | 48 | F | USA | 12.32 | 11.70 | 105.30 | — | 6.0 | 4 |
| HAM-9 | 56 | F | USA | 13.62 | 24.76 | 55.01 | — | 6.0 | 13 |
| HAM-10 | 64 | M | USA | 17.78 | 20.67 | 86.02 | — | 6.0 | 12 |
| HAM-11 | 59 | M | Jamaica | 20.08 | 21.99 | 91.31 | 4.71 | 7.0 | 10 |
| HAM-12 | 45 | F | USA | 25.44 | 23.09 | 110.18 | — | 6.0 | 3 |
| HAM-13 | 59 | M | USA | 71.18 | 22.89 | 310.97 | 17.64 | 7.0 | 14 |
| HAM-14 | 44 | M | USA | 109.10 | 47.14 | 231.44 | — | 5.5 | 2 |
| HAM-15 | 79 | M | USA | 137.70 | 69.49 | 198.16 | — | 8.0 | 12 |
| HAM-16 | 49 | M | USA | 187.90 | 60.16 | 312.33 | 24.35 | 8.0 | 6 |
| Mean | 39.48 | 25.41 | 109.82 | 8.67 | |||||
| Median | 12.97* | 22.44† | 88.67‡ | ||||||
| AC-1 | 61 | F | USA | 0.00 | 0.00 | 0.00 | — | — | — |
| AC-2 | 62 | M | USA | 0.00 | 0.07 | 0.00 | — | — | — |
| AC-3 | 49 | F | USA | 0.01 | 0.02 | 50.00 | — | — | — |
| AC-4 | 62 | F | USA | 0.11 | 4.35 | 2.53 | 0.06 | — | — |
| AC-5 | 37 | M | USA | 0.51 | 2.92 | 17.47 | — | — | — |
| AC-6 | 66 | F | USA | 2.87 | 7.97 | 36.01 | 0.45 | — | — |
| AC-7 | 23 | M | USA | 2.94 | 3.61 | 81.44 | 1.64 | — | — |
| AC-8 | 44 | F | Japan | 5.58 | 3.10 | 180.00 | — | — | — |
| Mean | 1.50 | 2.76 | 45.93 | 0.72 | |||||
| Median | 0.31* | 3.01† | 26.74‡ |
| Patients . | Age, y . | Sex . | Origin . | tax mRNA load . | Proviral DNA load . | mRNA/DNA ratio . | Tax-specific CD8+ T-cell frequency, % . | EDSS . | Duration of illness, y . |
|---|---|---|---|---|---|---|---|---|---|
| HAM-1 | 38 | F | USA | 0.06 | 1.62 | 3.70 | — | 5.5 | 6 |
| HAM-2 | 47 | F | USA | 1.61 | 11.92 | 13.51 | 0.52 | 6.0 | 10 |
| HAM-3 | 63 | M | USA | 2.32 | 25.16 | 9.22 | 5.70 | 1.5 | 18 |
| HAM-4 | 78 | F | USA | 5.00 | 23.20 | 21.55 | 3.53 | 2.0 | 12 |
| HAM-5 | 46 | F | USA | 5.95 | 12.38 | 48.06 | 4.21 | 3.5 | 12 |
| HAM-6 | 47 | F | Caribbean | 9.76 | 9.37 | 104.16 | — | 6.5 | 2 |
| HAM-7 | 57 | F | USA | 11.79 | 20.98 | 56.20 | — | 6.5 | 3 |
| HAM-8 | 48 | F | USA | 12.32 | 11.70 | 105.30 | — | 6.0 | 4 |
| HAM-9 | 56 | F | USA | 13.62 | 24.76 | 55.01 | — | 6.0 | 13 |
| HAM-10 | 64 | M | USA | 17.78 | 20.67 | 86.02 | — | 6.0 | 12 |
| HAM-11 | 59 | M | Jamaica | 20.08 | 21.99 | 91.31 | 4.71 | 7.0 | 10 |
| HAM-12 | 45 | F | USA | 25.44 | 23.09 | 110.18 | — | 6.0 | 3 |
| HAM-13 | 59 | M | USA | 71.18 | 22.89 | 310.97 | 17.64 | 7.0 | 14 |
| HAM-14 | 44 | M | USA | 109.10 | 47.14 | 231.44 | — | 5.5 | 2 |
| HAM-15 | 79 | M | USA | 137.70 | 69.49 | 198.16 | — | 8.0 | 12 |
| HAM-16 | 49 | M | USA | 187.90 | 60.16 | 312.33 | 24.35 | 8.0 | 6 |
| Mean | 39.48 | 25.41 | 109.82 | 8.67 | |||||
| Median | 12.97* | 22.44† | 88.67‡ | ||||||
| AC-1 | 61 | F | USA | 0.00 | 0.00 | 0.00 | — | — | — |
| AC-2 | 62 | M | USA | 0.00 | 0.07 | 0.00 | — | — | — |
| AC-3 | 49 | F | USA | 0.01 | 0.02 | 50.00 | — | — | — |
| AC-4 | 62 | F | USA | 0.11 | 4.35 | 2.53 | 0.06 | — | — |
| AC-5 | 37 | M | USA | 0.51 | 2.92 | 17.47 | — | — | — |
| AC-6 | 66 | F | USA | 2.87 | 7.97 | 36.01 | 0.45 | — | — |
| AC-7 | 23 | M | USA | 2.94 | 3.61 | 81.44 | 1.64 | — | — |
| AC-8 | 44 | F | Japan | 5.58 | 3.10 | 180.00 | — | — | — |
| Mean | 1.50 | 2.76 | 45.93 | 0.72 | |||||
| Median | 0.31* | 3.01† | 26.74‡ |
The mRNA/DNA ratio was calculated by the following formula: mRNA/DNA ratio = (HTLV-1 tax mRNA load/HTLV-1 proviral DNA load) × 100.
The Tax-specific CD8+ T-cell frequency is expressed as a percentages of HTLV-1 Tax11-19/HLA-A*0201 tetramer-specific CD8+ T cells in the total amount of CD8+ T cells.
— indicates subjects who are not HLA-A*0201.
HTLV-1 tax mRNA load, HTLV-1 proviral DNA load, and mRNA/DNA ratio were higher in HAM/TSP patients than in ACs with statistical significance by the Mann-Whitney U test:
P = .0014;
P = .0003;
P = .0433. The EDSS score significantly correlated with both HTLV-1 tax mRNA load and mRNA/DNA ratio (P = .0109 and .0089, respectively), but not with HTLV-1 proviral DNA load (P = .5236) by Spearman rank correlation analysis.
Real-time RT-PCR of complementary DNA
RNA was extracted from PBMCs using RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized using TaqMan Gold RT-PCR Kit (Applied Biosystems, Foster City, CA). For cDNA synthesis from extracted mRNA, 3 μg mRNA, 10 μL 10 × TaqMan RT buffer, 22 μL MgCl2 (25 mM), 20 μL dNTPs mixture (at a final concentration of 500 μM each), 5μL random hexamers (50 μM), 2 μL RNase inhibitor (20 U/μL), and 2.5 μL (50 U/μL) Moloney murine leukemia virus reverse transcriptase were added to a total volume of 100 μL. Samples were incubated at 25°C for 10 minutes and 48°C for 30 minutes. Reactions were stopped by heating to 95°C for 5 minutes.
Primers used for the amplification of HTLV-1 tax cDNA were slightly modified from the sequence previously reported18 to be suitable for this system. The forward primer RPX-3 (5096-5115; 5′-ATCCCGTGGAGACTCCTCAA-3′) and the reverse primer RPX-4 + 3 (7360-7338; 5′-CCAAACACGTAGACTGGGTATCC-3′) were located upstream and downstream of the second splice junction site of HTLV-1 pX (tax/rex) mRNA. The amplified DNA was 147 base pairs (bp). The probe (5172-5183, 7302-7312; 5′-TCCAACACCATG-GCCCACTTCCC-3′) surrounded the second splice junction site of HTLV-1 pX (tax/rex) mRNA. We used the human housekeeping gene hypoxanthine ribosyl transferase (HPRT) primers and probe set (Applied Biosystems) for internal calibration. This internal control appeared to be most stable within PBMCs from HDs, ACs, HAM/TSP patients, and ATL patients by using the TaqMan Human Endogenous Control Plate (Applied Biosystems). PCR conditions were as follows: 10 μL cDNA solution synthesized from 300 ng RNA was added to 40 μL reaction mixture containing10 mM Tris-HCl (pH 8.3), 50 mM KCl, 10 mM EDTA, 60 nM ROX (passive reference dye to normalize receptor signal), 5.5 mM MgCl2, 0.3 μM each primer, 0.2 μM TaqMan probe, 200 μM each of dATP, dGTP, and dCTP, 400 μM dUTP, 0.5 U AmpErase uracil-N-glycosylase (UNG), and 0.25 U AmpliTaq Gold DNA polymerase (AmpliTaq Gold; Applied Biosystems). The thermal cycler conditions were 2 minutes at 50°C to activate AmpErase UNG enzyme, 10 minutes at 95°C to inactivate UNG and activate AmpliTaq Gold DNA polymerase, and then 45 cycles of 15 seconds at 95°C (denaturation) followed by 1 minute at 60°C (annealing and extension).
Standard curves for the value of HTLV-1 tax mRNA and HPRT mRNA were generated using cDNA from MT-2 cells. MT-2 cDNA was serially diluted 10-fold with diethyl pyrocarbonate (DEPC) H2O down to a 10−4 dilution, and sample cDNA from 300 ng RNA per well was applied and analyzed by this system. The amplification of standard cDNA and sample cDNA was carried out in a 96-well reaction plate. All standards and samples were assayed in triplicate. Each plate always contained the same standard. When a serial dilution of 100 to 10−4 of the MT-2 cell cDNA was used as the template for the real-time PCR, a specific signal of each increased in accordance with the increase of PCR cycles, but not in the negative control. The threshold cycle (Ct) values were used to plot a standard curve in which Ct decreased in linear proportion to the log of the template copy number. The correlation values of standard curves were always more than 99%.
The relative HTLV-1 tax mRNA load was calculated by the following formula: HTLV-1 tax mRNA load = (value oftax)/(value of HPRT) × 10 000.
Real-time PCR of DNA
The HTLV-1 proviral DNA load was measured using this same TaqMan system as previously described.10 21 DNA was extracted from 1 × 106 cells using Puregene DNA Isolation Kit (Gentra, Minneapolis, MN) and 100 ng of the sample DNA solution per well was analyzed by this system. The HTLV-1 proviral DNA load was calculated by the following formula: copy number of HTLV-1 (pX) per 100 cells = (copy number of pX)/(copy number of β-actin/2) × 100.
HTLV-1 Tax protein expression in PBMCs
The PBMCs (5 × 105) were placed on a culture well (round bottom 96-well plate) in 200 μL RPMI 1640 supplemented withl-glutamine, penicillin, streptomycin, and 5% human AB serum. Harvested cells were washed with phosphate-buffered saline (PBS) containing 1% fetal calf serum (FCS) and 0.1% NaN3 and incubated with antihuman CD4-PE (Caltag Laboratories, Burlingame, CA) and antihuman CD8-Tricolor monoclonal antibodies (mAbs; Caltag Laboratories) for 20 minutes at 4°C. Following washing with PBS containing 1% FCS and 0.1% NaN3, cells were fixed and permeabilized with 4% formaldehyde and 0.1% saponin (CytoFix/Cytoperm kits, Pharmingen, SanDiego, CA) for 20 minutes at 4°C. After washing with 0.1% saponin buffer (Perm/Wash solution, Pharmingen), cells were incubated with anti-HTLV-1 Tax mAb (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HTLV-1 Tax hybridoma 168A51-42 from Dr Beatrice Langton) for 30 minutes at 4°C. Followed by washing unbound antibody with 0.1% saponin buffer, fluorescein isothiocyanate (FITC)–conjugated goat F(ab′)2antimouse IgG2a was used as second antibody for labeling anti-HTLV-1 Tax mAb. After a 20-minute incubation, cells underwent a final wash. Flow cytometric analyses were performed using a FACS Calibur (Becton Dickinson, Mountain View, CA).
Identification of HTLV-1 Tax-specific CD8+ T cells
The HTLV-1 Tax 11-19 peptide is a dominant epitope recognized by HLA-A2–restricted CD8+ T cells in HAM/TSP patients.13 28 HTLV-1 Tax11-19/HLA A*0201-specific CD8+ T cells were analyzed using a phycoerythrin (PE)–conjugated HLA-A*0201 tetramer (provided by NIAID MHC Tetramer Core Facility, Atlanta, GA, and NIH AIDS Research and Reference Reagent Program) for HLA A*0201 subjects. The tetramer was loaded with HTLV-1 Tax 11-19 peptide (LLFGYPVYV; single-letter amino acid codes), which was synthesized and 95% purified by high-performance liquid chromatography (HPLC; New England Peptide, Fitchburg, MA). Tetramer loaded with HIV Gag 77-85 peptide (SLYNTVATL) was used as a negative control. PBMCs from HLA-A*0201 subjects were washed and incubated with PE-conjugated HLA-A*0201 tetramer, antihuman CD4-FITC (Caltag Laboratories), and antihuman CD8-Tricolor mAbs (Caltag Laboratories) for 30 minutes at 4°C, and then analyzed by flow cytometry. The frequencies of HTLV-1 Tax-specific CD8+ T cells were expressed as a percentage of total CD8+ T cells.
Statistical analysis
The Mann-Whitney U test was used to compare the data between patients with HAM/TSP and ACs. Simple regression analysis was used to test the correlation between HTLV-1 proviral DNA load, HTLV-1tax mRNA load, mRNA/DNA ratio, and HTLV-1 Tax-specific CD8+ T-cell frequency. To test the correlation of EDSS with HTLV-1 proviral DNA load, HTLV-1 tax mRNA load and mRNA/DNA ratio, the Spearman rank correlation was used.
Results
Accuracy of real-time RT-PCR
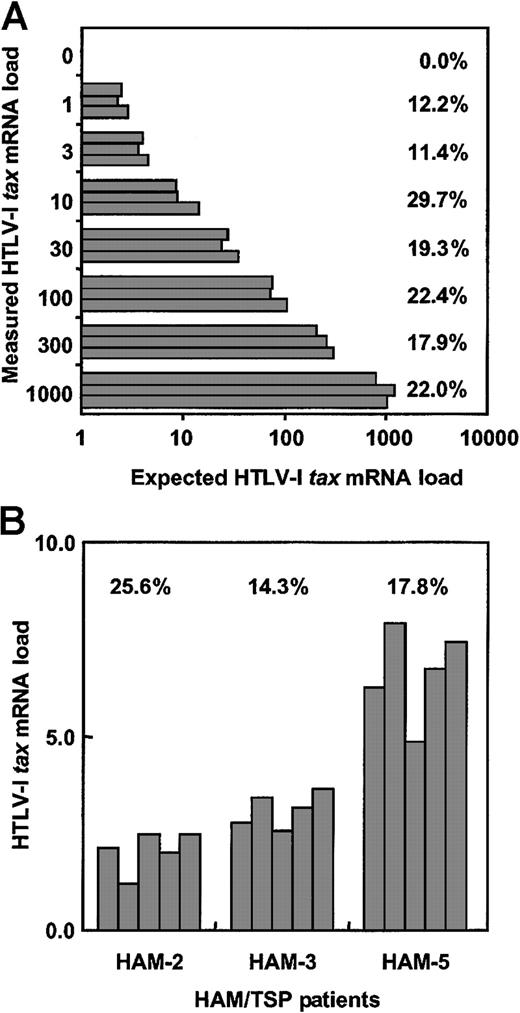
To determine the interassay coefficient of variance of this real-time HTLV-1 tax RT-PCR system, cDNA from the HTLV-1–infected MT-2 cell was diluted serially with cDNA from the HTLV-1–noninfected Jurkat cell, and HTLV-1 tax mRNA load of each dilution was measured 5 times (Figure1A). In this study, the HTLV-1tax mRNA load was calculated as the value of HTLV-1tax mRNA divided by the value of HPRT as described in “Patients, materials, and methods.” To estimate the accuracy of this system, we calculated the intercoefficient of variance (CV%) in each amount. The mean interassay CV% was 19.3%, which is consistent with the CV% measuring HTLV-1 proviral DNA using real-time PCR methods.10 29
Validation of real-time quantitative RT-PCR assay.
(A) To determine interassay CV% for this real-time RT-PCR assay, cDNA from the HTLV-1–infected MT-2 cell line was serially diluted with cDNA from the uninfected Jurkat cell line and HTLV-1 tax mRNA load was measured 5 times per dilution. (B) The intra-assay CV% was determined by measuring mRNA load from the PBMCs of 3 HAM/TSP patients in 5 separate experiments. The CV% represents (SD of HTLV-Itax mRNA load/average of HTLV-I tax mRNA load) × 100.
Validation of real-time quantitative RT-PCR assay.
(A) To determine interassay CV% for this real-time RT-PCR assay, cDNA from the HTLV-1–infected MT-2 cell line was serially diluted with cDNA from the uninfected Jurkat cell line and HTLV-1 tax mRNA load was measured 5 times per dilution. (B) The intra-assay CV% was determined by measuring mRNA load from the PBMCs of 3 HAM/TSP patients in 5 separate experiments. The CV% represents (SD of HTLV-Itax mRNA load/average of HTLV-I tax mRNA load) × 100.
To test the intra-assay coefficient of variance of this real-time HTLV-1 tax RT-PCR system, PBMCs from 3 patients with HAM/TSP were separated into 5 samples per individual. RNA was extracted from each sample, cDNA was synthesized, and HTLV-1 tax mRNA load for each sample was determined (Figure 1B). The CV% was calculated for each patient. The mean value of intra-assay CV% was 19.2%, which is consistent with the CV% measuring HIV-1 RNA.30 These data demonstrate that this real-time HTLV-1 tax RT-PCR assay is accurate and reliable for quantitation of HTLV-1 tax mRNA.
Sensitivity and specificity of real-time RT-PCR
To determine the sensitivity of this real-time RT-PCR assay, the HTLV-1 tax mRNA load was determined from MT-2 cells diluted serially with PBMCs from a healthy donor not infected with HTLV-1. As shown in Figure 2A, the HTLV-1tax mRNA signal could be detected in a dose-dependent manner with a sensitivity limit as low as one MT-2 cell in 106 PBMCs.
Sensitivity and specificity of real-time RT-PCR assay.
(A) To determine the sensitivity of this real-time RT-PCR assay, HTLV-1 tax mRNA load was determined from MT-2 cells serially diluted with PBMCs from an HTLV-1− healthy donor. HTLV-Itax mRNA could be detected from as little as a 10−6-fold dilution of MT-2 cells. (B) The specificity of this assay was determined by measuring HTLV-1 tax mRNA expression of PBMCs from 5 HTLV-1–noninfected HDs, HTLV-1–uninfected Jurkat cells, and 2 HTLV-1–infected cell lines (MT-2 and Hut 102). HTLV-I tax mRNA was detected in the 2 infected cell lines, but not in uninfected cells.
Sensitivity and specificity of real-time RT-PCR assay.
(A) To determine the sensitivity of this real-time RT-PCR assay, HTLV-1 tax mRNA load was determined from MT-2 cells serially diluted with PBMCs from an HTLV-1− healthy donor. HTLV-Itax mRNA could be detected from as little as a 10−6-fold dilution of MT-2 cells. (B) The specificity of this assay was determined by measuring HTLV-1 tax mRNA expression of PBMCs from 5 HTLV-1–noninfected HDs, HTLV-1–uninfected Jurkat cells, and 2 HTLV-1–infected cell lines (MT-2 and Hut 102). HTLV-I tax mRNA was detected in the 2 infected cell lines, but not in uninfected cells.
To test the specificity of this assay, cDNA from HTLV-1–noninfected Jurkat cells and PBMCs from HTLV-1–noninfected HDs (n = 5) were analyzed. No signal was observed from any of these HTLV-1−cells (Figure 2B). MT-2 cDNA without RT also showed no signal. Two HTLV-1–infected cell lines were tested for expression of HTLV-1tax mRNA by this system (Figure 2B). The HTLV-1tax mRNA was detected in these 2 cell lines, with Hut 102 cells expressing 20 times more HTLV-1 tax mRNA than MT-2 cells. This difference is consistent with previous reports demonstrating that Hut 102 cells have 10 to 17 HTLV-1 proviral copies per cell compared to MT-2 cells, which have 2 to 8 HTLV-1 proviral copies per cell.29 31-33
HTLV-1 tax mRNA load in HAM/TSP patients and ACs
Using this novel method, HTLV-1 tax mRNA load was analyzed in PBMCs from HAM/TSP patients and ACs. As demonstrated in Table 1 and Figure 3, the HTLV-1tax mRNA load ranged from 0.06 to 187.90 in HAM/TSP patients and 0.00 to 5.58 in ACs. The HTLV-1 tax mRNA load in HAM/TSP patients was significantly higher than that in ACs (P = .0014, Mann Whitney U test) as reported previously.34 The absolute amount of HTLV-1 taxmRNA before adjusting by the HPRT value in HTLV-1–infected individuals was 103 to 106 times less than that in MT-2 cells, which is consistent with previous reports.18 34
HTLV-I
tax mRNA load in HAM/TSP patients and ACs. HTLV-1 tax mRNA load was assessed in PBMCs from 16 HAM/TSP patients (HAM) and 8 asymptomatic HTLV-I carriers (AC). HTLV-1tax mRNA load was significantly higher (P = .0014) in HAM/TSP patients compared to ACs.
HTLV-I
tax mRNA load in HAM/TSP patients and ACs. HTLV-1 tax mRNA load was assessed in PBMCs from 16 HAM/TSP patients (HAM) and 8 asymptomatic HTLV-I carriers (AC). HTLV-1tax mRNA load was significantly higher (P = .0014) in HAM/TSP patients compared to ACs.
Comparison of HTLV-1 tax mRNA load with HTLV-1 proviral DNA load ex vivo
To determine if there was a correlation between the HTLV-1 proviral DNA load and the HTLV-1 tax mRNA load in HTLV-1–infected individuals, a real-time quantitative HTLV-1 DNA PCR assay21 was used to measure the HTLV-1 proviral DNA load in PBMCs from HAM/TSP patients and ACs. As previously reported,10 21 the HTLV-1 proviral DNA load, which represented a population of infected cells in PBMCs, was greater in HAM/TSP patients than ACs (P = .0003, Mann WhitneyU test, Table 1). Furthermore, the HTLV-1 proviral DNA load was correlated significantly with the HTLV-1 tax mRNA load in HAM/TSP patients (P < .0001, r2 = 0.812, simple regression analysis, Figure 4).
Correlation between HTLV-1 proviral DNA load and HTLV-1
tax mRNA load in HAM/TSP patients. A statistically significant correlation (P < .0001, r2 = 0.812) between the HTLV-1 proviral DNA load and HTLV-1 tax mRNA load was observed in PBMCs from HAM/TSP patients.
Correlation between HTLV-1 proviral DNA load and HTLV-1
tax mRNA load in HAM/TSP patients. A statistically significant correlation (P < .0001, r2 = 0.812) between the HTLV-1 proviral DNA load and HTLV-1 tax mRNA load was observed in PBMCs from HAM/TSP patients.
To adjust the HTLV-1 tax mRNA expression level in HTLV-1–infected PBMCs, we calculated the mRNA/DNA ratio by dividing the HTLV-1 tax mRNA load by HTLV-1 proviral DNA load. The mean value of the mRNA/DNA ratio was 109.82 for HAM/TSP patients and 45.93 for ACs. The mRNA/DNA ratio of HAM/TSP patients was statistically higher than that of AC (P = .0433, Mann WhitneyU test, Table 1).
Comparison of HTLV-1 tax mRNA load with HTLV-1 Tax protein expression in vitro
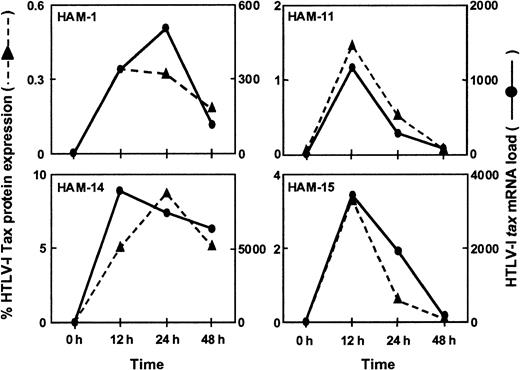
Because increased levels of mRNA may be associated with increased protein production, it was important to determine if there was a correlation between the HTLV-1 tax mRNA load and the amount of HTLV-1 Tax protein expression. PBMCs from 4 HAM/TSP patients (HAM-1, -11, -14, -15) were cultured for 48 hours in culture medium without mitogenic stimuli. PBMC samples were taken after 0, 12, 24, and 48 hours, respectively, and the HTLV-1 tax mRNA load and HTLV-1 Tax protein expression level as measured by FACS analysis using monoclonal anti-Tax antibodies were determined. As shown in Figure5, a close correlation was found between the HTLV-1 tax mRNA load and the percentage of cells expressing HTLV-1 Tax protein. The detection of HTLV-1 tax mRNA and the expression of HTLV-1 Tax protein were coincident and peaked at approximately 12 to 24 hours of in vitro culture (Figure 5).
Correlation between HTLV-1
tax mRNA load and HTLV-1 Tax protein expression in vitro. PBMCs from 4 HAM/TSP patients were cultured for 48 hours. Cultured PBMC samples were harvested after 0, 12, 24, and 48 hours to estimate HTLV-1tax mRNA load and the percentage of HTLV-1 Tax protein expressing cells. HTLV-1 Tax protein expression closely followed HTLV-1tax mRNA expression in the PBMCs from these individuals.
Correlation between HTLV-1
tax mRNA load and HTLV-1 Tax protein expression in vitro. PBMCs from 4 HAM/TSP patients were cultured for 48 hours. Cultured PBMC samples were harvested after 0, 12, 24, and 48 hours to estimate HTLV-1tax mRNA load and the percentage of HTLV-1 Tax protein expressing cells. HTLV-1 Tax protein expression closely followed HTLV-1tax mRNA expression in the PBMCs from these individuals.
Comparison of HTLV-1 tax mRNA load with HTLV-1 Tax-specific CD8+T-cell frequency ex vivo
The high levels of HTLV-1 proviral DNA load detected in PBMCs from HAM/TSP patients have been shown previously to correlate with high frequencies of virus-specific CD8+ T cells.20,21 It was therefore of interest to determine if HTLV-1 tax mRNA load also correlated with the frequency of HTLV-1 Tax-specific CD8+ T cells. The frequency of HTLV-1 Tax 11-19/HLA A*0201-specific CD8+ T cells was analyzed using a PE-conjugated HLA-A*0201 tetramer for HLA-A*0201 subjects (HAM-2-5, -11, -13, -16 and AC-4, -6, -7). The percentages of HTLV-1 Tax 11-19/HLA A*0201-specific CD8+ T cells in the total amount of CD8+ T cells are summarized in Table 1. As reported previously,20,21 the HTLV-1 proviral DNA load significantly correlated with the frequency of HTLV-1 Tax-specific CD8+ T cells in HAM/TSP patients (P = .0191, r2 = 0.699, simple regression analysis, Table 1). Furthermore, the HTLV-1 tax mRNA load was also significantly correlated with the frequency of HTLV-1 Tax-specific CD8+ T cells in HAM/TSP patients (P = .0013, r2 = 0.893, simple regression analysis; Table 1 and Figure 6). HTLV-1 Tax 11-19/HLA A*0201-specific CD8+ T cells have also been reported in PBMCs of ACs35 and although in this study the sample size of HLA A*0201+ ACs was too small to compare statistically, increased mRNA load in this group was associated with a higher frequency of HTLV-1 Tax-specific CD8+ T cells (Table1).
Correlation of HTLV-1 Tax-specific CD8+ T-cell frequency with HTLV-1 tax mRNA load.
The frequency of HTLV-1 Tax 11-19/HLA-A*0201-specific CD8+ T cells (expressed as a percentage of total CD8+ T cells) was analyzed using a PE-conjugated HLA-A*0201 tetramer for HLA-A*0201 subjects. A significant correlation (P = .0013, r2 = 0.893) was demonstrated between HTLV-1 tax mRNA load and the frequency of HTLV-1 Tax-specific CD8+ T cells in HAM/TSP patients.
Correlation of HTLV-1 Tax-specific CD8+ T-cell frequency with HTLV-1 tax mRNA load.
The frequency of HTLV-1 Tax 11-19/HLA-A*0201-specific CD8+ T cells (expressed as a percentage of total CD8+ T cells) was analyzed using a PE-conjugated HLA-A*0201 tetramer for HLA-A*0201 subjects. A significant correlation (P = .0013, r2 = 0.893) was demonstrated between HTLV-1 tax mRNA load and the frequency of HTLV-1 Tax-specific CD8+ T cells in HAM/TSP patients.
Comparison of disease severity with HTLV-1 proviral DNA load, HTLV-1 tax mRNA load, and mRNA/DNA ratio in HAM/TSP patients
Clinical features of HAM/TSP patients included in this study are shown in Table 1. Because HAM/TSP is a chronic progressive disease, patients with a long duration of illness are usually severely affected.36 However, there were 3 patients (HAM-3, -4, -5) who showed relatively mild progression and had a long duration of illness as reflected by relatively low scores on the EDSS (Table 1). The EDSS index is a neurologic measure of disability that is weighted toward ambulation.25 Of interest is the observation that the HTLV-1 tax mRNA loads of all 3 of these patients were low, even though 2 of the 3 patients (HAM-3, -4) harbored average levels of HTLV-1 proviral DNA load. Furthermore, patients with high HTLV-1 tax mRNA load and a long duration of illness (HAM-11, -13, -15, -16) had more severe disease with EDSS scores above 7 (patients essentially restricted to a wheelchair).
To examine the possibility that quantitative real-time PCR for HTLV-1 DNA and RNA determinations might have prognostic value in HAM/TSP disease progression, the EDSS values of these patients were statistically compared with HTLV-1 proviral DNA load, HTLV-1tax mRNA load, and mRNA/DNA ratio by Spearman rank correlation analysis (Table 1). The EDSS score significantly correlated with both HTLV-1 tax mRNA load and mRNA/DNA ratio (P = .0109 and .0089, respectively), but not with HTLV-1 proviral DNA load (P = .5236). Thus, HTLV-1 taxmRNA load and mRNA/DNA ratio may have relative prognostic value for the assessment of disease progression in HAM/TSP.
Discussion
Using a newly established real-time RT-PCR assay to quantitate HTLV-1 tax mRNA load, this study demonstrated a significant difference in the HTLV-1 tax mRNA load between patients with HAM/TSP and ACs. Furthermore, HTLV-1 tax mRNA load significantly correlated with the HTLV-1 proviral DNA load, the HTLV-1 Tax protein expression, and the frequency of HTLV-1 Tax 11-19/HLA A*0201-specific CD8+ T cells. Collectively, these results support previous studies demonstrating a relationship between the HTLV-1 proviral DNA load and the frequency of HTLV-1 Tax-specific CD8+ T cells in HAM/TSP patients,20 the existence of chronically activated HTLV-1 Tax-specific CD8+T cells,21 and the existence of high anti-HTLV-1 IgM antibodies suggesting continuous immune stimulation in vivo.37 These results indicate that HAM/TSP patients have more HTLV-1 proviral DNA load than ACs, which may be associated with higher mRNA expression resulting in heightened virus-specific immune responses. This profile does not characterize all retroviruses. For example, in individuals infected with HIV-1, the plasma viral RNA load, which is known to be significantly correlated with HIV-1 RNA load in PBMCs,38,39 is inversely proportional to the frequency of HIV-1–specific CD8+ T cells,40 41 which suggests an effective immunologic elimination of HIV-1 antigens. However, in HTLV-1 infection, as the virus antigen expression increases, HTLV-1–specific CD8+ T cells are generated, which appear to be unable to eliminate infected cells. Therefore, these results support the hypothesis that expansion of HTLV-1 can occur in the presence of an activated immune system that is trying to control the infection. This novel assay is also useful to define the role of HTLV-1 viral expression in patients with ATL. In preliminary data, HTLV-1 tax mRNA load was generally low in patients with ATL even in individuals with a high proviral DNA load (data not shown).
Quantitative measurements of HTLV-1 tax mRNA load in PBMCs from HAM/TSP patients and ACs continue to highlight differences between these 2 groups. This study demonstrated that adjusting the HTLV-1 tax mRNA load with HTLV-1 proviral DNA load (mRNA/DNA ratio) was higher in HAM/TSP patients than in ACs. These results indicate that in HAM/TSP patients a higher proportion of HTLV-1–infected cells express HTLV-1 tax mRNA at higher levels than in ACs. These observations support and extend previous reports describing a number of host genetic and virologic differences between HAM/TSP patients and ACs including differences in human leukocyte antigen haplotypes,35,42 differences in the amount of soluble suppressive factors16,43 and CD8+ T-cell responses,19-21,44 and differences in HTLV-1 tax genomic sequences.45 Many of these observations distinguishing HAM/TSP patients from carriers may also be related to higher proviral DNA load, higher mRNA load (as demonstrated in this study), and higher immune response against HTLV-1.10-13 The mRNA/DNA ratios as determined by quantitative DNA and RNA PCR assays presented here differed from a previous report34 that showed that the mRNA/DNA ratio was similar in both HAM/TSP patients and ACs although the total HTLV-1tax mRNA load was higher in HAM/TSP patients than in ACs (also demonstrated in the present study). This may be due to differences in the technology and accuracy of the assays used. Therefore, it is important to extend the observations on differences in the mRNA/DNA ratio between HAM/TSP patients and ACs to larger groups of individuals infected with HTLV-1.
Because HAM/TSP is a chronic progressive disorder,36clinical end points are cumbersome when evaluating disease activity. Thus, a clinical and laboratory goal is to define surrogate markers of HTLV-1 infection that can be used to monitor HAM/TSP disease activity. Surrogate markers of HAM/TSP disease activity have been reported. Neopterin levels in cerebrospinal fluid (CSF), a marker of cell-mediated immune activation,46,47 have been used as an immunologic marker for monitoring disease activity and treatment efficacy.48 Monitoring of ex vivo spontaneous lymphoproliferation and the frequency of HTLV-1 Tax-specific CD8+ T cells, which have reported to be associated with the pathogenesis of HAM/TSP,12,20,21 have also been used as potential surrogate immunologic markers of disease progression.49 In this present report, HTLV-1tax mRNA load was correlated with HTLV-1 Tax-specific CD8+ T-cell frequencies in HAM/TSP patients, suggesting that the quantitative analysis of HTLV-1 tax mRNA might also be used to monitor HAM/TSP disease activity. In this study, the frequency of CD8+ T cells specific only to the HTLV-1 Tax region was analyzed; however, other HTLV-1 regions have also been shown to be important as targets for CD8+ T-cell immune responses.50 51
In the assessment of any new HTLV-1–associated immunologic assay, it is of interest to determine if it can be used as a prognostic indicator of disease progression in HAM/TSP. For example, it has been reported that the anti-HTLV-1 antibody titers and the neopterin levels in CSF are high in patients with acute progressive HAM/TSP and low in those with slowly progressive disease.36 The HTLV-1tax mRNA load measured by this novel real-time RT-PCR method was demonstrated to generate a quantitative prognostic index of clinical progression in a limited cohort of HAM/TSP patients. The EDSS value is a commonly used neurologic measure of disability25 and significantly correlated with both HTLV-1tax mRNA load and mRNA/DNA ratio (P = .0109 and .0089, respectively), but interestingly not with HTLV-1 proviral DNA load (P = .5236). This tendency has been also observed in HIV-1 infection.22 23 These findings suggest that the amounts of expressed antigen and induced immune response may be more sensitive than proviral DNA load as a prognostic indicator in virus-associated diseases. As with any chronic, progressive neurologic disease, it is difficult to predict clinical progression. However, the HTLV-1 tax mRNA load was demonstrated to reflect viral activity and could be an important marker to be used to predict long-term outcome in HAM/TSP patients. We are currently evaluating virologic and immunologic markers described here over time in HAM/TSP patients with or without treatment. HTLV-1 proviral DNA load, HTLV-1tax mRNA load, and HTLV-1 Tax-specific CD8+T-cell frequency are generally stable over time in untreated patients (data not shown). Further longitudinal and controlled studies enrolling a large number of patients must be conducted.
In summary, a real-time quantitative RT-PCR of HTLV-1tax mRNA has been developed. This approach proved to be a sensitive and specific technique for precise quantification of HTLV-1tax mRNA. The novel assay demonstrated a significant correlation between HTLV-1 DNA, RNA, protein, and HTLV-1–specific T-cell immune response. Additionally, HTLV-1 expression levels in virus-infected cells (mRNA/DNA ratio) were higher in HAM/TSP patients than in ACs, suggesting the existence of genetic or virologic differences. Furthermore, comparison of molecular results with clinical features suggested that HTLV-1 tax mRNA load and mRNA/DNA ratio may be valid predictors of disease progression. These data support the hypothesis that higher HTLV-1 expression plays an important role in the pathogenesis of HAM/TSP.
We thank Nazli Azimi, National Cancer Institute, for her kind help and James M. Dambrosia, Biostatistics Branch, National Institute of Neurological Disorders and Stroke, for his support in statistical analysis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Steven Jacobson, NIH/NINDS/NIB Bldg 10, Rm 5B-16, Bethesda, MD 20892; e-mail: jacobsons@ninds.nih.gov.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal