Alloimmunization to the neutrophil antigen NB1 (HNA-2a, CD177) can result in immune neutropenia and transfusion-related acute lung injury. Recently, we were able to elucidate the primary structure of NB1. To shed light also on the molecular basis of the NB1-negative phenotype, we studied the neutrophils of 2 women with NB1-specific alloantibodies for intracellular and extracellular NB1 expression, NB1-specific mRNA production, and the presence of the NB1 gene. No antibody binding to neutrophils was observed by immunofluorescence and immunoblot using a variety of human and monoclonal NB1-specific antibodies. By reverse transcription–polymerase chain reaction with NB1-specific primers we could not detect NB1 cDNAs without accessory sequences, which were found to be introns. The NB1 gene was present in the genome of both patients. Our data indicate that the NB1-negative phenotype is the result of different off-frame insertions on RNA level, resulting in NB1 deficiency on neutrophils.
Introduction
NB1 was first described in 1971 as a neutrophil-specific antigen,1 and it is included as HNA-2a in the new nomenclature for human neutrophil antigens.2Recently, the glycoprotein has been clustered as CD 177. NB1 is expressed on neutrophils of 97% of whites, 95% of African Americans, and 88% of Japanese.3-9 A special feature of NB1 is its expression on a neutrophil subpopulation.10,11 Recently, we elucidated the primary structure of the NB1 glycoprotein.12 A 1614–base pair (bp) cDNA codes for 437 amino acids (aa), of which 416 residues form an NH2-terminal extracellular protein with 2 cysteine-rich domains, 3 potential N-linked glycosylation sites, and a short segment including a glycosyl-phosphatidylinositol (GPI) attachment site. NB1 sequence differs from deduced PRV-1 protein (polycythemia rubra vera-1)13 in 4 amino acids.12 Database searches revealed homology to the Ly-6 (uPAR) domain, suggesting that NB1 glycoprotein belongs to the uPAR/CD59/Ly-6 snake toxin superfamily. In this study, we shed light on the molecular basis of the NB1-negative phenotype by analyzing the neutrophils of 2 women who had formed NB1 alloantibodies, probably during pregnancy. NB1 alloimmunization is clinically important because NB1 alloantibodies can cause immune neutropenia and transfusion-related acute lung injury.1 14-19
Study design
Sera containing NB1-specific alloantibodies were obtained from mothers who gave birth to neonates with alloimmune neonatal neutropenia. Approval was obtained from the institutional review board at University Hospital of Giessen for this study. Informed consent was provided according to the Declaration of Helsinki. NB1-specific monoclonal antibodies (mAbs) 7D8, TAG 4, MEM 166, and 1B5 were kindly provided by Dr D. Stroncek (Bethesda, MD), Dr K. Taniguchi (Hiroshima, Japan), Dr V. Horesji (Praha, Czech Republic), and Dr L. Clement (Los Angeles, CA). An mAb specific for CD16, 3G8, was purchased from Immunotech (Marseilles, France). Granulocyte isolation, phenotyping, solubilization, sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and immunoblotting were performed as previously described.20 Antibody binding to granulocytes was determined by flow cytometry and fluorescence microscopy. Isolation of granulocyte mRNA and amplification of NB1 3′, 5′ untranslated regions and coding region were performed using sense and antisense primers based on NB1 cDNA sequence and amplification protocol.12cDNA was subcloned into pGEMT vector (Stratagene, La Jolla, CA), sequenced according to the manufacturer's protocols, and analyzed using Internet programs.12 For amplification of genomic DNA fragments to establish NB1 fragments encompassing exons 5 to 7 (patient 1) and 3 to 4 (patient 2), 5 μL genomic DNA was amplified with 200 μM of each dNTP, 0.5 μM sense and antisense primers, 4 U Taq polymerase XL (Perkin Elmer, Weiterstadt, Germany), 15 μL 10× polymerase chain reaction (PCR) buffer, and 1.25 mM Mg(OAC)2 in a total volume of 50 μL. Initial denaturation was at 94°C, followed by 35 cycles of 15 seconds at 94°C and 10 minutes at 65°C to 68°C. Final elongation was 10 minutes at 72°C (GeneAmp Systems 2400; Perkin Elmer). NB1 exon–intron borders were established by comparison with human genome sequences in GenBank and experimental data (not shown).
Results and discussion
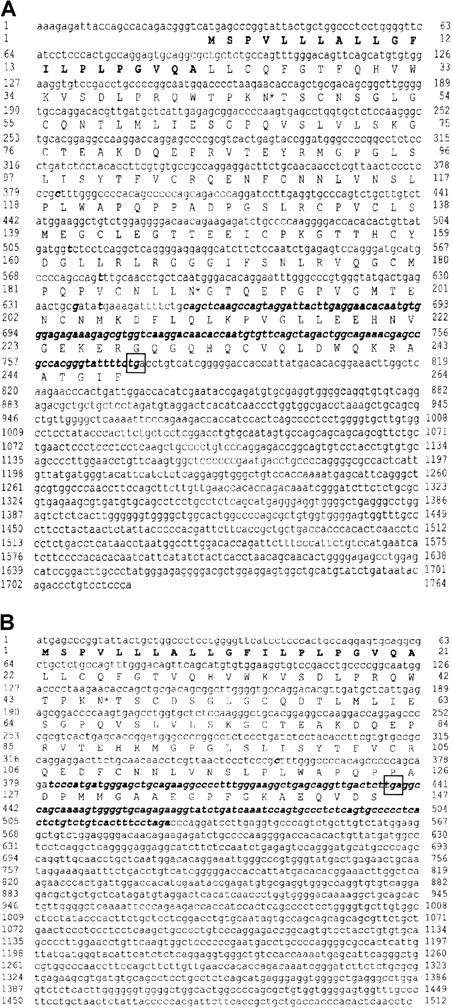
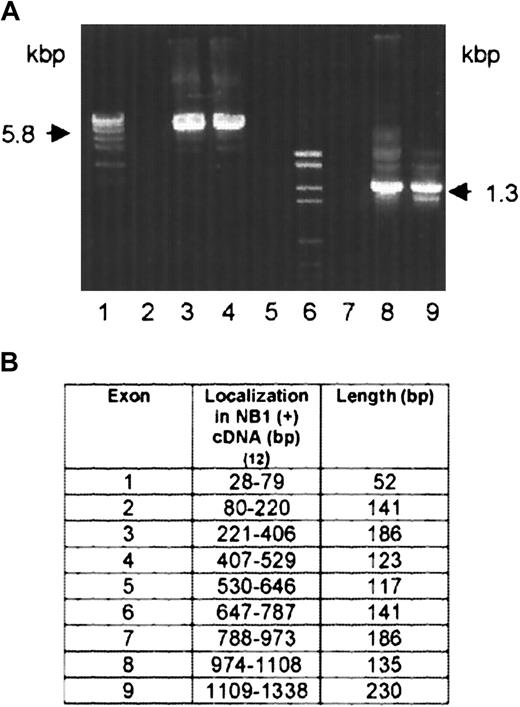
On the neutrophils of 2 NB1-alloimmunized women (patients 1 and 2), NB1 glycoprotein was not detectable by immunofluorescence using NB1-specific mAbs 7D8 and 1B5 or human alloantibodies. The neutrophils of their siblings, one sister each, were also typed serologically for NB1 and were found to be NB1−. Testing neutrophil lysates of the 2 women for the presence of NB1 glycoprotein in immunoblot, none of the 12 polyclonal or 3 monoclonal (TAG 4, MEM 166, and 7D8) blotting anti-NB1 antibodies used showed the 50- to 64-kd band typical for NB1 (data not shown). Based on these findings, we analyzed whether NB1-specific mRNA is synthesized by the neutrophils of the 2 NB1− patients. For that we isolated neutrophil RNA from the NB1− neutrophils and transcribed it in cDNA, which was amplified and subcloned. In patient 1, we identified 3 clones among 27 tested containing NB1-specific cDNA. This cDNA (GenBank accession number AC AJ310433) spans 1717 bp, consisting of a 27-bp 5′ coding region, a 946-bp 3′-untranslated coding region, and a 744-bp coding region. In patient 2, we identified 2 clones among 18 tested containing NB1-specific cDNA spanning 1511 bp (AC AJ305326) consisting of a 435-bp coding region and a 1076-bp 3′ untranslated region (Figure1B). All clones had identical accessory sequences (specific for each donor) and were of identical length. Compared with NB1 reference sequence,12 both cDNAs showed off-frame insertions of 118 bp (position 655-773, patient 1) and 145 bp (position 381-526, patient 2), respectively. Both insertions showed the stop codon tga. In patient 1, the predicted protein (Figure 1A) consisted of 248 amino acids, of which the first 21 form the signal sequence. The remaining 227 residues code for an NH2-terminal protein, with one domain homologous to Ly-6 (uPAR) and 2 potential N-linked glycosylation sites. In patient 2, a 145-aa protein can be predicted, consisting of a 21-aa signal peptide and a 124-aa NH2-terminal protein part containing one Ly-6 (uPAR) homologous domain and one potential N-linked glycosylation site (Figure 1B). In both patients the deduced proteins would lack GPI linkage sites and transmembrane segments. Although the presence of such soluble NB1 glycoprotein fragments, which are not recognized by the polyclonal and monoclonal antibodies used in immunoblot, cannot be excluded, these putative fragments do not prevent NB1 alloimmunization. Thus, our findings indicate that the NB1− phenotype is the result of common NB1 glycoprotein deficiency. Analyzing the genomic organization of the NB1gene in an NB1-expressing patient, we identified 9 exons (Figure2B). Based on this finding, we analyzed the genomic DNA sequence encompassing cDNA nucleotides 655-773 (patient 1) and 381-526 (patient 2) in NB1-expressing and NB1− patients. DNA fragments of 5.8 kbp (patient 1) and 1.3 kbp (patient 2) were amplified (Figure 2A). Length of genomic fragments from NB1− and NB1-expressing donors were identical. Sequence analysis of the 4 genomic DNA fragments showed that the accessory fragments were present in the NB1 gene of NB1-expressing and -deficient donors. Analysis of the regions encompassing the dinucleotides gt/ag at the intron termini did not detect differences between NB1+ and NB1−(patient 2) so that the found off-frame insertions are not caused by mutations in these splicing sites. Because it is known that the recognition of splicing sites requires cross-talk between multiple components with relatively weak interactions,21 it is not unlikely that NB1 deficiency is the result of incorrect splicing complex formation. However, this warrants further investigation. Our results show that in contrast to FcγRIIIb-deficiency, which is caused by FcγRIIIB gene deletion and leads to the HNA (NA) null phenotype,22 NB1 deficiency is the result of a gene expression defect.
cDNA nucleotide sequence of human NB1− phenotypes and deduced amino acid sequences.
(A) Patient 1 (AC AJ310433). (B) Patient 2 (AC AJ305326). Signal sequence is shown in bold letters. Potential N-linked glycosylation sites (*) are indicated. Stop codons are framed. Differences to NB1 reference sequence12 are indicated in italic and bold letters. Ly-6 (uPAR)–domains comprise aa 131-218 in patient 1 and aa 43-124 in patient 2.
cDNA nucleotide sequence of human NB1− phenotypes and deduced amino acid sequences.
(A) Patient 1 (AC AJ310433). (B) Patient 2 (AC AJ305326). Signal sequence is shown in bold letters. Potential N-linked glycosylation sites (*) are indicated. Stop codons are framed. Differences to NB1 reference sequence12 are indicated in italic and bold letters. Ly-6 (uPAR)–domains comprise aa 131-218 in patient 1 and aa 43-124 in patient 2.
Amplification of NB1-genomic fragments by PCR and exon organization of NB1 cDNA.
(A) Primers were numbered according to the NB1 cDNA reference sequence.12 5.8 kbp fragment: FP1 (409-426) and reverse primer RP1 (859-879) (lanes 2-4). 1.3 kbp fragment: FP2 (329-348) and RP 2 (416-436) (lanes 7-9). Lanes 4 and 9: NB1− patient 1 and patient 2, respectively. Lanes 3 and 8: NB1-expressing healthy persons. Lanes 1 and 6: molecular weight standards. Lanes 2 and 7: negative controls. (B) Genomic DNA from NB1+ and NB1− patients was amplified by PCR and sequenced. Exon–intron borders were determined according to the NB1 cDNA reference sequence12 and dinucleotides gt/ag at the intron termini.
Amplification of NB1-genomic fragments by PCR and exon organization of NB1 cDNA.
(A) Primers were numbered according to the NB1 cDNA reference sequence.12 5.8 kbp fragment: FP1 (409-426) and reverse primer RP1 (859-879) (lanes 2-4). 1.3 kbp fragment: FP2 (329-348) and RP 2 (416-436) (lanes 7-9). Lanes 4 and 9: NB1− patient 1 and patient 2, respectively. Lanes 3 and 8: NB1-expressing healthy persons. Lanes 1 and 6: molecular weight standards. Lanes 2 and 7: negative controls. (B) Genomic DNA from NB1+ and NB1− patients was amplified by PCR and sequenced. Exon–intron borders were determined according to the NB1 cDNA reference sequence12 and dinucleotides gt/ag at the intron termini.
Supported by a research grant from the Deutsche Forschungsgemeinschaft DFG BU 770/3-4.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Juergen Bux, Institute for Clinical Immunology and Transfusion Medicine, Langhansstrasse 7, 35385 Giessen, Germany; e-mail: juergen.bux@immunologie.med.uni-giessen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal