Chemokines and chemokine receptors play important roles in migration and tissue localization of various lymphocyte subsets. Here, we report the highly frequent expression of CCR4 in adult T-cell leukemia (ATL) and human T-cell leukemia virus type 1 (HTLV-1)–immortalized T cells. Flow cytometric analysis revealed that ATL and HTLV-1–immortalized T-cell lines consistently expressed CCR4. Inducible expression of HTLV-1 transcriptional activator tax in a human T-cell line Jurkat did not, however, up-regulate CCR4 mRNA. In vitro immortalization of peripheral blood T cells led to preferential outgrowth of CD4+ T cells expressing CCR4. We further demonstrated highly frequent expression of CCR4 in fresh ATL cells by (1) reverse transcriptase–polymerase chain reaction (RT-PCR) analysis of CCR4 expression in peripheral blood mononuclear cells (PBMCs) from patients with ATL and healthy controls; (2) flow cytometric analysis of CCR4-expressing cells in PBMCs from patients with ATL and healthy controls; (3) CCR4 staining of routine blood smears from patients with ATL; and (4) an efficient migration of fresh ATL cells to the CCR4 ligands, TARC/CCL17 and MDC/CCL22, in chemotaxis assays. Furthermore, we detected strong signals for CCR4, TARC, and MDC in ATL skin lesions by RT-PCR. Collectively, most ATL cases have apparently derived from CD4+ T cells expressing CCR4. It is now known that circulating CCR4+ T cells are mostly polarized to Th2 and also contain essentially all skin-seeking memory T cells. Thus, HTLV-1–infected CCR4+ T cells may have growth advantages by deviating host immune responses to Th2. CCR4 expression may also account for frequent infiltration of ATL into tissues such as skin and lymph nodes.
Introduction
Human T-cell leukemia virus type 1 (HTLV-1) is the causative agent of adult T-cell leukemia (ATL), a unique malignancy of mature CD4+ T cells with characteristic lobular nuclei and surface activation markers such as CD25.1,2 In vitro, HTLV-1 efficiently immortalizes CD4+ T cells into continuously growing T-cell blasts.3 HTLV-1 is vertically transmitted from mother to child through breast feeding and also horizontally by sexual contact. Asymptomatic carriers contain a large number of HTLV-1–infected cells which are mostly CD4+CD45RO+ T cells.4 With about 5% life risk, ATL develops in HTLV-1 carriers after 40 to 50 years of latency.2 This implies that ATL develops from HTLV-1–infected T cells after accumulation of multiple critical mutations.5 HTLV-1 is also etiologically associated with various chronic inflammatory diseases such as HTLV-1–associated myelopathy/tropical spastic paraparesis, HTLV-1–associated uveitis, HTLV-1–associated bronchopneumopathy and HTLV-1–associated arthrophathy.2
Chemokines are a group of structurally related cytokines that induce directed migration of various leukocyte populations.6 In humans, more than 40 chemokines have been identified. Based on the arrangement of the N-terminal conserved cysteine residues, chemokines are classified into 4 subfamilies: CC, CXC, C, and CX3C. All chemokines transduce signals through 7 transmembrane G protein–coupled receptors. At present, 18 chemokine receptors have been identified.6 It is now known that chemokines are crucially involved in migration and tissue localization of various lymphocyte subpopulations, which express specific chemokine receptors in accordance with their differentiation pathways and maturation stages.6 In ATL, leukemic cells frequently infiltrate into organs such as lymph nodes, spleen, liver, and skin.2Various HTLV-1–associated inflammatory diseases are also commonly characterized by infiltration of HTLV-1+ T cells into target organs.2 It is thus conceivable that chemokines and their receptors play important roles in tissue infiltration of ATL cells and HTLV-1+ T cells. Previously, we have shown that increased surface expression of CCR7, the shared receptor for SLC/CCL21 and ELC/CCL19, which are both expressed in the secondary lymphoid organs,7,8 correlates well with lymphoid organ involvement in ATL.9 In the present study, we have extended this observation and demonstrated highly frequent expression of CCR4 in HTLV-immortalized T cells and fresh ATL. It is now known that peripheral blood T cells expressing CCR4 are mostly polarized to Th2 and also include essentially all skin-seeking memory T cells positive for cutaneous lymphocyte antigen (CLA).10-12Thus, our findings may provide new clues to understanding various aspects of ATL and HTLV-1–associated diseases such as suppression of cell-mediated immunity and frequent involvement of tissues such as skin and lymph nodes.
Materials and methods
Chemokines and monoclonal antibodies
Recombinant TARC/CCL17 and MDC/CCL22 were purchased from R&D Systems (Minneapolis, MN). Phycoerythrin (PE)–labeled anti-CD4 and PE-labeled anti-CD25 were purchased from DAKO Japan (Kyoto, Japan). PerCP-labeled CD4 was purchased from Becton Dickinson Japan (Tokyo, Japan). Monoclonal anti-CXCR4 (12G5) was also purchased from DAKO Japan. Murine monoclonal antibodies (mAbs) to CCR4 (KM-2160) and CCR7 (6B3) were described previously.9 10 For double staining experiments, fluorescein isothiocyanate (FITC)–labeled anti-CCR4 was used with PerCP-labeled anti-CD4 and PE-labeled anti-CD25.
Cells and tissues
Human T-cell lines used in the present study were all described previously.13 JPX-9 was a Jurkat subline carrying HTLV-1 Tax under the control of the metallothionine gene promoter.14 All the cell lines were maintained in RPMI-1640 supplemented with 10% fetal bovine serum. Heparinized venous blood was obtained from patients with ATL and healthy adult donors upon informed consent. Peripheral blood mononuclear cells (PBMCs) were isolated using the standard method using Ficoll-Paque (Pharmacia, Uppsala, Sweden). Normal skin tissues were obtained from patients undergoing mammectomy. Lesional skin tissues were obtained from patients with ATL. This study was approved by the ethical committees of the Kinki University School of Medicine and the Nagasaki University School of Medicine. Informed consent was obtained from all blood and tissue donors according to the Declaration of Helsinki.
Flow cytometry
Cells were washed with phosphate buffered saline (PBS) containing 2% fetal calf serum (FCS) and incubated for 30 minutes with mAbs or control isotype-matched murine IgG. The primary antibodies were detected by FITC-conjugated sheep (Fab')2 antimouse IgG (Sigma, St Louis, MO). For double staining, cells were incubated with FITC-labeled anti-CCR4 and PerCP-labeled anti-CD4 or PE-labeled anti-CD25. If necessary, dead cells were stained with propidium iodide (PI) and gated out. Cells were analyzed on a FACS Calibur (Becton Dickinson, Mountain View, CA), and the data were collected in the log mode. In each sample, 10 000 cells were analyzed.
In vitro immortalization
Immortalization of peripheral blood T cells with HTLV-1 was carried out by the coculture method.3 In brief, PBMCs from adult donors and MT-2 cells pretreated with mitomycin C (Sigma) at 50 μg/mL at 37°C for 1 hour were cocultured in 96-well microtest plates at 100 cells/well and 2 × 104 cells/well, respectively. After 10 days, interleukin 2 (IL-2) was added to each culture at 100 U/mL. After about a month, growing cultures were individually expanded and examined for expression of surface molecules by flow cytometry.
CCR4 staining in blood smear
Routine blood smears were fixed with ice-cold acetone and blocked with PBS containing 3% normal horse serum. After washing, slides were incubated first with anti-CCR4 (KM2160) or isotype-matched control mouse IgG and then with biotinylated horse anti–mouse IgG (Vector, Burlingame, CA). After treatment with 0.3% H2O2, slides were incubated with avidin-biotin complex (Vector) and developed with diaminobenzidine (DAB) (Sigma). Slides were counterstained by Gill hematoxylin.
RT-PCR analysis
Total RNA was prepared from cells or tissues by using Trizol reagent (GIBCO-BRL, Gaithersburg, MD). RNA was further purified by using RNeasy (Qiagen, Hilden, Germany). Genome DNA was also prepared from the same tissues following the instruction manual for Trizol reagent. Total RNA (1 μg) was reverse transcribed using oligo(dT)18 primer and SuperScript II reverse transcriptase (GIBCO-BRL). Resulting first-strand DNA (20 ng total RNA equivalent), original total RNA (20 ng), and genome DNA (200 ng) were amplified in a final volume of 20 μL containing 10 pmol of each primer and 1 unit of Ex-Taq polymerase (Takara Shuzo, Kyoto, Japan).The primers used were: +5′-ACTGCTCCAGGGATGCCATCGTTTTT-3′ and -5′-ACAAGGGGATGGGATCTCCCTCACTG-3′ for TARC; +5′-AGGACAGAGCATGGCTCGCCTACAGA-3′ and -5′-TAATGGCAGGGAGGTAGGGCTCCTGA-3′ for MDC; +5′-AAGAAGAACAAGGCGGTGAAGATG-3′ and -5′-AGGCCCCTGCAGGTTTTGAAG-3′ for CCR4; +5′-GTGCCCGCGTCCTTCTCATCAG-3′ and -5′-GGCCAGGACCACCCCATTGTAG-3′ for CCR7; +5′-AAAAAGCGGGTCACTCTATATGCTC-3′ and -5′-CCACTGCTACCTGGTACTCTGTTGT-3′ for IL-2Rα (CD25); +5′-ATCCCGTGGAGACTCCTCAA-3′ and -5′-AACACGTAGACTGGGTATCC-3′ for Tax; +5′-GCCAAGGTCATCCATGACAACTTTGG-3′ and -5′-GCCTGCTTCACCACCTTCTTGATGTC-3′ for glyceraldehyde 3-phosphate dehydrogenase (G3PDH). Amplification conditions were denaturation at 94°C for 30 seconds (5 minutes for the first cycle), annealing at 60°C for 30 seconds and extension at 72°C for 30 seconds (5 minutes for the last cycle) for 34 cycles for MDC and TARC, 35 cycles for CCR4 and CCR7, 37 cycles for IL-2Rα, 38 cycles for Tax, and 27 cycles for G3PDH. Amplification products (10 μL each) were subjected to electrophoresis on 2% agarose and stained with ethidium bromide.
Chemotaxis assay
Chemotaxis assay was carried out using Transwell plates with 3-μm pore size (Coster, Cambridge, MA). Cells were suspended at 2 × 107/mL in RPMI-1640 containing 1 mg/mL bovine serum albumin (BSA) and 20 mM HEPES, pH 7.4. Cells were applied to upper wells (200 μL/well). Lower wells contained chemokines at various concentrations (600 μL/well). After 3 hours at 37°C, cells migrated into lower chambers were counted by flow cytometry.
Results
Flow cytometric analysis on surface expression of chemokine receptors
Using a panel of 24 human T-cell lines, we examined the surface expression of CXCR4, CCR4, and CCR7 by flow cytometry. The results are summarized in Table 1. Based on the expression of CD4 and CD8, HTLV-1− leukemic T-cell lines were further divided into 3 groups: double-negative, double-positive and CD4+.13 On the other hand, ATL-derived leukemic cell lines and HTLV-1–transformed T-cell lines were consistently CD4+. CXCR4 was consistently expressed in all HTLV-1− T-cell lines (15/15) and HTLV-1–immortalized T-cell lines (5/5). However, only a fraction of ATL-derived leukemic cell lines were positive for CXCR4 (2/4). CCR7 was mostly negative in HTLV-1− T-cell lines (4/15). On the other hand, most ATL-derived leukemic cell lines (3/4) and all HTLV-1–immortalized T-cell lines were positive for CCR7 (5/5). Concerning CCR4, some double-negative T-cell lines were clearly positive (2/4), whereas double-positive T-cell lines were all negative (0/6). Some HTLV-1− CD4+ T-cell lines were marginally positive (3/5). However, HUT78, a mature T-cell line derived from a patient with cutaneous T-cell lymphoma (Sezary syndrome), was strongly positive. Rather strikingly, ATL-derived leukemic T-cell lines and HTLV-1–immortalized T-cell lines were consistently found to express CCR4 at high levels (4/4 and 5/5, respectively).
Summary of surface expression of chemokine receptors
| Cell type . | Cell line . | CXCR4 . | CCR4 . | CCR7 . |
|---|---|---|---|---|
| Double-negative | HSB-2 | P | N | M |
| MOLT-15 | P | N | N | |
| MOLT-17 | P | P | N | |
| PEER | P | P | P | |
| Double-positive | HPB-ALL | P | N | N |
| JM | P | N | N | |
| KOPT-K1 | P | N | N | |
| MOLT-3 | P | N | N | |
| MOLT-4 | P | N | N | |
| SALT-12 | P | N | N | |
| CD4+ | CEM | P | M | M |
| DND4.1 | P | N | N | |
| Jurkat | P | M | N | |
| MKB-1 | P | M | N | |
| HUT78 | P | P | P | |
| ATL | HUT102 | M | P | P |
| H582 | N | P | P | |
| MT-1 | P | P | N | |
| TL-Om1 | N | P | P | |
| HTLV-1+ | C8166 | P | P | P |
| C91/PL | P | P | P | |
| MT-2 | P | P | P | |
| MT-4 | P | P | M | |
| TCL-Kan | P | P | P |
| Cell type . | Cell line . | CXCR4 . | CCR4 . | CCR7 . |
|---|---|---|---|---|
| Double-negative | HSB-2 | P | N | M |
| MOLT-15 | P | N | N | |
| MOLT-17 | P | P | N | |
| PEER | P | P | P | |
| Double-positive | HPB-ALL | P | N | N |
| JM | P | N | N | |
| KOPT-K1 | P | N | N | |
| MOLT-3 | P | N | N | |
| MOLT-4 | P | N | N | |
| SALT-12 | P | N | N | |
| CD4+ | CEM | P | M | M |
| DND4.1 | P | N | N | |
| Jurkat | P | M | N | |
| MKB-1 | P | M | N | |
| HUT78 | P | P | P | |
| ATL | HUT102 | M | P | P |
| H582 | N | P | P | |
| MT-1 | P | P | N | |
| TL-Om1 | N | P | P | |
| HTLV-1+ | C8166 | P | P | P |
| C91/PL | P | P | P | |
| MT-2 | P | P | P | |
| MT-4 | P | P | M | |
| TCL-Kan | P | P | P |
P indicates positive; N, negative; M, marginal.
No induction of CCR4 or CCR7 by Tax
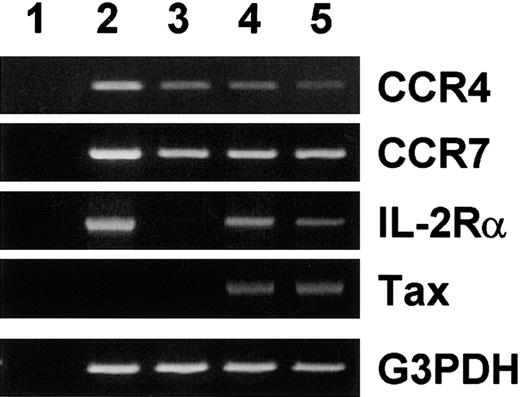
Consistent expression of CCR4 in HTLV-1+ T-cell lines may be due to induction of CCR4 by the viral transactivator tax. To test this possibility, we used JPX-9, a Jurkat subline carrying HTLV-1 tax under the control of the metallothionine gene promoter.14 This cell line has been widely used to demonstrate induction of various cellular target genes by tax.14 As shown in Figure 1, JPX-9 constitutively expressed CCR4 and CCR7 mRNA. Upon treatment with Cd2+, tax mRNA was induced in JPX-9. Accordingly, IL-2Rα (CD25), a tax-responsive cellular gene,2 was induced. However, there was no up-regulation of CCR4 or CCR7 mRNA even after 7 days of treatment. In fact, CCR4 mRNA was slightly down-regulated upon induction of tax with Cd2+. Thus, at least in the cellular background of Jurkat, tax does not up-regulate expression of CCR4 or CCR7.
Effect of tax on expression of CCR4 and CCR7 mRNA.
Total RNA was prepared from PBMCs stimulated with phytohemaggutinin (PHA) for 3 days and JPX-9 treated without (control) or with 10 μM of Cd2+ for 3 and 7 days. RT-PCR was carried out for CCR4, CCR7, IL-2 receptor α chain (IL-2Rα), Tax, and G3PDH as described in “Materials and methods.” (1) No RNA. (2) PHA-stimulated PBMCs (positive control). (3) Untreated JPX-9. (4) JPX-9 treated with Cd2+ for 3 days. (5) JPX-9 treated with Cd2+ for 7 days.
Effect of tax on expression of CCR4 and CCR7 mRNA.
Total RNA was prepared from PBMCs stimulated with phytohemaggutinin (PHA) for 3 days and JPX-9 treated without (control) or with 10 μM of Cd2+ for 3 and 7 days. RT-PCR was carried out for CCR4, CCR7, IL-2 receptor α chain (IL-2Rα), Tax, and G3PDH as described in “Materials and methods.” (1) No RNA. (2) PHA-stimulated PBMCs (positive control). (3) Untreated JPX-9. (4) JPX-9 treated with Cd2+ for 3 days. (5) JPX-9 treated with Cd2+ for 7 days.
Preferential outgrowth of CCR4+ T cells upon immortalization with HTLV-1
We next examined whether T cells freshly immortalized by HTLV-1 expressed CCR4. For this purpose, we carried out immortalization of PBMCs by HTLV-1 at a limiting cell concentration (100 PBMCs cocultured with MMC-treated MT-2/well). By this way, rare outgrowing cultures might be mostly monoclonal. A total of 17 immortalized CD4+T-cell lines were thus obtained starting with 960 wells, and all turned out to be positive for CCR4 by flow cytometric analysis (data not shown). Thus, CD4+ T cells expressing CCR4 may be either preferentially infected by HTLV-1 and/or, upon HTLV-1 infection, may have some in vitro growth advantages over CCR4− T cells.
Expression of CCR4 on fresh ATL
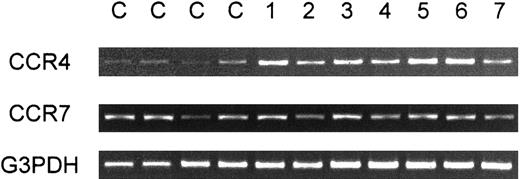
We next examined expression of CCR4 in fresh leukemic cells from patients with ATL. We first carried out RT-PCR analysis on CCR4 expression in PBMCs from a total of 13 healthy adult donors and 24 patients with ATL (16 acute and 8 chronic cases) (Table2). Representative results are shown in Figure 2. Whereas PBMCs from healthy donors gave only weak signals for CCR4, strong signals were consistently observed for PBMCs from most patients with ATL (22/24). In the case of CCR7, however, no such consistent differences between healthy donors and patients with ATL were observed (Figure 2). Since peripheral blood samples from all these patients with ATL contained leukemic cells at high frequencies (15%-97%, average 62%), these results supported that leukemic cells from most ATL cases were positive for CCR4. Two patients with ATL (one acute and one chronic), however, gave only weak CCR4 signals. Since PBMCs from these 2 patients also contained leukemic cells at high frequencies (60%-70%), their ATL cells were likely to be negative for CCR4. Table 2 summarizes the results together with the clinical profiles of each patient.
Summary of patients examined for CCR4 expression with RT-PCR
| Case . | Sex . | Age . | Type . | Tissue involvement . | CCR4 (RT-PCR) . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Skin . | LN . | Liver . | Spleen . | Lung . | |||||
| 1 | M | 70 | Acute | − | + | − | − | + | + |
| 2 | M | 61 | Acute | − | + | − | − | − | + |
| 3 | M | 61 | Acute | − | − | − | − | − | + |
| 4 | M | 65 | Acute | +++ | − | − | − | + | + |
| 5 | M | 73 | Acute | + | + | + | − | + | + |
| 6 | M | 83 | Acute | +++ | − | − | − | − | + |
| 7 | M | 63 | Acute | − | − | − | − | − | − |
| 8 | M | 58 | Acute | − | + | − | + | − | + |
| 9 | M | 70 | Acute | − | − | − | − | + | + |
| 10 | M | 75 | Acute | + | + | − | − | − | + |
| 11 | F | 69 | Acute | + | + | + | + | − | + |
| 12 | M | 51 | Acute | + | − | − | − | + | + |
| 13 | F | 70 | Acute | − | + | − | − | − | + |
| 14 | F | 80 | Acute | − | + | − | − | − | + |
| 15 | M | 62 | Acute | − | − | − | − | − | + |
| 16 | M | 88 | Acute | − | + | − | − | − | + |
| 17 | M | 76 | Chronic | − | − | − | − | − | − |
| 18 | M | 69 | Chronic | +/− | + | − | + | − | + |
| 19 | M | 34 | Chronic | + | − | − | − | − | + |
| 20 | F | 64 | Chronic | − | + | − | − | − | + |
| 21 | M | 74 | Chronic | +++ | − | − | − | + | + |
| 22 | F | 44 | Chronic | + | + | − | − | − | + |
| 23 | M | 75 | Chronic | − | − | − | − | − | + |
| 24 | M | 68 | Chronic | − | − | − | − | − | + |
| Case . | Sex . | Age . | Type . | Tissue involvement . | CCR4 (RT-PCR) . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Skin . | LN . | Liver . | Spleen . | Lung . | |||||
| 1 | M | 70 | Acute | − | + | − | − | + | + |
| 2 | M | 61 | Acute | − | + | − | − | − | + |
| 3 | M | 61 | Acute | − | − | − | − | − | + |
| 4 | M | 65 | Acute | +++ | − | − | − | + | + |
| 5 | M | 73 | Acute | + | + | + | − | + | + |
| 6 | M | 83 | Acute | +++ | − | − | − | − | + |
| 7 | M | 63 | Acute | − | − | − | − | − | − |
| 8 | M | 58 | Acute | − | + | − | + | − | + |
| 9 | M | 70 | Acute | − | − | − | − | + | + |
| 10 | M | 75 | Acute | + | + | − | − | − | + |
| 11 | F | 69 | Acute | + | + | + | + | − | + |
| 12 | M | 51 | Acute | + | − | − | − | + | + |
| 13 | F | 70 | Acute | − | + | − | − | − | + |
| 14 | F | 80 | Acute | − | + | − | − | − | + |
| 15 | M | 62 | Acute | − | − | − | − | − | + |
| 16 | M | 88 | Acute | − | + | − | − | − | + |
| 17 | M | 76 | Chronic | − | − | − | − | − | − |
| 18 | M | 69 | Chronic | +/− | + | − | + | − | + |
| 19 | M | 34 | Chronic | + | − | − | − | − | + |
| 20 | F | 64 | Chronic | − | + | − | − | − | + |
| 21 | M | 74 | Chronic | +++ | − | − | − | + | + |
| 22 | F | 44 | Chronic | + | + | − | − | − | + |
| 23 | M | 75 | Chronic | − | − | − | − | − | + |
| 24 | M | 68 | Chronic | − | − | − | − | − | + |
RT-PCR analysis of PBMCs for expression of CCR4.
Total RNA was prepared from PBMCs obtained from 4 healthy adult donors (C) and 7 patients with ATL (1-7). RT-PCR was carried out for CCR4, CCR7, and G3PDH as described in “Materials and methods.”
RT-PCR analysis of PBMCs for expression of CCR4.
Total RNA was prepared from PBMCs obtained from 4 healthy adult donors (C) and 7 patients with ATL (1-7). RT-PCR was carried out for CCR4, CCR7, and G3PDH as described in “Materials and methods.”
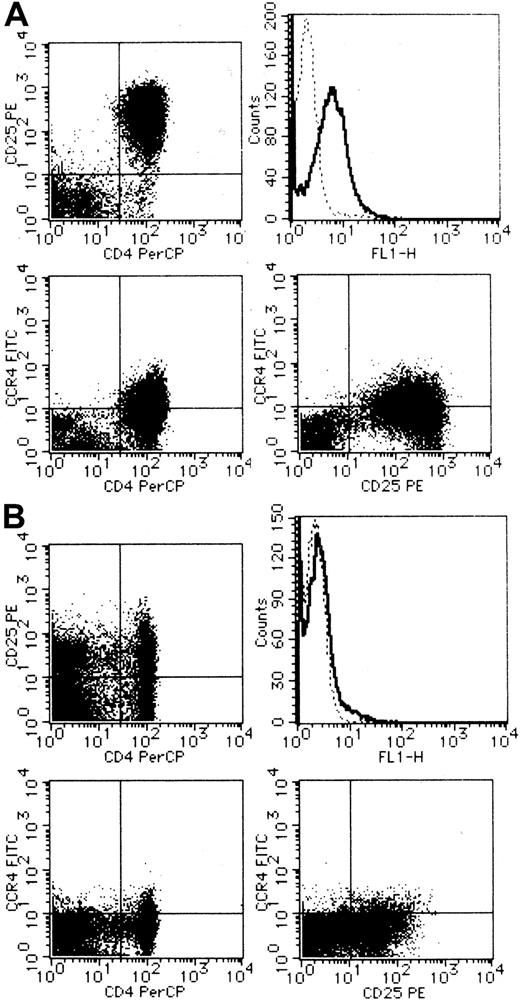
We next carried out CCR4 staining and flow cytometric analysis of PBMCs from a total of 4 healthy adult donors and 10 patients with ATL. Representative results are shown in Figure3 (A, ATL; B, healthy). Compared with PBMCs from healthy adult donors, PBMCs from most ATL cases (8/10) indeed contained a large fraction of cells expressing CCR4 (right upper panel). Double staining with anti-CD25 or anti-CD4 demonstrated that CCR4-expressing cells were largely positive for CD25 (right lower panel) and CD4 (left lower panel). Thus, the majority of CCR4-expressing cells in PBMCs from patients with ATL were likely to be leukemic cells. However, 2 patients showed little significant increases in CCR4-expressing cells in their peripheral blood. These 2 patients also gave only a weak signal for CCR4 by RT-PCR analysis (data not shown). Thus, ATL cells from these 2 patients were likely to be negative for CCR4.
Flow cytometric analysis on surface expression of CCR4.
PBMCs were obtained from a patient with ATL (A) and a healthy adult (B). CCR4 was stained singly (upper right) or in combination with CD25 (lower right) or CD4 (lower left). Double staining for CD4 and CD25 was also carried out (upper left). See “Materials and methods” for details.
Flow cytometric analysis on surface expression of CCR4.
PBMCs were obtained from a patient with ATL (A) and a healthy adult (B). CCR4 was stained singly (upper right) or in combination with CD25 (lower right) or CD4 (lower left). Double staining for CD4 and CD25 was also carried out (upper left). See “Materials and methods” for details.
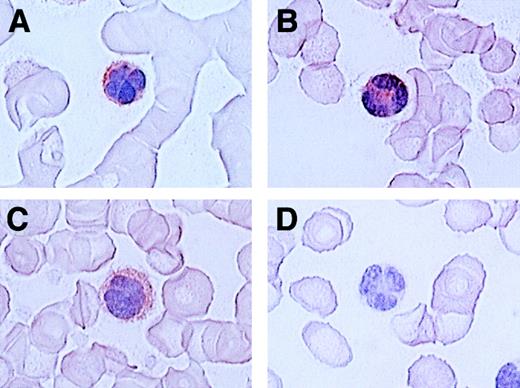
Since ATL cells have characteristic flowerlike nuclei, it is rather easy to identify ATL cells in blood smears. We therefore further carried out immunologic staining of CCR4 using routine blood smears from a total of 7 patients with ATL. Representative results are shown in Figure 4. Again, most ATL cases (6/7) were clearly stained positive for CCR4 (A, B, C: positive; D: negative).
Immunocytologic staining of CCR4 in blood smears.
Routine blood smears obtained from 4 patients with ATL (A-D) were stained for CCR4 as described in “Materials and methods.” Leukemic cells are positive (A, B, C) or negative (D) for CCR4. No staining was seen by an isotype-matched control mouse IgG (not shown).The slides were counterstained by hematoxylin and the original magnification was × 1000.
Immunocytologic staining of CCR4 in blood smears.
Routine blood smears obtained from 4 patients with ATL (A-D) were stained for CCR4 as described in “Materials and methods.” Leukemic cells are positive (A, B, C) or negative (D) for CCR4. No staining was seen by an isotype-matched control mouse IgG (not shown).The slides were counterstained by hematoxylin and the original magnification was × 1000.
Collectively, a total of 40 ATL cases were examined by at least one of the 3 different techniques described above and 36 cases were concluded to be positive for CCR4.
Chemotactic response of fresh ATL cells to CCR4 ligands
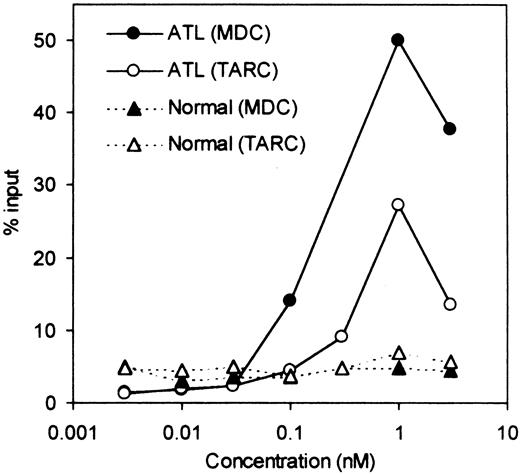
We next examined whether fresh ATL cells were capable of responding to the CCR4 ligands, TARC/CCL17 and MDC/CCL22,15 16 in a chemotactic assay using a Transwell plate. A total of 8 ATL cases (4 acute and 4 chronic) were studied. Representative results are shown in Figure5. We observed an efficient migration of PBMCs from all patients with ATL (8/8) toward TARC and MDC with a typical bell-shaped dose-response curve and an optimal concentration of about 1 nM. No such vigorous migration was observed by using fresh PBMCs from healthy adult donors in the present conditions. Thus, it is likely that the responding cells were mostly leukemic cells. To confirm this, we carried out a cytologic evaluation of cells migrated into the lower chambers. As shown in Figure 6, Giemsa staining of cytospin samples confirmed that migrated cells from PBMCs of patients with ATL were indeed mostly abnormal cells with characteristic flowerlike nuclei. Thus, fresh ATL cells expressing CCR4 are capable of efficiently responding to the CCR4 ligands TARC and MDC.
Chemotactic response to TARC/CCL17 and MDC/CCL22.
PBMCs were obtained from a patient with ATL and a healthy adult donor. Chemotactic responses to TARC and MDC (the ligands for CCR4) were determined as described in “Materials and methods.”
Chemotactic response to TARC/CCL17 and MDC/CCL22.
PBMCs were obtained from a patient with ATL and a healthy adult donor. Chemotactic responses to TARC and MDC (the ligands for CCR4) were determined as described in “Materials and methods.”
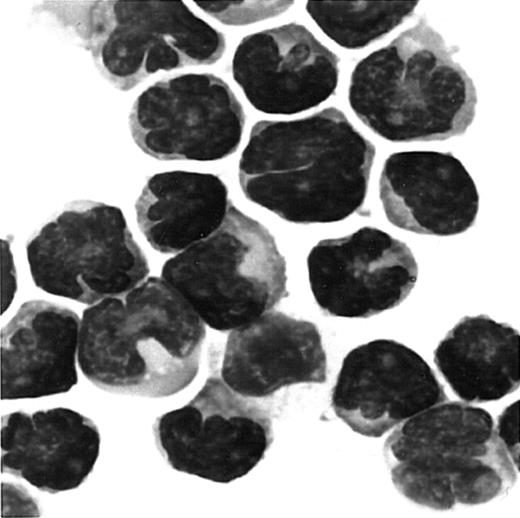
Giemsa staining of cells migrated to MDC/CCL22.
PBMCs were obtained from a patient with ATL. Cells migrated to MDC in chemotaxis chambers were collected and stained with Giemsa as described in “Materials and methods.” Original magnification, × 1000.
Giemsa staining of cells migrated to MDC/CCL22.
PBMCs were obtained from a patient with ATL. Cells migrated to MDC in chemotaxis chambers were collected and stained with Giemsa as described in “Materials and methods.” Original magnification, × 1000.
ATL skin lesions
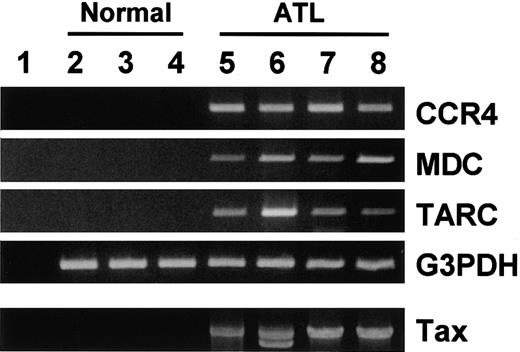
ATL frequently invades skin.2,17 Accumulating evidence supports that CCR4 and its ligands TARC and MDC play important roles in T-cell infiltration into skin.12 18-20 It is thus possible that frequent skin invasion in ATL may be facilitated by expression of CCR4 on ATL cells and production of TARC and/or MDC in lesional skin tissues. To test this notion, we examined expression of CCR4, TARC, and MDC in normal skin tissues from 3 donors and lesional skin tissues from 4 patients with ATL by RT-PCR. As shown in Figure 7, normal skin tissues hardly contained mRNA for TARC, MDC, or CCR4. On the other hand, ATL skin lesions gave strong signals of TARC, MDC, and CCR4. Detection of tax DNA by PCR supported that these ATL skin lesions were indeed invaded by cells carrying HTLV-1. Thus, expression of CCR4 by ATL cells and production of the CCR4 ligands in local skin tissues may facilitate invasion of ATL cells into lesional skin tissues.
RT-PCR analysis for CCR4 and its ligands TARC/CCL17 and MDC/CCL22 in normal and ATL skin tissues.
Total RNA and DNA were prepared from normal skin tissues from 3 adult donors (2, 3, 4) and lesional skin tissues from 4 patients with ATL (5, 6, 7, 8). RT-PCR was carried out for CCR4, MDC, TARC, and G3PDH as described in “Materials and methods.” Genomic PCR was carried out for tax as described in “Materials and methods.” (1) No RNA. (2-4) Normal skin tissues. (5-8) Lesional skin tissues from patients with ATL.
RT-PCR analysis for CCR4 and its ligands TARC/CCL17 and MDC/CCL22 in normal and ATL skin tissues.
Total RNA and DNA were prepared from normal skin tissues from 3 adult donors (2, 3, 4) and lesional skin tissues from 4 patients with ATL (5, 6, 7, 8). RT-PCR was carried out for CCR4, MDC, TARC, and G3PDH as described in “Materials and methods.” Genomic PCR was carried out for tax as described in “Materials and methods.” (1) No RNA. (2-4) Normal skin tissues. (5-8) Lesional skin tissues from patients with ATL.
Discussion
Besides cell adhesion molecules, chemokines and chemokine receptors are likely to play important roles in in vivo spread and tissue localization of various tumors.21 Therefore, it is of great interest to determine the patterns of chemokine receptor expression in various types of leukemia and lymphoma. Jones et al have reported that all types of B-cell lymphoma express CXCR5 whereas certain distinct subtypes of B-cell lymphoma (ie, chronic lymphocytic leukemia/small lymphocytic lymphoma and splenic marginal zone lymphoma) also express CXCR3.22 Trentin et al have also demonstrated frequent expression of CXCR3 in malignant B cells, especially chronic lymphocytic leukemia.23 CXCR5 is the receptor for an important B-cell attractant, BLC/CXCL13, which is expressed in the lymphoid follicles.24,25 During normal B-cell differentiation and maturation, CXCR5 has been shown to be expressed by pro-B cells and all stages of mature B cells.26 On the other hand, CXCR3 is the shared receptor for IP-10/CXCL10, Mig/CXCL9, and I-TAC/CXCL11, all commonly induced by interferon gamma (IFN-γ).27,28 CXCR3 is known to be expressed by T cells, especially upon activation.28,29 Only a small proportion of B cells in peripheral blood express CXCR3.23,29 Thus, the findings that certain types of B-cell malignancy consistently express CXCR3 are rather striking and may indicate their origin from a specific subset of B cells. In the case of T-cell lymphoma, Jones et al have reported that CXCR3 is typically expressed in angioimmunoblastic lymphoma, angiocentric lymphoma, histiocyte-rich tumors, and many unspecified T-cell lymphomas, whereas expression of CCR4 is associated especially with ALK-positive anaplastic large-cell lymphomas and mycosis fungoides in large-cell transformation.30 CCR4 is the receptor for TARC/CCL17 and MDC/CCL2215,16 and is selectively expressed by CD4+ memory T cells.10 Importantly, it is now a consensus that CXCR3 is a selective marker of Th1 whereas CCR4 is a selective marker of Th2.10,11,31 In this context, ALK-positive anaplastic large-cell lymphoma and mycosis fungoides expressing CCR4 frequently coexpress CD30, a marker also associated with Th2.30However, coexpression of CXCR3 and CCR4 has also been observed in some T-cell lymphomas22 and in primary CD4+ memory T cells.11 Furthermore, whereas CCR4 expression clearly correlated with selective production of Th2-type cytokines, a significant fraction of primary CCR4+ memory T cells were nevertheless observed to produce IFN-γ upon activation.11 Thus, CXCR3 and CCR4 are selective for Th1 and Th2, respectively, but are not mutually exclusive. At any rate, the unique expression patterns of chemokine receptors in various types of B- and T-cell lymphomas and leukemias are likely to reflect their corresponding stages of differentiation and maturation, and may also have important roles in their in vivo spread and tissue localization.
ATL is a malignancy of mature T cells and is particularly notorious for its highly frequent invasion into various organs.2Previously, various cell adhesion molecules have been implicated in tissue infiltration of ATL cells.32-35 Recently, we have shown that high levels of expression of CCR7 in ATL, the receptor for SLC/CCL21 and ELC/CCL19 commonly expressed in the secondary lymphoid organs,7,8 correlates positively with lymphoid organ involvement.9 In the present study, we have further shown that HTLV-1–immortalized T cells and fresh ATL cells frequently express CCR4. The HTLV-1–encoded transactivator tax is a potent inducer of cellular target genes including various cell adhesion molecules and chemokines.34,36-40 However, it does not up-regulate CCR4 or CCR7 at least in the cellular background of Jurkat (Figure 1). Thus, highly frequent expression of CCR4 in ATL and HTLV-1–immortalized T cells may be due to preferential infection of HTLV-1 to CCR4+ T cells and/or some growth advantages of HTLV-1–infected CCR4+ T cells in vivo as well as in vitro. Since CCR4+ T cells are considered to be mostly polarized to Th2,10,11 the production of Th2-type cytokines by HTLV-1–infected T cells may help them to avoid immunologic attacks in vivo by shifting the host Th1/Th2 balance to Th2. In fact, suppression of cell-mediated immunity has been well documented in HTLV-1 carriers and patients with ATL.41-43 Even though previous studies on the cytokine profiles of ATL cell lines and HTLV-1–immortalized T cells do not particularly support a shift to Th2,44 45similar studies using freshly isolated cells from carriers and patients with ATL may be necessary to test this possibility. It should also be noted that not all ATL cases are CCR4+. This may be due to down-regulation of CCR4 during development of ATL in some patients or development of ATL from CCR4− HTLV-1–infected T cells. It remains to be seen whether there are any phenotypic differences between CCR4+ and CCR4− ATL cases.
As stated above, CCR4 is the receptor for TARC/CCL17 and MDC/CCL22.15,16 Consistent with Th2-selective expression of CCR4,10,11 TARC has been shown to be produced by epidermal keratinocytes in atopic dermatitis and by bronchial epithelium in asthma, both typical Th2-type diseases.18,46On the other hand, MDC has been shown to be produced by dermal dendritic cells and also by mature dendritic cells in draining regional lymph nodes.18,20,47 Furthermore, Campbell et al have reported that essentially all CLA+ skin-seeking memory T cells are CCR4+, whereas microvascular endothelium in inflammed skin tissues express TARC.12 Andrew et al have also demonstrated that CCR4 is expressed by the majority of circulating nonintestinal (α4β7−) memory CD4+ T cells, including almost all CLA+ skin-homing memory T cells.11 Thus, CCR4-expressing CLA+ T cells may emigrate into inflamed skin tissue by the guidance of TARC and MDC.20 Consistently, we have detected strong signals of TARC and MDC as well as those of CCR4 and Tax in ATL skin lesions (Figure 7). Thus, the frequent involvement of skin in ATL may be accounted for, in part, by frequent expression of CCR4 by ATL. In this context, CCR4 is also frequently expressed by skin-associated lymphomas such as mycosis fungoides.30
In spite of highly frequent expression of CCR4 on ATL cells, however, only a fraction of patients have skin lesions (Table 2). It is thus evident that CCR4 positivity of leukemic cells per se is not sufficient for skin invasion. Other factors such as expression of CLA by leukemic cells and inflammatory responses in lesional skin leading to up-regulation of TARC and MDC are also likely to contribute to skin involvement of ATL.20,48 It should also be pointed out that together with CCR7,9 CCR4 may also facilitate ATL cells to invade the secondary lymphoid organs where mature dendritic cells are known to produce MDC.20 47
In conclusion, we have demonstrated a highly frequent expression of CCR4 on ATL and HTLV-1–infected T cells. This suggests that HTLV-1 is preferentially transmitted to CCR4-expressing Th2 cells and/or HTLV-1–infected Th2 cells have certain growth advantages. Our findings may also provide a new clue to understand certain aspects of ATL and HTLV-1–associated diseases such as frequent tissue involvement and suppression of Th1-mediated immune responses.
We are grateful to Kazuyuki Sugahara and Kazuto Tsuruda for their excellent technical assistance.
Supported by Grants-in-Aid for Science Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Osamu Yoshie, Department of Microbiology, Kinki University School of Medicine, 377-2 Ohno-Higashi, Osaka-Sayama, Osaka 589-8511, Japan; e-mail: o.yoshie@med.kindai.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal