Bone marrow plasma cells (PCs) from 74 patients with newly diagnosed multiple myeloma (MM), 5 with monoclonal gammopathy of undetermined significance (MGUS), and 31 healthy volunteers (normal PCs) were purified by CD138+ selection. Gene expression of purified PCs and 7 MM cell lines were profiled using high-density oligonucleotide microarrays interrogating about 6800 genes. On hierarchical clustering analysis, normal and MM PCs were differentiated and 4 distinct subgroups of MM (MM1, MM2, MM3, and MM4) were identified. The expression pattern of MM1 was similar to normal PCs and MGUS, whereas MM4 was similar to MM cell lines. Clinical parameters linked to poor prognosis, abnormal karyotype (P = .002) and high serum β2-microglobulin levels (P = .0005), were most prevalent in MM4. Also, genes involved in DNA metabolism and cell cycle control were overexpressed in a comparison of MM1 and MM4. In addition, using χ2 and Wilcoxon rank sum tests, 120 novel candidate disease genes were identified that discriminate normal and malignant PCs (P < .0001); many are involved in adhesion, apoptosis, cell cycle, drug resistance, growth arrest, oncogenesis, signaling, and transcription. A total of 156 genes, including FGFR3 andCCND1, exhibited highly elevated (“spiked”) expression in at least 4 of the 74 MM cases (range, 4-25 spikes). Elevated expression of these 2 genes was caused by the translocation t(4;14)(p16;q32) or t(11;14)(q13;q32). Thus, novel candidate MM disease genes have been identified using gene expression profiling and this profiling has led to the development of a gene-based classification system for MM.
Introduction
Progress in understanding the biology and genetics of and advancing therapy for multiple myeloma (MM) has been slow. MM cells are endowed with a multiplicity of antiapoptotic signaling mechanisms, which account for their resistance to current chemotherapy and thus the ultimately fatal outcome for most patients.1Although aneuploidy by interphase fluorescence in situ hybridization (FISH)2 and DNA flow cytometry3 is observed in more than 90% of cases, cytogenetic abnormalities in this typically hypoproliferative tumor are informative in only about 30% of cases and are typically complex, involving on average 7 different chromosomes. Given this “genetic chaos,” it has been difficult to establish correlations between genetic abnormalities and clinical outcomes.4,5 Only recently has chromosome 13 deletion been identified as a distinct clinical entity with a grave prognosis.6-8 However, even with the most comprehensive analysis of laboratory parameters, such as β2-microglobulin (β2M),9C-reactive protein,10 plasma cell labeling index,11 metaphase karyotyping,7,8 and FISH,12-14 the clinical course of patients afflicted with MM can only be approximated, because no more than more than 20% of the clinical heterogeneity can be accounted for.7
The advent of high-density oligonucleotide DNA microarray has made possible a simultaneous analysis of messenger RNA (mRNA) expression patterns of thousands of genes pertinent to various biologic functions.15 Here we report that, in a comparison with normal plasma cells (PCs), MM PCs are distinctly different. Furthermore, using hierarchical clustering, 4 distinct subgroups of MM PCs were established that reveal significant correlations with clinical characteristics known to be associated with poor prognosis. This system represents the framework for a new classification system and identifies the genetic differences associated with these distinct subgroups.
Materials and methods
Cell collection and total RNA purification
Samples included PCs from 74 newly diagnosed cases of MM, 5 patients with monoclonal gammopathy of undetermined significance (MGUS), and 31 healthy donors (normal PCs). Written informed consent was obtained in keeping with institutional policies. PC isolation from mononuclear cell fraction was performed by immunomagnetic bead selection with monoclonal mouse antihuman CD138 antibodies using the AutoMACs automated separation system (Miltenyi-Biotec, Auburn, CA). PC purity of more than 95% homogeneity was confirmed by 2-color flow cytometry using CD138+/CD45− and CD38+/CD45− criteria (Becton Dickinson, San Jose, CA), immunocytochemistry for cytoplasmic light-chain immunoglobulin (Ig), and morphology by Wright-Giemsa staining. MM cell lines (U266, ARP1, RPMI-8226, UUN, ANBL-6, CAG, and H929 [courtesy of P. L. Bergsagel]) and an Epstein-Barr virus (EBV)–transformed B-lymphoblastoid cell line (ARH-77) were grown as recommended (American Type Culture Collection, Chantilly, VA). Total RNA was isolated with RNeasy Mini Kit (Qiagen, Valencia, CA). The entire Affymetrix data set of all 118 PC samples can be found athttp://lambertlab.uams.edu.
Preparation of labeled complementary RNA and hybridization to high-density microarray
Double-stranded complementary DNA (cDNA) and biotinylated complementary RNA (cRNA) were synthesized from total RNA and hybridized to HuGeneFL GeneChip microarrays (Affymetrix, Santa Clara, CA), which were washed and scanned according to procedures developed by the manufacturer. The arrays were scanned using Hewlett Packard confocal laser scanner and visualized using Affymetrix 3.3 software (Affymetrix). Arrays were scaled to an average intensity of 1500 and analyzed independently. Technical details of this analysis are available via the Internet athttp://lambertlab.uams.edu.
GeneChip data analysis
To efficiently manage and mine high-density oligonucleotide DNA microarray data, a new data-handling tool was developed. GeneChip-derived expression data were stored on an MS SQL Server. This database was linked, via an MS Access interface called Clinical Gene-Organizer to multiple clinical parameter databases for patients with MM. This Data Mart concept allows gene expression profiles to be directly correlated with clinical parameters and clinical outcomes using standard statistical software. An MS JET version of Clinical Gene-Organizer can be downloaded from our Web page athttp://lambertlab.uams.edu. All data used in our analysis were derived from Affymetrix 3.3 software. GeneChip 3.3 output files are given (1) as an average difference (AD) that represents the difference between the intensities of the sequence-specific perfect match probe set and the mismatch probe set, or (2) as an absolute call (AC) of present or absent as determined by the GeneChip 3.3 algorithm. AD calls were transformed by the natural log after substituting any sample with an AD of less than 60 with the value 60 (2.5 times the average Raw Q). Statistical analyses of the data were performed with software packages SPSS 10.0 (SPSS, Chicago, IL), S-Plus 2000 (Insightful, Seattle, WA), and Gene Cluster/Treeview.16
Hierarchical clustering of average linkage clustering with the centered correlation metric was used.16 The clustering was done on the AD data of 5483 genes. Either χ2 or Fisher exact test was used to find significant differences between cluster groups with the AC data. To compare the expression levels, the nonparametric Wilcoxon rank sum (WRS) test was used. This test uses a null hypothesis that is based on ranks rather than on normally distributed data. Before the above tests were performed, genes that were absent (AC) across all samples were removed; 5483 genes were used in the analyses. Genes that were significant (P < .0001) for both the χ2 test and the WRS test were considered to be significantly differentially expressed.
Clinical parameters were tested across MM cluster groups. To test the continuous variables, we used an ANOVA test; to test discrete variables, a χ2 test of independence or Fisher exact test was applied.
The natural logs of the AD data were used to find genes with a “spiked profile” of expression in MM. Genes were identified that had low to undetectable expression in the majority of patients and normal samples (no more than 4 present absolute calls [P-ACs]). A total of 2030 genes fit the criteria of this analysis. The median expression value of each of the genes across all patient samples was determined. For the ith gene, we called this value medgene (i). We called the ith gene a “spiked” gene if it had at least 4 patient expression values more than 2.5 + medgene (i). The constant 2.5 was based on the log of the AD data. These genes that were “spiked” were further divided into subsets according to whether or not the largest spike had an AD expression value more than 10 000.
Reverse transcription–polymerase chain reaction
Reverse transcription–polymerase chain reaction (RT-PCR) for the FGFR3 MMSET was performed on the same cDNAs used in the microarray analysis. Briefly, cDNA was mixed with the IGJH2 (5′-CAATGGTCACCGTCTCTTCA-3′) primer and the MMSET primer (5′-CCTCAATTTCCTGAAATTGGTT-3′). PCR reactions consisted of 30 cycles with a 58°C annealing temperature and 1-minute extension time at 72°C using a Perkin-Elmer GeneAmp 2400 thermocycler (Wellesley, MA). PCR products were visualized by ethidium bromide staining after agarose gel electrophoresis.
Immunohistochemistry
Immunohistochemical staining was performed on a Ventana ES (Ventana Medical Systems, Tucson, AZ) using Zenker-fixed paraffin-embedded bone marrow sections, an avidin-biotin peroxidase complex technique (Ventana Medical Systems), and the antibody L26 (CD20, Ventana Medical Systems). Heat-induced epitope retrieval was performed by microwaving the sections for 28 minutes in a 1.0-mmol/L concentration of citrate buffer at pH 6.0.
Interphase FISH
For interphase detection of the t(11;14)(q13;q32) translocation fusion signal, we used a the LSI IGH/CCND1 dual-color, dual-fusion translocation probe (Vysis, Downers Grove, IL). The TRI-FISH procedure used to analyze the samples has been previously described.12 Briefly, at least 100 clonotypic PCs, identified by cytoplasmic immunoglobulin (cIg) staining were counted for the presence or absence of the translocation fusion signal in all samples except one, which yielded only 35 PCs. An MM sample was defined as having the translocation when more than 25% of the cells contained the fusion.
Flow cytometry
For flow cytometric analysis of CD marker expression, a panel of antibodies directly conjugated to fluorescein isothiocyanate (FITC) or phycoerythrin (PE) was used: FITC-labeled CD19, CD20, and CD22 (Becton Dickinson); CD38 and CD45 (BD Pharmingen, San Diego, CA); CD52 and CD138 (Serotec, Raleigh, NC), and PE-labeled CD21 (BD Pharmingen). Cells were harvested from culture, washed in phosphate-buffered saline (PBS) and stained at 4°C with CD antibodies or isotype-matched control antibodies. After staining, cells were fixed in 1% paraformaldehyde and analyzed using a FACSscan flow cytometer (Becton Dickinson).
Results
Hierarchical clustering of PC gene expression demonstrates class distinction
As a result of 656 000 measurements of gene expression in 118 PC samples, altered gene expression in the MM samples was identified. Two-dimensional hierarchical clustering differentiated cell types by gene expression when performed on 5483 genes when expression was present in at least one of the 118 samples (Figure1A). The sample dendrogram derived 2 major branches (Figure 1A,D). One branch contained all 31 normal samples and a single MGUS case, whereas the second branch contained all 74 MM and 4 MGUS cases and the 8 cell lines. The MM-containing branch was further divided into 2 sub-branches, one containing the 4 MGUS and the other the 8 MM cell lines, which were all clustered next to one another, thus showing a high degree of similarity in gene expression among the cell lines. This suggested that MM could be differentiated from normal PCs and that at least 2 different classes of MM could be identified, one more similar to MGUS and the other similar to MM cell lines. To show reproducibility of the technique and analysis, we repeated the hierarchical clustering analysis with all 118 samples, including duplicate samples from 12 patients (PCs taken 24 hours or 48 hours after initial sample). All samples from the 12 patients studied longitudinally were found to cluster adjacent to one another. This indicated that gene expression in samples from the same patient were more similar to each other than they were to all other samples (data not shown).
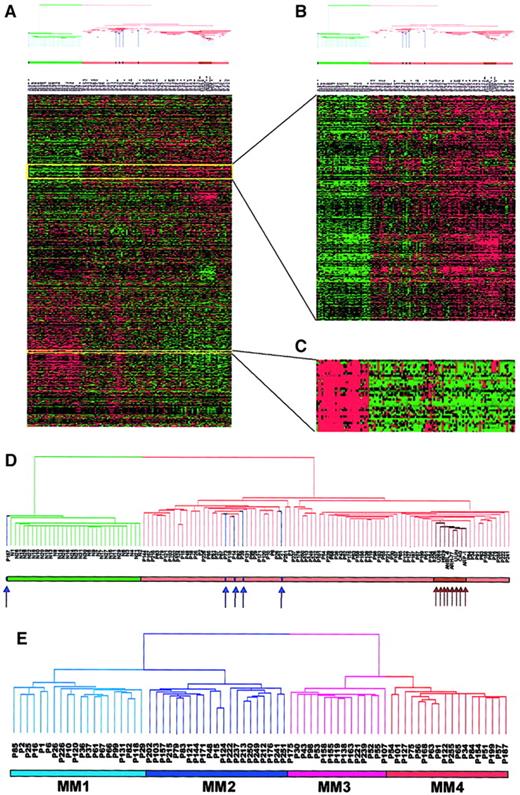
Two-dimensional hierarchical cluster analysis of experimental expression profiles and gene behavior.
(A) Cluster-ordered data table. The clustering is presented graphically as a colored image. Along the vertical axis, the analyzed genes are arranged as ordered by the clustering algorithm. The genes with the most similar patterns of expression are placed adjacent to each other. Likewise, along the horizontal axis, experimental samples are arranged; those with the most similar patterns of expression across all genes are placed adjacent to each other. Both sample and gene groupings can be further described by following the solid lines (branches) that connect the individual components with the larger groups. The color of each cell in the tabular image represents the expression level of each gene, with red representing an expression greater than the mean, green representing an expression less than the mean, and the deeper color intensity representing a greater magnitude of deviation from the mean. (B) Amplified gene cluster showing genes down-regulated in MM. Most of the characterized and sequence-verified cDNA-encoded genes are known to be immunoglobulins. (C) Cluster enriched with genes whose expression level was correlated with tumorigenesis, cell cycle, and proliferation rate. Many of these genes were also statistically significantly up-regulated in MM (χ2 and WRS test) (Table 5). (D) Dendrogram of hierarchical cluster; 74 cases of newly diagnosed untreated MM, 5 MGUS, 8 MM cell lines, and 31 normal bone marrow PC samples clustered based on the correlation of 5483 genes (probe sets). Different-colored branches represent normal PC (green), MGUS (blue arrow), MM (tan), and MM cell lines (brown arrow). (E) Dendrogram of a hierarchical cluster analysis of 74 cases of newly diagnosed untreated MM alone (clustergram not shown). Two major branches contained 2 distinct cluster groups. The subgroups under the right branch, designated MM1 (light blue) and MM2 (blue) were more related to the MGUS cases in Figure 1D. The 2 subgroups under the left branch, designated MM3 (violet) and MM4 (red) represent samples that were more related to the MM cell lines in Figure 1D.
Two-dimensional hierarchical cluster analysis of experimental expression profiles and gene behavior.
(A) Cluster-ordered data table. The clustering is presented graphically as a colored image. Along the vertical axis, the analyzed genes are arranged as ordered by the clustering algorithm. The genes with the most similar patterns of expression are placed adjacent to each other. Likewise, along the horizontal axis, experimental samples are arranged; those with the most similar patterns of expression across all genes are placed adjacent to each other. Both sample and gene groupings can be further described by following the solid lines (branches) that connect the individual components with the larger groups. The color of each cell in the tabular image represents the expression level of each gene, with red representing an expression greater than the mean, green representing an expression less than the mean, and the deeper color intensity representing a greater magnitude of deviation from the mean. (B) Amplified gene cluster showing genes down-regulated in MM. Most of the characterized and sequence-verified cDNA-encoded genes are known to be immunoglobulins. (C) Cluster enriched with genes whose expression level was correlated with tumorigenesis, cell cycle, and proliferation rate. Many of these genes were also statistically significantly up-regulated in MM (χ2 and WRS test) (Table 5). (D) Dendrogram of hierarchical cluster; 74 cases of newly diagnosed untreated MM, 5 MGUS, 8 MM cell lines, and 31 normal bone marrow PC samples clustered based on the correlation of 5483 genes (probe sets). Different-colored branches represent normal PC (green), MGUS (blue arrow), MM (tan), and MM cell lines (brown arrow). (E) Dendrogram of a hierarchical cluster analysis of 74 cases of newly diagnosed untreated MM alone (clustergram not shown). Two major branches contained 2 distinct cluster groups. The subgroups under the right branch, designated MM1 (light blue) and MM2 (blue) were more related to the MGUS cases in Figure 1D. The 2 subgroups under the left branch, designated MM3 (violet) and MM4 (red) represent samples that were more related to the MM cell lines in Figure 1D.
The clustergram (Figure 1A) showed that genes of unrelated sequence but similar function clustered tightly together along the vertical axis. For example, a particular cluster of 22 genes, primarily those encoding immunoglobulin molecules and major histocompatibility genes, had relatively low expression in MM PCs and high expression in normal PCs (Figure 1B). This was anticipated, given that the PCs isolated from MM are clonal and hence only express single immunoglobulin light-chain and heavy-chain variable and constant region genes, whereas PCs from healthy donors are polyclonal and express many different genes of these 2 classes. Another cluster of 195 genes was highly enriched for numerous oncogenes/growth-related genes (eg,MYC, ABL1, PHB, and EXT2), cell cycle–related genes (eg, CDC37, CDK4, andCKS2), and translation machinery genes (EIF2,EIF3, HTF4A, and TFIIA) (Figure 1C). These genes were all highly expressed in MM, especially in MM cell lines, but had low expression levels in normal PCs.
Hierarchical clustering of newly diagnosed MM identifies 4 distinct subgroups
We performed 2-dimensional cluster analysis of the 74 MM cases alone. The sample dendrogram identified 2 major branches with 2 distinct subgroups within each branch (Figure 1E). We designated the 4 subgroups MM1, MM2, MM3, and MM4 containing 20, 21, 15, and 18 patients, respectively. The MM1 subgroup represented the patients whose PCs were most closely related to the MGUS PCs and whose MM4 were most like the MM cell lines (Figure 1D). These data suggested that the 4 gene expression subgroups were authentic and might represent 4 distinct clinical entities. We then examined differences in gene expression across the 4 subgroups using the χ2 and WRS tests (Table1). As expected the largest difference was between MM1 and MM4 (205 genes) and the smallest between MM1 and MM2 (24 genes). We then looked at the top 30 genes turned on, or up-regulated, in MM4 compared with MM1 (Table2). These data demonstrated that 13 of the 30 most significant genes (10 of the top 15 genes) were involved in DNA replication/repair or cell cycle control. Thymidylate synthase (TYMS), which was present in all 18 samples comprising the MM4 subgroup, was only present in 3 of the 20 MM1 samples and represented the most significant gene in the χ2 test. The DNA mismatch repair gene, mutS (Escherichia coli) homolog 2 (MSH2) with a WRSP value of 2.8 × 10−6 was the most significant gene in the WRS test. Other notable genes in the list included the CAAX farnesyltransferase (FNTA), the transcription factors enhancer of zeste homolog 2 (EZH2) andMYC-associated zinc finger protein (MAZ), eukaryotic translation initiation factors (EIF2S1 andEIF2B1), as well as the mitochondrial translation initiation factor 2 (MTIF2), the chaperone (CCT4), the UDP-glucose pyrophosphorylase 2 (IUGP2), and the 26S proteasome–associated pad1 homolog (POH1).
Differences in gene expression among MM subgroups
| Comparison . | Range of WRS*P . | Number of genes . |
|---|---|---|
| MM1 vs MM4 | .00097 to 9.58 × 10−7 | 205 |
| MM2 vs MM4 | .00095 to 1.04 × 10−6 | 162 |
| MM3 vs MM4 | .00098 to 3.75 × 10−6 | 119 |
| MM1 vs MM3 | .00091 to 6.27 × 10−6 | 68 |
| MM2 vs MM3 | .00097 to 1.98 × 10−5 | 44 |
| MM1 vs MM2 | .00083 to 2.93 × 10−5 | 24 |
| Comparison . | Range of WRS*P . | Number of genes . |
|---|---|---|
| MM1 vs MM4 | .00097 to 9.58 × 10−7 | 205 |
| MM2 vs MM4 | .00095 to 1.04 × 10−6 | 162 |
| MM3 vs MM4 | .00098 to 3.75 × 10−6 | 119 |
| MM1 vs MM3 | .00091 to 6.27 × 10−6 | 68 |
| MM2 vs MM3 | .00097 to 1.98 × 10−5 | 44 |
| MM1 vs MM2 | .00083 to 2.93 × 10−5 | 24 |
Wilcoxon rank sum test. Comparisons are ordered based on the number of significant genes. All comparisons have a WRSP < .001.
The 30 most differentially expressed genes in a comparison of MM1 and MM4 subgroups
| Accession* . | Function . | Gene symbol† . | MM1 (N = 20) P . | MM4 (N = 18)P . | χ2 . | WRS‡Pvalue . |
|---|---|---|---|---|---|---|
| D00596 | DNA replication | TYMS | 3 | 18 | 24.35 | 1.26 × 10−4 |
| U35835 | DNA repair | PRKDC | 2 | 17 | 23.75 | 4.55 × 10−6 |
| U77949 | DNA replication | CDC6 | 1 | 13 | 15.62 | 5.14 × 10−6 |
| U91985 | DNA fragmentation | DFFA | 1 | 12 | 13.38 | 6.26 × 10−5 |
| U61145 | Transcription | EZH2 | 4 | 15 | 12.77 | 1.67 × 10−4 |
| U20979 | DNA replication | CHAF1A | 2 | 12 | 10.75 | 1.10 × 10−4 |
| U03911 | DNA repair | MSH2 | 0 | 9 | 10.48 | 2.88 × 10−6 |
| X74330 | DNA replication | PRIM1 | 0 | 9 | 10.48 | 9.36 × 10−6 |
| X12517 | SnRNP | SNRPC | 0 | 9 | 10.48 | 5.26 × 10−6 |
| D85131 | Transcription | MAZ | 0 | 9 | 10.48 | 1.08 × 10−5 |
| L00634 | Farnesyltransferase | FNTA | 10 | 18 | 9.77 | 7.28 × 10−5 |
| U21090 | DNA replication | POLD2 | 11 | 18 | 8.27 | 8.05 × 10−5 |
| X54941 | Cell cycle | CKS1 | 10 | 17 | 7.07 | 1.26 × 10−4 |
| U62136 | Cell cycle | UBE2V2 | 13 | 18 | 5.57 | 4.96 × 10−6 |
| D38076 | Cell cycle | RANBP1 | 13 | 18 | 5.57 | 7.34 × 10−6 |
| X95592 | Unknown | C1D† | 13 | 18 | 5.57 | 1.10 × 10−4 |
| X66899 | Cell cycle | EWSR1 | 14 | 18 | 4.35 | 1.89 × 10−4 |
| L34600 | Translation initiation | MTIF2 | 14 | 18 | 4.35 | 3.09 × 10−5 |
| U27460 | Metabolism | IUGP2 | 15 | 18 | 3.22 | 1.65 × 10−4 |
| U15009 | SnRNP | SNRPD3 | 15 | 18 | 3.22 | 1.47 × 10−5 |
| J02645 | Translation initiation | EIF2S1 | 16 | 18 | 2.18 | 7.29 × 10−5 |
| X95648 | Translation initiation | EIF2B1 | 16 | 18 | 2.18 | 1.45 × 10−4 |
| M34539 | Calcium signaling | FKBP1A | 18 | 18 | 0.42 | 1.71 × 10−5 |
| J04611 | DNA repair | G22P1 | 18 | 18 | 0.42 | 7.29 × 10−5 |
| U67122 | Antiapoptosis | UBL1 | 20 | 18 | 0.00 | 7.29 × 10−5 |
| U38846 | Chaperon | CCT4 | 20 | 18 | 0.00 | 1.26 × 10−4 |
| U80040 | Metabolism | ACO2 | 20 | 18 | 0.00 | 8.38 × 10−5 |
| U86782 | Proteasome | POH† | 20 | 18 | 0.00 | 5.90 × 10−5 |
| X57152 | Signaling | CSNK2B | 20 | 18 | 0.00 | 7.29 × 10−5 |
| D87446 | Unknown | KIAA0257† | 20 | 18 | 0.00 | 1.26 × 10−5 |
| Accession* . | Function . | Gene symbol† . | MM1 (N = 20) P . | MM4 (N = 18)P . | χ2 . | WRS‡Pvalue . |
|---|---|---|---|---|---|---|
| D00596 | DNA replication | TYMS | 3 | 18 | 24.35 | 1.26 × 10−4 |
| U35835 | DNA repair | PRKDC | 2 | 17 | 23.75 | 4.55 × 10−6 |
| U77949 | DNA replication | CDC6 | 1 | 13 | 15.62 | 5.14 × 10−6 |
| U91985 | DNA fragmentation | DFFA | 1 | 12 | 13.38 | 6.26 × 10−5 |
| U61145 | Transcription | EZH2 | 4 | 15 | 12.77 | 1.67 × 10−4 |
| U20979 | DNA replication | CHAF1A | 2 | 12 | 10.75 | 1.10 × 10−4 |
| U03911 | DNA repair | MSH2 | 0 | 9 | 10.48 | 2.88 × 10−6 |
| X74330 | DNA replication | PRIM1 | 0 | 9 | 10.48 | 9.36 × 10−6 |
| X12517 | SnRNP | SNRPC | 0 | 9 | 10.48 | 5.26 × 10−6 |
| D85131 | Transcription | MAZ | 0 | 9 | 10.48 | 1.08 × 10−5 |
| L00634 | Farnesyltransferase | FNTA | 10 | 18 | 9.77 | 7.28 × 10−5 |
| U21090 | DNA replication | POLD2 | 11 | 18 | 8.27 | 8.05 × 10−5 |
| X54941 | Cell cycle | CKS1 | 10 | 17 | 7.07 | 1.26 × 10−4 |
| U62136 | Cell cycle | UBE2V2 | 13 | 18 | 5.57 | 4.96 × 10−6 |
| D38076 | Cell cycle | RANBP1 | 13 | 18 | 5.57 | 7.34 × 10−6 |
| X95592 | Unknown | C1D† | 13 | 18 | 5.57 | 1.10 × 10−4 |
| X66899 | Cell cycle | EWSR1 | 14 | 18 | 4.35 | 1.89 × 10−4 |
| L34600 | Translation initiation | MTIF2 | 14 | 18 | 4.35 | 3.09 × 10−5 |
| U27460 | Metabolism | IUGP2 | 15 | 18 | 3.22 | 1.65 × 10−4 |
| U15009 | SnRNP | SNRPD3 | 15 | 18 | 3.22 | 1.47 × 10−5 |
| J02645 | Translation initiation | EIF2S1 | 16 | 18 | 2.18 | 7.29 × 10−5 |
| X95648 | Translation initiation | EIF2B1 | 16 | 18 | 2.18 | 1.45 × 10−4 |
| M34539 | Calcium signaling | FKBP1A | 18 | 18 | 0.42 | 1.71 × 10−5 |
| J04611 | DNA repair | G22P1 | 18 | 18 | 0.42 | 7.29 × 10−5 |
| U67122 | Antiapoptosis | UBL1 | 20 | 18 | 0.00 | 7.29 × 10−5 |
| U38846 | Chaperon | CCT4 | 20 | 18 | 0.00 | 1.26 × 10−4 |
| U80040 | Metabolism | ACO2 | 20 | 18 | 0.00 | 8.38 × 10−5 |
| U86782 | Proteasome | POH† | 20 | 18 | 0.00 | 5.90 × 10−5 |
| X57152 | Signaling | CSNK2B | 20 | 18 | 0.00 | 7.29 × 10−5 |
| D87446 | Unknown | KIAA0257† | 20 | 18 | 0.00 | 1.26 × 10−5 |
Accession numbers listed are GenBank numbers, except those beginning with “HT,” which are provided by the Institute of Genomic Research (TIGR).
Gene symbol not Human Genome Organisation (HUGO) approved.
Wilcoxon rank sum test.
To assess the validity of the clusters with respect to clinical features, correlations of various clinical parameters across the 4 subgroups were analyzed (Table 3). Of 17 clinical variables tested, the presence of an abnormal karyotype (P = .0003) and serum β2M levels (P = .0005) were significantly different among the 4 subgroups and increased creatinine (P = .06) and cytogenetic deletion of chromosome 13 (P = .09) were marginally significant. The trend was to have higher β2M and creatinine as well as an abnormal karyotype and deletion 13 in the MM4 subgroup, as compared with the other 3 subgroups.
Clinical parameters linked to MM subgroups
| Clinical parameter . | MM subgroups . | P . | |||
|---|---|---|---|---|---|
| 1 . | 2 . | 3 . | 4 . | ||
| Abnormal cytogenetics | 40.0% | 10.5% | 53.3% | 72.2% | .0017 |
| Average β2M (mg/L) | 2.81 | 2.73 | 4.62 | 8.81 | .00047 |
| Average creatinine (mg/dL) | 0.96 | 1.02 | 1.32 | 1.77 | .06019 |
| Clinical parameter . | MM subgroups . | P . | |||
|---|---|---|---|---|---|
| 1 . | 2 . | 3 . | 4 . | ||
| Abnormal cytogenetics | 40.0% | 10.5% | 53.3% | 72.2% | .0017 |
| Average β2M (mg/L) | 2.81 | 2.73 | 4.62 | 8.81 | .00047 |
| Average creatinine (mg/dL) | 0.96 | 1.02 | 1.32 | 1.77 | .06019 |
ANOVA, χ2, and Fisher exact tests were used to determine significance.
Altered expression of 120 genes differentiates malignant from normal PCs
Our hierarchical cluster analysis showed that MM PCs could be differentiated from normal PCs. Genes distinguishing the MM from normal PCs were identified as significant by χ2 analysis and the WRS test (P < .0001). A statistical analysis showed that 120 genes distinguished MM from normal PCs. Pearson correlation analyses of the 120 differentially expressed genes were used to identify whether the genes were up-regulated or down-regulated in MM.
When genes associated with immune function (eg, IGH,IGL, HLA), representing the majority of significantly down-regulated genes, were filtered out, 50 genes showed significant down-regulation in MM (Table4). The P values for the WRS test ranged from 9.80 × 10−5 to 1.56 × 10−14 and the χ2 test of the absence or presence of the expression of the gene in the groups ranged from 18.83 to 48.45. The gene representing the most significant difference in the χ2 test was the CXC chemokineSDF1. It is important to note that a comparison of MM PCs to tonsil-derived PCs showed that, like MM PCs, tonsil PCs also do not express SDF1 (J.S., manuscript in preparation). Two additional CXC chemokines, PF4 and PF4V1, were also absent in MM PCs. The second most significant gene was the tumor necrosis factor receptor (TNFR) super family member TNFRF7coding for CD27, a molecule that has been linked to controlling maturation and apoptosis of plasma cells.17-19 The largest group of genes, 20 of 50, were linked to signaling cascades. MM PCs have reduced or no expression of genes associated with calcium signaling (S100A9 and S100A12) or lipoprotein signaling (LIPA, LCN2, PLA2G7, APOE, APOC1).LCN2, also known as 24p3, codes for secreted lipocalin, which has recently been shown to induce apoptosis in pro B-cells after growth factor deprivation.20 Another major class absent in MM PCs was adhesion-associated genes (ITGA2B, IGTB2, GP5, VCAM, and MIC2).
The 50 most significantly down-regulated genes in MM in comparison with normal bone marrow PCs
| Accession4-150 . | Function . | Gene symbol4-151 . | χ2 . | WRS‡P value . |
|---|---|---|---|---|
| L36033 | CXC chemokine | SDF1 | 48.45 | 3.05 × 10−12 |
| M63928 | Signaling | TNFRSF7 | 48.45 | 6.35 × 10−11 |
| U64998 | Ribonuclease | RNASE6 | 46.44 | 2.82 × 10−9 |
| M20902 | Lipoprotein signaling | APOC1 | 45.62 | 4.63 × 10−10 |
| M26602 | Immunity | DEFA1 | 40.75 | 1.06 × 10−12 |
| M21119 | Immunity | LYZ | 40.73 | 6.24 × 10−10 |
| M14636 | Metabolism | PYGL | 39.84 | 1.15 × 10−10 |
| M26311 | Calcium signaling | S100A9 | 38.96 | 3.60 × 10−13 |
| M54992 | Signaling | CD72 | 36.14 | 2.40 × 10−9 |
| X16832 | Protein degradation | CTSH | 35.26 | 1.81 × 10−12 |
| M12529 | Lipoprotein signaling | APOE | 34.50 | 3.95 × 10−14 |
| M15395 | Adhesion | ITGB2 | 34.02 | 1.74 × 10−13 |
| Z74616 | Extracellular matrix | COL1A2 | 34.02 | 8.06 × 10−11 |
| HT2152 | Receptor signaling | CD163 | 33.01 | 1.66 × 10−12 |
| U97105 | Pyrimidine metabolism | DPYSL2 | 32.52 | 2.22 × 10−10 |
| U81787 | Signaling | WNT10B | 32.50 | 1.77 × 10−5 |
| HT3165 | Receptor tyrosine kinase | AXL | 31.36 | 5.26 × 10−11 |
| M83667 | Transcription | CEBPD | 31.19 | 4.69 × 10−10 |
| L33930 | Receptor signaling | CD24 | 30.33 | 1.56 × 10−14 |
| D83657 | Calcium signaling | S100A12 | 29.91 | 6.58 × 10−8 |
| M11313 | Proteinase inhibitor | A2M | 29.91 | 1.07 × 10−10 |
| M31158 | Signaling | PRKAR2B | 29.91 | 2.20 × 10−9 |
| U24577 | Lipoprotein signaling | PLA2G7 | 29.78 | 2.08 × 10−10 |
| M16279 | Adhesion | MIC2 | 28.75 | 8.01 × 10−11 |
| HT2811 | Cell cycle | CDK8 | 28.32 | 6.53 × 10−9 |
| M26167 | CXC chemokine | PF4V1 | 27.35 | 4.68 × 10−11 |
| U44111 | Metabolism | HNMT | 27.24 | 2.07 × 10−11 |
| X59871 | Transcription | TCF7 | 26.79 | 7.16 × 10−10 |
| X67235 | Transcription | HHEX | 25.21 | 2.07 × 10−10 |
| U19713 | Calcium signaling | AIF1 | 25.21 | 2.57 × 10−10 |
| Y08136 | Apoptosis | ASM3A4-151 | 24.74 | 3.30 × 10−8 |
| M97676 | Transcription | MSX1 | 24.58 | 9.80 × 10−5 |
| M64590 | Housekeeping | GLDC | 24.27 | 4.10 × 10−8 |
| M20203 | Protease | ELA2 | 24.03 | 6.36 × 10−12 |
| M30257 | Adhesion | VCAM1 | 23.42 | 1.71 × 10−10 |
| M93221 | Mediates endocytosis | MRC1 | 23.30 | 1.15 × 10−7 |
| S75256 | Lipoprotein signaling | LCN2 | 23.30 | 4.17 × 10−7 |
| U97188 | RNA binding | KOC14-151 | 22.47 | 5.86 × 10−9 |
| Z23091 | Adhesion | GP5 | 22.47 | 7.58 × 10−7 |
| M34344 | Adhesion | ITGA2B | 21.99 | 8.00 × 10−8 |
| M25897 | CXC chemokine | PF4 | 21.89 | 1.12 × 10−8 |
| M31994 | Housekeeping | ALDH1A1 | 21.36 | 4.86 × 10−9 |
| Z31690 | Lipoprotein signaling | LIPA | 20.67 | 1.50 × 10−9 |
| S80267 | Signaling | SYK | 20.42 | 5.90 × 10−5 |
| U00921 | Signaling | LY117 | 18.83 | 1.57 × 10−8 |
| Accession4-150 . | Function . | Gene symbol4-151 . | χ2 . | WRS‡P value . |
|---|---|---|---|---|
| L36033 | CXC chemokine | SDF1 | 48.45 | 3.05 × 10−12 |
| M63928 | Signaling | TNFRSF7 | 48.45 | 6.35 × 10−11 |
| U64998 | Ribonuclease | RNASE6 | 46.44 | 2.82 × 10−9 |
| M20902 | Lipoprotein signaling | APOC1 | 45.62 | 4.63 × 10−10 |
| M26602 | Immunity | DEFA1 | 40.75 | 1.06 × 10−12 |
| M21119 | Immunity | LYZ | 40.73 | 6.24 × 10−10 |
| M14636 | Metabolism | PYGL | 39.84 | 1.15 × 10−10 |
| M26311 | Calcium signaling | S100A9 | 38.96 | 3.60 × 10−13 |
| M54992 | Signaling | CD72 | 36.14 | 2.40 × 10−9 |
| X16832 | Protein degradation | CTSH | 35.26 | 1.81 × 10−12 |
| M12529 | Lipoprotein signaling | APOE | 34.50 | 3.95 × 10−14 |
| M15395 | Adhesion | ITGB2 | 34.02 | 1.74 × 10−13 |
| Z74616 | Extracellular matrix | COL1A2 | 34.02 | 8.06 × 10−11 |
| HT2152 | Receptor signaling | CD163 | 33.01 | 1.66 × 10−12 |
| U97105 | Pyrimidine metabolism | DPYSL2 | 32.52 | 2.22 × 10−10 |
| U81787 | Signaling | WNT10B | 32.50 | 1.77 × 10−5 |
| HT3165 | Receptor tyrosine kinase | AXL | 31.36 | 5.26 × 10−11 |
| M83667 | Transcription | CEBPD | 31.19 | 4.69 × 10−10 |
| L33930 | Receptor signaling | CD24 | 30.33 | 1.56 × 10−14 |
| D83657 | Calcium signaling | S100A12 | 29.91 | 6.58 × 10−8 |
| M11313 | Proteinase inhibitor | A2M | 29.91 | 1.07 × 10−10 |
| M31158 | Signaling | PRKAR2B | 29.91 | 2.20 × 10−9 |
| U24577 | Lipoprotein signaling | PLA2G7 | 29.78 | 2.08 × 10−10 |
| M16279 | Adhesion | MIC2 | 28.75 | 8.01 × 10−11 |
| HT2811 | Cell cycle | CDK8 | 28.32 | 6.53 × 10−9 |
| M26167 | CXC chemokine | PF4V1 | 27.35 | 4.68 × 10−11 |
| U44111 | Metabolism | HNMT | 27.24 | 2.07 × 10−11 |
| X59871 | Transcription | TCF7 | 26.79 | 7.16 × 10−10 |
| X67235 | Transcription | HHEX | 25.21 | 2.07 × 10−10 |
| U19713 | Calcium signaling | AIF1 | 25.21 | 2.57 × 10−10 |
| Y08136 | Apoptosis | ASM3A4-151 | 24.74 | 3.30 × 10−8 |
| M97676 | Transcription | MSX1 | 24.58 | 9.80 × 10−5 |
| M64590 | Housekeeping | GLDC | 24.27 | 4.10 × 10−8 |
| M20203 | Protease | ELA2 | 24.03 | 6.36 × 10−12 |
| M30257 | Adhesion | VCAM1 | 23.42 | 1.71 × 10−10 |
| M93221 | Mediates endocytosis | MRC1 | 23.30 | 1.15 × 10−7 |
| S75256 | Lipoprotein signaling | LCN2 | 23.30 | 4.17 × 10−7 |
| U97188 | RNA binding | KOC14-151 | 22.47 | 5.86 × 10−9 |
| Z23091 | Adhesion | GP5 | 22.47 | 7.58 × 10−7 |
| M34344 | Adhesion | ITGA2B | 21.99 | 8.00 × 10−8 |
| M25897 | CXC chemokine | PF4 | 21.89 | 1.12 × 10−8 |
| M31994 | Housekeeping | ALDH1A1 | 21.36 | 4.86 × 10−9 |
| Z31690 | Lipoprotein signaling | LIPA | 20.67 | 1.50 × 10−9 |
| S80267 | Signaling | SYK | 20.42 | 5.90 × 10−5 |
| U00921 | Signaling | LY117 | 18.83 | 1.57 × 10−8 |
Accession numbers listed are GenBank numbers, except those beginning with “HT,” which are provided by the Institute of Genomic Research (TIGR).
Gene symbol not HUGO approved.
Wilcoxon rank sum test.
Correlation analysis showed that 70 genes were either turned on or up-regulated in MM (Table5). When considering the χ2 test of whether expression is present or absent, the cyclin-dependent inhibitor, CDKN1A, was the most significantly differentially expressed gene (χ2 = 53.33, WRS = 3.65 × 10−11). When considering a quantitative change using the WRS test, the tyrosine kinase oncogene ABL1 was the most significant (χ2 = 43.10, WRS = 3.96 × 10−14). Other oncogenes in the list included USF2, USP4, MLLT3, andMYC. The largest class of genes represented those whose products are involved in protein metabolism (12 genes), including amino acid synthesis, translation initiation, protein folding, glycosylation, trafficking, and protein degradation. Other multiple-member classes included transcription (11 genes), signaling (9 genes), DNA synthesis and modification (6 genes), and histone synthesis and modification (5 genes). Members of the signaling group included genes whose overexpression has been linked to growth arrest, QSCN6 andPHB, as well as phosphatases, PTPRK andPPP2R4, and the kinase MAPKAPK3. The only secreted growth factor in the signaling class was HGF, a factor known to play a role in MM biology.21 TheMOX2 gene, whose product is normally expressed as an integral membrane protein on activated T cells and CD19+ B cells and is involved in inhibiting macrophage activation, was in the signaling class. The tumor suppressor gene and negative regulator of β-catenin signaling, APC, was another member of the signaling class. Classes containing 2 members included RNA binding, mitochondrial respiration, cytoskeletal matrix, metabolism, cell cycle, and adhesion. Single member classes included complement cascade (MASP1), drug resistance (MVP), glycosaminoglycan catabolism, heparin sulfate synthesis (EXTL2), and vesicular transport (TSC1). Four genes of unknown function were also identified as significantly up-regulated in MM.
The 70 most significantly up-regulated genes in MM in comparison with normal bone marrow PCs
| Accession5-150 . | Function . | Gene symbol5-151 . | χ2 . | WRS5-152P . |
|---|---|---|---|---|
| U09579 | Cell cycle | CDKN1A | 53.33 | 3.65 × 10−11 |
| U78525 | Amino acid synthesis | EIF3S9 | 49.99 | 2.25 × 10−12 |
| HT5158 | DNA synthesis | GMPS | 47.11 | 4.30 × 10−12 |
| X57129 | Histone | H1F2 | 46.59 | 5.78 × 10−13 |
| M55210 | Adhesion | LAMC1 | 45.63 | 1.34 × 10−9 |
| L77886 | Signaling, phosphatase | PTPRK | 45.62 | 5.42 × 10−10 |
| U73167 | Glycosaminoglycan catabolism | HYAL3 | 44.78 | 1.07 × 10−10 |
| X16416 | Oncogene, kinase | ABL1 | 43.10 | 3.96 × 10−14 |
| U57316 | Transcription | GCN5L2 | 43.04 | 1.36 × 10−12 |
| Y09022 | Protein glycosylation | NOT56L5-151 | 42.05 | 5.53 × 10−10 |
| M25077 | RNA binding | SSA2 | 41.26 | 1.69 × 10−7 |
| AC002115 | Mitochondrial respiration | COX6B | 41.16 | 2.16 × 10−8 |
| Y07707 | Transcription | NRF5-151 | 37.59 | 4.79 × 10−10 |
| L22005 | Protein ubiquination | CDC34 | 34.50 | 2.89 × 10−6 |
| X66899 | Transcription | EWSR1 | 34.39 | 4.23 × 10−8 |
| D50912 | RNA binding | RBM10 | 33.93 | 2.61 × 10−6 |
| HT4824 | Amino acid synthesis | CBS | 33.77 | 1.49 × 10−6 |
| U10324 | Transcription | ILF3 | 33.33 | 3.66 × 10−11 |
| AD000684 | Oncogene, transcription | USF2 | 32.18 | 7.41 × 10−11 |
| U68723 | Cell cycle | CHES1 | 31.68 | 1.03 × 10−6 |
| X16323 | Signaling, growth factor | HGF | 30.67 | 4.82 × 10−9 |
| U24183 | Metabolism | PFKM | 30.47 | 8.92 × 10−10 |
| D13645 | Unknown | KIAA00205-151 | 30.47 | 7.40 × 10−6 |
| S85655 | Signaling, growth arrest | PHB | 29.37 | 1.32 × 10−8 |
| X73478 | Signaling, phosphatase | PPP2R4 | 28.32 | 6.92 × 10−9 |
| L77701 | Mitochondrial respiration | COX17 | 27.81 | 1.33 × 10−6 |
| U20657 | Oncogene, proteasome | USP4 | 27.71 | 2.31 × 10−6 |
| M59916 | Signaling, DAG signaling | SMPD1 | 27.49 | 3.52 × 10−8 |
| D16688 | Oncogene, DNA binding | MLLT3 | 27.24 | 6.97 × 10−13 |
| X90392 | DNA endonuclease | DNASE1L1 | 26.98 | 4.72 × 10−7 |
| U07424 | Amino acid synthesis | FARSL | 26.93 | 1.66 × 10−6 |
| X54199 | DNA synthesis | GART | 26.57 | 9.61 × 10−11 |
| L06175 | Unknown | P5-15-151 | 26.57 | 5.16 × 10−7 |
| M55267 | Unknown | EVI2A | 25.92 | 3.79 × 10−6 |
| M87507 | Protein degradation | CASP1 | 25.78 | 5.46 × 10−7 |
| M90356 | Transcription | BTF3L2 | 25.78 | 9.68 × 10−8 |
| U35637 | Cytoskeletal matrix | NEB | 25.40 | 9.15 × 10−6 |
| L06845 | Amino acid synthesis | CARS | 25.34 | 5.39 × 10−8 |
| U81001 | DNA, nuclear matrix attachment | SNURF | 24.58 | 4.54 × 10−5 |
| U76189 | Heparan sulfate synthesis | EXTL2 | 24.58 | 7.28 × 10−6 |
| U53225 | Protein trafficking | SNX1 | 24.48 | 5.53 × 10−7 |
| X04366 | Protein degradation | CAPN1 | 24.35 | 1.26 × 10−9 |
| U77456 | Protein folding | NAP1L4 | 24.27 | 4.23 × 10−10 |
| L42379 | Signaling, growth arrest | QSCN6 | 24.27 | 1.28 × 10−10 |
| U09578 | Signaling, kinase | MAPKAPK3 | 24.27 | 2.35 × 10−9 |
| Z80780 | Histone | H2BFH | 24.27 | 3.44 × 10−12 |
| HT4899 | Oncogene, transcription | MYC | 24.27 | 1.77 × 10−5 |
| M74088 | Signaling, b-catenin regulator | APC | 23.94 | 1.50 × 10−5 |
| X57985 | Histone | H2BFQ | 23.90 | 3.25 × 10−12 |
| X79882 | Drug resistance | MVP | 23.47 | 1.77 × 10−11 |
| X77383 | Protein degradation | CTSO | 23.18 | 4.68 × 10−7 |
| M91592 | Transcription | ZNF76 | 23.16 | 1.12 × 10−8 |
| X63692 | DNA methyltransferase | DNMT1 | 23.12 | 5.15 × 10−11 |
| M60752 | Histone | H2AFO | 21.60 | 1.46 × 10−8 |
| M96684 | Transcription | PURA | 21.25 | 4.54 × 10−5 |
| U16660 | Metabolism | ECH1 | 21.18 | 5.52 × 10−5 |
| M86737 | DNA repair | SSRP1 | 20.60 | 2.62 × 10−5 |
| U35113 | Histone deacetylase | MTA1 | 20.60 | 6.67 × 10−10 |
| X81788 | Unknown | ICT1 | 20.42 | 2.97 × 10−7 |
| HT2217 | Signaling | MUC2A | 20.33 | 2.61 × 10−7 |
| M62324 | Unknown | MRF-15-151 | 20.31 | 3.98 × 10−9 |
| U09367 | Transcription | ZNF136 | 20.30 | 7.72 × 10−9 |
| X89985 | Cytoskeletal matrix | BCL7B | 19.81 | 5.50 × 10−9 |
| L19871 | Transcription repression | ATF3 | 19.43 | 1.13 × 10−6 |
| X69398 | Adhesion | CD47 | 19.16 | 6.44 × 10−7 |
| X05323 | Signaling, macrophage inhibitor | MOX2 | 19.16 | 8.58 × 10−6 |
| X04741 | Protein ubiquination | UCHL1 | 19.14 | 9.76 × 10−5 |
| D87683 | Vesicular transport | TSC1 | 19.12 | 6.81 × 10−6 |
| D17525 | Complement cascade | MASP1 | 18.81 | 4.05 × 10−7 |
| Accession5-150 . | Function . | Gene symbol5-151 . | χ2 . | WRS5-152P . |
|---|---|---|---|---|
| U09579 | Cell cycle | CDKN1A | 53.33 | 3.65 × 10−11 |
| U78525 | Amino acid synthesis | EIF3S9 | 49.99 | 2.25 × 10−12 |
| HT5158 | DNA synthesis | GMPS | 47.11 | 4.30 × 10−12 |
| X57129 | Histone | H1F2 | 46.59 | 5.78 × 10−13 |
| M55210 | Adhesion | LAMC1 | 45.63 | 1.34 × 10−9 |
| L77886 | Signaling, phosphatase | PTPRK | 45.62 | 5.42 × 10−10 |
| U73167 | Glycosaminoglycan catabolism | HYAL3 | 44.78 | 1.07 × 10−10 |
| X16416 | Oncogene, kinase | ABL1 | 43.10 | 3.96 × 10−14 |
| U57316 | Transcription | GCN5L2 | 43.04 | 1.36 × 10−12 |
| Y09022 | Protein glycosylation | NOT56L5-151 | 42.05 | 5.53 × 10−10 |
| M25077 | RNA binding | SSA2 | 41.26 | 1.69 × 10−7 |
| AC002115 | Mitochondrial respiration | COX6B | 41.16 | 2.16 × 10−8 |
| Y07707 | Transcription | NRF5-151 | 37.59 | 4.79 × 10−10 |
| L22005 | Protein ubiquination | CDC34 | 34.50 | 2.89 × 10−6 |
| X66899 | Transcription | EWSR1 | 34.39 | 4.23 × 10−8 |
| D50912 | RNA binding | RBM10 | 33.93 | 2.61 × 10−6 |
| HT4824 | Amino acid synthesis | CBS | 33.77 | 1.49 × 10−6 |
| U10324 | Transcription | ILF3 | 33.33 | 3.66 × 10−11 |
| AD000684 | Oncogene, transcription | USF2 | 32.18 | 7.41 × 10−11 |
| U68723 | Cell cycle | CHES1 | 31.68 | 1.03 × 10−6 |
| X16323 | Signaling, growth factor | HGF | 30.67 | 4.82 × 10−9 |
| U24183 | Metabolism | PFKM | 30.47 | 8.92 × 10−10 |
| D13645 | Unknown | KIAA00205-151 | 30.47 | 7.40 × 10−6 |
| S85655 | Signaling, growth arrest | PHB | 29.37 | 1.32 × 10−8 |
| X73478 | Signaling, phosphatase | PPP2R4 | 28.32 | 6.92 × 10−9 |
| L77701 | Mitochondrial respiration | COX17 | 27.81 | 1.33 × 10−6 |
| U20657 | Oncogene, proteasome | USP4 | 27.71 | 2.31 × 10−6 |
| M59916 | Signaling, DAG signaling | SMPD1 | 27.49 | 3.52 × 10−8 |
| D16688 | Oncogene, DNA binding | MLLT3 | 27.24 | 6.97 × 10−13 |
| X90392 | DNA endonuclease | DNASE1L1 | 26.98 | 4.72 × 10−7 |
| U07424 | Amino acid synthesis | FARSL | 26.93 | 1.66 × 10−6 |
| X54199 | DNA synthesis | GART | 26.57 | 9.61 × 10−11 |
| L06175 | Unknown | P5-15-151 | 26.57 | 5.16 × 10−7 |
| M55267 | Unknown | EVI2A | 25.92 | 3.79 × 10−6 |
| M87507 | Protein degradation | CASP1 | 25.78 | 5.46 × 10−7 |
| M90356 | Transcription | BTF3L2 | 25.78 | 9.68 × 10−8 |
| U35637 | Cytoskeletal matrix | NEB | 25.40 | 9.15 × 10−6 |
| L06845 | Amino acid synthesis | CARS | 25.34 | 5.39 × 10−8 |
| U81001 | DNA, nuclear matrix attachment | SNURF | 24.58 | 4.54 × 10−5 |
| U76189 | Heparan sulfate synthesis | EXTL2 | 24.58 | 7.28 × 10−6 |
| U53225 | Protein trafficking | SNX1 | 24.48 | 5.53 × 10−7 |
| X04366 | Protein degradation | CAPN1 | 24.35 | 1.26 × 10−9 |
| U77456 | Protein folding | NAP1L4 | 24.27 | 4.23 × 10−10 |
| L42379 | Signaling, growth arrest | QSCN6 | 24.27 | 1.28 × 10−10 |
| U09578 | Signaling, kinase | MAPKAPK3 | 24.27 | 2.35 × 10−9 |
| Z80780 | Histone | H2BFH | 24.27 | 3.44 × 10−12 |
| HT4899 | Oncogene, transcription | MYC | 24.27 | 1.77 × 10−5 |
| M74088 | Signaling, b-catenin regulator | APC | 23.94 | 1.50 × 10−5 |
| X57985 | Histone | H2BFQ | 23.90 | 3.25 × 10−12 |
| X79882 | Drug resistance | MVP | 23.47 | 1.77 × 10−11 |
| X77383 | Protein degradation | CTSO | 23.18 | 4.68 × 10−7 |
| M91592 | Transcription | ZNF76 | 23.16 | 1.12 × 10−8 |
| X63692 | DNA methyltransferase | DNMT1 | 23.12 | 5.15 × 10−11 |
| M60752 | Histone | H2AFO | 21.60 | 1.46 × 10−8 |
| M96684 | Transcription | PURA | 21.25 | 4.54 × 10−5 |
| U16660 | Metabolism | ECH1 | 21.18 | 5.52 × 10−5 |
| M86737 | DNA repair | SSRP1 | 20.60 | 2.62 × 10−5 |
| U35113 | Histone deacetylase | MTA1 | 20.60 | 6.67 × 10−10 |
| X81788 | Unknown | ICT1 | 20.42 | 2.97 × 10−7 |
| HT2217 | Signaling | MUC2A | 20.33 | 2.61 × 10−7 |
| M62324 | Unknown | MRF-15-151 | 20.31 | 3.98 × 10−9 |
| U09367 | Transcription | ZNF136 | 20.30 | 7.72 × 10−9 |
| X89985 | Cytoskeletal matrix | BCL7B | 19.81 | 5.50 × 10−9 |
| L19871 | Transcription repression | ATF3 | 19.43 | 1.13 × 10−6 |
| X69398 | Adhesion | CD47 | 19.16 | 6.44 × 10−7 |
| X05323 | Signaling, macrophage inhibitor | MOX2 | 19.16 | 8.58 × 10−6 |
| X04741 | Protein ubiquination | UCHL1 | 19.14 | 9.76 × 10−5 |
| D87683 | Vesicular transport | TSC1 | 19.12 | 6.81 × 10−6 |
| D17525 | Complement cascade | MASP1 | 18.81 | 4.05 × 10−7 |
Accession numbers listed are GenBank numbers, except those beginning with “HT,” which are provided by the Institute of Genomic Research (TIGR).
Gene symbol not HUGO approved.
Wilcoxon rank sum test.
Gene expression “spikes” in subsets of MM
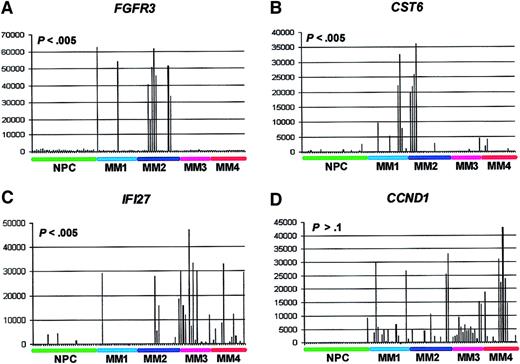
A total of 156 genes not identified as differently expressed in the statistical analysis of MM versus normal PCs, yet highly overexpressed in subsets of MM, were also identified. A total of 25 genes with an AD spike more than 10 000 in at least one sample are shown (Table 6). With 27 spikes, the adhesion-associated gene FBLN2 was the most frequently spiked. The gene for the interferon induced protein 27,IFI27, with 25 spikes was the second most frequently spiked gene and contained the highest number of spikes over 10 000 (n = 14). The FGFR3 gene was spiked in 9 of the 74 cases (Figure 2A). It was the only gene for which all spikes were more than 10 000 AD. In fact, the lowest AD value was 18 961 and the highest 62 515, which represented the highest of all spikes. The finding of FGFR3 spikes suggested that these spikes were induced by the MM-specific,FGFR3-activating t(4;14)(p21;q32) translocation.22 To test this hypothesis, we performed RT-PCR for a t(4;14)(p21;q32) translocation-specific fusion transcript between the IGH locus and the gene MMSET (data not shown). The translocation-specific transcript was present in all 9FGFR3 spike samples but was absent in 5 samples that did not express FGFR3. These data suggested that the spike was caused by the t(4;14)(p21;q32) translocation. The CCND1 gene was spiked with AD values of more than 10 000 in 13 cases. We performed TRI-FISH analysis for the t(11;14)(q13;q32) translocation (Table 7). All 11 evaluable samples were positive for the t(11;14)(q13;q32) translocation by TRI-FISH; 2 samples were not analyzable due to loss of cell integrity during storage. Thus, all FGFR3 and CCND1 spikes could be accounted for by the presence of either the t(4;14)(p21;q32) translocation or the t(11;14)(q13;q32) translocation, respectively.
Genes with “spiked” expression in subsets of MM PCs from newly diagnosed patients
| Accession6-150 . | Function . | Gene symbol6-151 . | No. of spikes . | Spikes greater than 10K . | Maximum spike . |
|---|---|---|---|---|---|
| M64347 | Signaling | FGFR3 | 9 | 9 | 62 515 |
| U89922 | Immunity | LTB | 4 | 2 | 49 261 |
| X67325 | Interferon signaling | IFI27 | 25 | 14 | 47 072 |
| X59798 | Cell cycle | CCND1 | 6 | 13 | 42 814 |
| U62800 | Cysteine protease inhibitor | CST6 | 17 | 6 | 36 081 |
| U35340 | Eye lens protein | CRYBB1 | 4 | 1 | 35 713 |
| X12530 | B-cell signaling | MS4A2 | 5 | 5 | 34 487 |
| X59766 | Unknown | AZGP1 | 18 | 4 | 28 523 |
| U58096 | Unknown | TSPY | 4 | 1 | 23 325 |
| U52513 | Interferon signaling | IFIT4 | 5 | 2 | 21 078 |
| X76223 | Vesicular trafficking | MAL | 19 | 5 | 20 432 |
| X92689 | O-linked glycosylation | GALNT3 | 4 | 1 | 18 344 |
| D17427 | Adhesion | DSC3 | 8 | 7 | 17 616 |
| L11329 | Signaling | DUSP2 | 14 | 1 | 15 962 |
| L13210 | Adhesion, macrophage lectin | LGALS3BP | 8 | 2 | 14 876 |
| U10991 | Unknown | G26-151 | 7 | 1 | 14 815 |
| L10373 | Integral membrane protein | TM4SF2 | 4 | 2 | 14 506 |
| U60873 | Unknown | 137308 | 12 | 1 | 12 751 |
| M65292 | Complement regulation | HFL1 | 9 | 1 | 12 718 |
| HT4215 | Phospholipid transport | PLTP | 23 | 1 | 12 031 |
| D13168 | Growth factor receptor | ENDRB | 18 | 1 | 11 707 |
| AC002077 | Signaling | GNAT1 | 21 | 1 | 11 469 |
| M92934 | Growth factor | CTGF | 4 | 1 | 11 201 |
| X82494 | Adhesion | FBLN2 | 27 | 7 | 10 648 |
| M30703 | Growth factor | AR | 5 | 1 | 10 163 |
| Accession6-150 . | Function . | Gene symbol6-151 . | No. of spikes . | Spikes greater than 10K . | Maximum spike . |
|---|---|---|---|---|---|
| M64347 | Signaling | FGFR3 | 9 | 9 | 62 515 |
| U89922 | Immunity | LTB | 4 | 2 | 49 261 |
| X67325 | Interferon signaling | IFI27 | 25 | 14 | 47 072 |
| X59798 | Cell cycle | CCND1 | 6 | 13 | 42 814 |
| U62800 | Cysteine protease inhibitor | CST6 | 17 | 6 | 36 081 |
| U35340 | Eye lens protein | CRYBB1 | 4 | 1 | 35 713 |
| X12530 | B-cell signaling | MS4A2 | 5 | 5 | 34 487 |
| X59766 | Unknown | AZGP1 | 18 | 4 | 28 523 |
| U58096 | Unknown | TSPY | 4 | 1 | 23 325 |
| U52513 | Interferon signaling | IFIT4 | 5 | 2 | 21 078 |
| X76223 | Vesicular trafficking | MAL | 19 | 5 | 20 432 |
| X92689 | O-linked glycosylation | GALNT3 | 4 | 1 | 18 344 |
| D17427 | Adhesion | DSC3 | 8 | 7 | 17 616 |
| L11329 | Signaling | DUSP2 | 14 | 1 | 15 962 |
| L13210 | Adhesion, macrophage lectin | LGALS3BP | 8 | 2 | 14 876 |
| U10991 | Unknown | G26-151 | 7 | 1 | 14 815 |
| L10373 | Integral membrane protein | TM4SF2 | 4 | 2 | 14 506 |
| U60873 | Unknown | 137308 | 12 | 1 | 12 751 |
| M65292 | Complement regulation | HFL1 | 9 | 1 | 12 718 |
| HT4215 | Phospholipid transport | PLTP | 23 | 1 | 12 031 |
| D13168 | Growth factor receptor | ENDRB | 18 | 1 | 11 707 |
| AC002077 | Signaling | GNAT1 | 21 | 1 | 11 469 |
| M92934 | Growth factor | CTGF | 4 | 1 | 11 201 |
| X82494 | Adhesion | FBLN2 | 27 | 7 | 10 648 |
| M30703 | Growth factor | AR | 5 | 1 | 10 163 |
Accession numbers listed are GenBank numbers, except those beginning with “HT,” which are provided by the Institute of Genomic Research (TIGR).
Gene symbol not HUGO approved.
Spike profile distributions among the gene expression–defined MM subgroups.
GeneChip HuGeneFL analysis of FGFR3, CST6,IFI27, and CCND1 gene expression. The normalized AD value of fluorescence intensity of streptavidin-PE–stained biotinylated cRNA as hybridized to probe sets is on the vertical axis and samples are on the horizontal axis. The samples are ordered from left to right: normal PCs (NPCs; green), MM1 (light blue), MM2 (dark blue), MM3 (violet), and MM4 (red). Note relatively low expression in 31 NPCs and spiked expression in subsets of MM samples. TheP values of the test for significant nonrandom spike distributions are noted.
Spike profile distributions among the gene expression–defined MM subgroups.
GeneChip HuGeneFL analysis of FGFR3, CST6,IFI27, and CCND1 gene expression. The normalized AD value of fluorescence intensity of streptavidin-PE–stained biotinylated cRNA as hybridized to probe sets is on the vertical axis and samples are on the horizontal axis. The samples are ordered from left to right: normal PCs (NPCs; green), MM1 (light blue), MM2 (dark blue), MM3 (violet), and MM4 (red). Note relatively low expression in 31 NPCs and spiked expression in subsets of MM samples. TheP values of the test for significant nonrandom spike distributions are noted.
Correlation of CCND1 spikes with FISH-defined t(11;14)(q13;q32)
| GC PT . | CCND1 spike (AD value) . | FISH t(11;14) . | % PCs with translocation . | Cells counted . |
|---|---|---|---|---|
| P168 | 42 813 | Yes | 59 | 113 |
| P251 | 33 042 | Yes | 80 | 124 |
| P91 | 31 030 | Not done | — | — |
| P99 | 29 862 | Yes | 65 | 111 |
| P85 | 26 737 | Yes | 92 | 124 |
| P241 | 25 611 | Yes | 96 | 114 |
| P56 | 23 654 | Yes | 100 | 106 |
| P63 | 22 358 | Yes | 98 | 104 |
| P199 | 18 761 | Yes | 60 | 35 |
| P107 | 15 205 | Yes | 100 | 147 |
| P75 | 14 642 | Yes | 100 | 105 |
| P187 | 14 295 | Yes | 25 | 133 |
| P124 | 10 594 | Not done | — | — |
| GC PT . | CCND1 spike (AD value) . | FISH t(11;14) . | % PCs with translocation . | Cells counted . |
|---|---|---|---|---|
| P168 | 42 813 | Yes | 59 | 113 |
| P251 | 33 042 | Yes | 80 | 124 |
| P91 | 31 030 | Not done | — | — |
| P99 | 29 862 | Yes | 65 | 111 |
| P85 | 26 737 | Yes | 92 | 124 |
| P241 | 25 611 | Yes | 96 | 114 |
| P56 | 23 654 | Yes | 100 | 106 |
| P63 | 22 358 | Yes | 98 | 104 |
| P199 | 18 761 | Yes | 60 | 35 |
| P107 | 15 205 | Yes | 100 | 147 |
| P75 | 14 642 | Yes | 100 | 105 |
| P187 | 14 295 | Yes | 25 | 133 |
| P124 | 10 594 | Not done | — | — |
GC PT indicates patient identifier; AD, average difference call.
We next determined the distribution of the FGFR3,CST6, IFI27, and CCND1 spikes within the gene expression–defined MM subgroups (Figure 2). The data showed that whereas FGFR3 and CST6 spikes were more likely to be found in MM1 or MM2 (P < .005), the spikes for IFI27 were associated with an MM3 and MM4 distribution (P < .005). CCND1 spikes were not associated with any specific subgroup (P > .1). It is noteworthy that both CST6 and CCND1 map to 11q13 and had no overlap in spikes. We are currently testing whether CST6overexpression is due to a variant t(11;14)(q13;q32) translocation. The 5 spikes for MS4A2 (CD20) were found in either the MM1 (3 spikes) or MM2 (2 spikes) subgroups (data not shown).
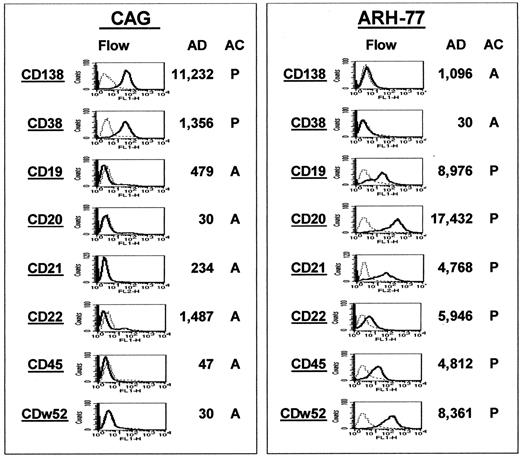
The gene MS4A2, which codes for the CD20 molecule, was also found as a spiked gene in 4 cases (Figure3A). To investigate whether spiked gene expression correlated with protein expression, we performed immunohistochemistry for CD20 on biopsies from 15 of the 74 MM samples (Figure 3B). All 4 cases that had spiked MS4A2 gene expression were also positive for CD20 protein expression, whereas 11 that had no MS4A2 gene expression were also negative for CD20 by immunohistochemistry. To add additional validation to the gene expression profiling, we performed a comparison of CD marker protein and gene expression in the MM cell line CAG and the EBV-transformed lymphoblastoid cell line ARH-77 (Figure4). The expression of CD138 and CD38 protein and gene expression was high in CAG but absent in ARH-77 cells. On the other hand, expression of CD19, CD20, CD21, CD22, CD45, and CDw52 was found to be strong in ARH-77 and absent in CAG cells. The nearly 100% coincidence of FGFR3 or CCND1 spiked gene expression with the presence of the t(4;14)(p14;q32) or t(11;14)(q13;q32) translocation, the strong correlation of CD20 andMS42A gene expression in primary MM, and CD marker protein and gene expression in B cells and PCs represent important validations of the accuracy of our gene expression profiling.
Spiked gene expression corresponds to protein expression in MM.
(A) GeneChip HuGeneFL analysis of MS4A2 (CD20)gene expression. The normalized AD value of fluorescence intensities of streptavidin-PE–stained biotinylated cRNA as hybridized to 2 independent probe sets (accession numbers M27394 [blue] andX12530 [red]) located in different regions of the MS4A2gene is on the vertical axis and samples are on the horizontal axis. Note relatively low expression in 31 normal PCs (NPCs) and spiked expression in 5 of 74 MM samples (MM PCs) Also note similarity in expression levels detected by the 2 different probe sets. (B) Immunohistochemistry for CD20 expression on clonal MM PCs: (i) bone marrow biopsy section showing asynchronous type MM cells (hematoxylin and eosin; original magnification × 500); (ii) CD20+ MM cells (original magnification × 100; inset, original magnification × 500); (iii) biopsy from a patient with mixed asynchronous and Marschalko-type MM cells (hematoxylin and eosin; original magnification × 500); and (iv) CD20+ single lymphocyte and CD20− MM cells (original magnification × 200). CD20 immunohistochemistry was examined without knowledge of clinical history or gene expression findings.
Spiked gene expression corresponds to protein expression in MM.
(A) GeneChip HuGeneFL analysis of MS4A2 (CD20)gene expression. The normalized AD value of fluorescence intensities of streptavidin-PE–stained biotinylated cRNA as hybridized to 2 independent probe sets (accession numbers M27394 [blue] andX12530 [red]) located in different regions of the MS4A2gene is on the vertical axis and samples are on the horizontal axis. Note relatively low expression in 31 normal PCs (NPCs) and spiked expression in 5 of 74 MM samples (MM PCs) Also note similarity in expression levels detected by the 2 different probe sets. (B) Immunohistochemistry for CD20 expression on clonal MM PCs: (i) bone marrow biopsy section showing asynchronous type MM cells (hematoxylin and eosin; original magnification × 500); (ii) CD20+ MM cells (original magnification × 100; inset, original magnification × 500); (iii) biopsy from a patient with mixed asynchronous and Marschalko-type MM cells (hematoxylin and eosin; original magnification × 500); and (iv) CD20+ single lymphocyte and CD20− MM cells (original magnification × 200). CD20 immunohistochemistry was examined without knowledge of clinical history or gene expression findings.
Gene expression correlates with protein expression.
Gene and protein expression of CD markers known to be differentially expressed during B-cell differentiation were compared between the MM cell line CAG (left panel) and the EBV-transformed B-lymphoblastoid line ARH-77 (right panel). In both panels, the 8 CD markers are listed in the left column of each panel. Flow cytometric analysis of protein expression is presented in the second column; the AD and AC values of gene expression, in the third and fourth columns. Note the strong expression of both the gene and protein for CD138 and CD38 in the CAG cells but the low expression in the ARH-77 cells. The opposite correlation is observed for the remaining markers.
Gene expression correlates with protein expression.
Gene and protein expression of CD markers known to be differentially expressed during B-cell differentiation were compared between the MM cell line CAG (left panel) and the EBV-transformed B-lymphoblastoid line ARH-77 (right panel). In both panels, the 8 CD markers are listed in the left column of each panel. Flow cytometric analysis of protein expression is presented in the second column; the AD and AC values of gene expression, in the third and fourth columns. Note the strong expression of both the gene and protein for CD138 and CD38 in the CAG cells but the low expression in the ARH-77 cells. The opposite correlation is observed for the remaining markers.
Discussion
In this report, we have shown that both normal and malignant PCs can be purified to homogeneity from bone marrow aspirates using anti-CD138–based immunomagnetic bead-positive selection. Using these cells we have provided the first comprehensive global gene expression profiling of newly diagnosed MM patients and contrasted these expression patterns with those of normal PCs.
Hierarchical cluster analysis of MM and normal PCs, as well as the benign PC dyscrasia MGUS and the end-stage–like MM cell lines, revealed that normal PCs are unique and that primary MM is either like MGUS or MM cell lines. In addition, MM cell line gene expression was homogeneous as evidenced by the tight clustering in the hierarchical analysis. The similarity of MM cell line expression patterns to primary newly diagnosed forms of MM support the validity of using MM cell lines as models for MM, in particular for our gene expression–defined MM4 subgroup.
On hierarchical clustering of MM alone, 4 MM subgroups were distinguished. Differences indicate that gene expression signatures distinguish distinct clinical entities as (1) the MM1 subgroup contained samples that were more like MGUS (in our first cluster analysis), whereas the MM4 subgroup contained samples more like MM cell lines; (2) the most significant gene expression patterns differentiating MM1 and MM4 were cell cycle control and DNA metabolism genes; and (3) the MM4 subgroup was more likely to have abnormal cytogenetics, elevated serum β2M, elevated creatinine, and deletions of chromosome 13—important variables that historically have been linked to poor prognosis.
We speculate that the MM4 subgroup thus likely represents the most high-risk clinical entity. Thus, knowledge of the molecular genetics of this particular subgroup should provide insight into its biology and possibly provide a rationale for appropriate subtype-specific therapeutic interventions. On analysis, the most significant gene expression changes differentiating the MM1 and MM4 subgroups code for activities that clearly implicate MM4 as having a more proliferative and autonomous phenotype. The most significantly altered gene in the comparison, TYMS (thymidylate synthase), which functions in the pyrimidine biosynthetic pathway, has been linked to resistance to fluoropyrimidine chemotherapy and also poor prognosis in colorectal carcinomas.23 Other notable genes up-regulated in MM4 were the CAAX farnesyltransferase gene,FTNA. Farnesyltransferase prenylates RAS, a posttranslational modification required to allow RAS to attach to the plasma membrane. These data suggest that farnesyltransferase inhibitors may be effective in treating patients with high levels of FTNAexpression. Two genes coding for components of the proteasome pathway, POH1 (26S proteasome–associated pad1 homolog) andUBL1 (ubiquitin-like protein 1) were also overexpressed in MM4. Overexpression of POH1 confers P-glycoprotein–independent, pleotropic drug resistance to mammalian cells.24,25 Given the uniform development of chemotherapy resistance in MM the combined overexpression of POH1 andMVP may have profound influences on this phenotype. In contrast to normal PCs, more than 75% of MM PCs express abundant mRNA for the multidrug resistance gene, lung resistance–related protein (MVP). These data are consistent with previous reports showing that expression of MVP in MM is a poor prognostic factor.26 Ubiquitin-like protein 1 (UBL1) also known as sentrin, is involved in many processes including associating with RAD51, RAD52, and p53 proteins in the double-strand repair pathway27-29; conjugating with RANGAP1, involved in nuclear protein import; and importantly for MM, protecting against both Fas/Apo-1 (TNFRSF6) or TNFR1-induced apoptosis.30 The deregulated expression of many genes whose products function in the proteasome pathway may be used in the pharmacogenomic analysis of efficacy of proteasome inhibitors like PS-341 (Millennium Pharmaceuticals, Cambridge, MA).
Another significantly up-regulated gene in MM4 was the single-stranded DNA-dependent adenosine triphosphate (ATP)–dependent helicase (G22P1) also known as Ku70 autoantigen. The DNA helicase II complex, made up of p70 and p80, binds preferentially to forklike ends of double-stranded DNA in a cell cycle–dependent manner. Binding to DNA is thought to be mediated by p70 and dimerization with p80 forms the ATP-dependent DNA-unwinding enzyme (helicase II) and acts as the regulatory component of a DNA-dependent protein kinase (DNPK), which was also significantly up-regulated in MM4. The involvement of the helicase II complex in DNA double-strand break repair, V(D)J recombination, and notably chromosomal translocations has been proposed. Another gene up-regulated was the DNA fragmentation factor, 45 kd, alpha (DFFA). Caspase-3 cleaves the DFFA-encoded 45-kd subunit at 2 sites to generate an active factor that produces DNA fragmentation during apoptosis signaling. We speculate that, in light of the many blocks to apoptosis in MM, DFFA activation could result in DNA fragmentation, which in turn would activate the helicase II complex that then may facilitate chromosomal translocations. It is of note that abnormal karyotypes, and thus chromosomal translocations, are associated with the MM4 subgroup, which tended to overexpress these 2 genes.
A direct comparison of gene expression patterns in MM and normal PCs identified novel genes with highly significant differences that could represent the fundamental changes associated with the malignant transformation of PCs.
The progression of MM as a hypoproliferative tumor is thought to be linked to a defect in programmed cell death rather than rapid cell replication.31 Two genes, prohibitin (PHB) and quiescin Q6 (QSCN6), overexpressed in MM are involved in growth arrest. The overexpression of these genes may be responsible for the typically low proliferation indices seen in MM. It is hence conceivable that therapeutic down-regulation of these genes possibly resulting in enhanced proliferation could render MM cells more susceptible to cell cycle–active chemotherapeutic agents.
The gene coding for CD27, TNFRSF7, the second most significantly underexpressed gene in MM, is a member of the TNFR superfamily that provides costimulatory signals for T- and B-cell proliferation and B-cell immunoglobulin production and apoptosis.18 Anti-CD27 significantly inhibits the induction of Blimp-1 and J-chain transcripts, which are turned on in cells committed to PC differentiation,19 suggesting that ligation of CD27 on B cells may prevent terminal differentiation. CD27 ligand (CD70) prevents apoptosis mediated by interleukin 10 (IL-10) and directs differentiation of CD27+ memory B cells toward PCs in cooperation with IL-10.17 Thus, it is possible that the down-regulation of CD27 gene expression in MM may block an apoptotic program.
The overexpression of CD47 on MM may be related to escape of MM cells from immune surveillance. Studies have shown that cells lacking CD47 are rapidly cleared from the bloodstream by splenic red pulp macrophages and CD47 on normal red blood cells prevents this elimination.32
The gene product of DNA methyltransferase 1, DNMT1, overexpressed in MM, is responsible for cytosine methylation in mammals and has an important role in epigenetic gene silencing. In fact, aberrant hypermethylation of tumor suppressor genes plays an important role in the development of many tumors (for a review, see Baylin33). De novo methylation of p16/INK4a is a frequent finding in primary MM.34,35 Also, recent studies have shown that up-regulated expression of DNMTs may contribute to the pathogenesis of leukemia by inducing aberrant regional hypermethylation.36 DNA methylation represses genes partly by recruitment of the methyl-CpG-binding protein MeCP2, which in turn recruits a histone deacetylase activity. Fuks et al37 have shown that the process of DNA methylation, mediated by Dnmt1, may depend on or generate an altered chromatin state via histone deacetylase activity. It is potentially significant that MM cases also demonstrate significant overexpression of the gene for metastasis-associated 1 (MTA1).MTA1 was originally identified as being highly expressed in metastatic cells.38MTA1 has more recently been discovered to be one subunit of the nucleosome remodeling and histone deacetylation (NURD) complex, which contains not only ATP-dependent nucleosome disruption activity, but also histone deacetylase activity.39 Thus, overexpression of DNMT1 andMTA1 may have dramatic effects on repressing gene expression in MM.
Oncogenes activated in MM included ABL and MYC. Although it is not clear whether ABL tyrosine kinase activity is present in MM, it is important to note that overexpression ofabl and c-myc results in the accelerated development of mouse plasmacytomas.40 Thus, it may be more than a coincidence that MM cells significantly overexpressMYC and ABL. Chromosomal translocations involving the MYC oncogene and IGH and IGLgenes, resulting in dysregulated MYC expression, are hallmarks of Burkitt lymphoma41 and experimentally induced mouse plasmacytomas42; however,MYC/IGH-associated translocations are rare in MM.43 44 Although high MYC expression was a common feature in our panel of MM, it was quite variable, ranging from little or no expression to highly elevated expression. It is also of note that the MAZ gene whose product is known to bind to and activate MYC expression was significantly up-regulated in the MM4 subgroup. Given the important role of MYC in B-cell neoplasia, we speculate that overexpression of MYC, and possibly ABL, in MM may have biologic and possibly prognostic significance.
EXT1 and EXT2, which are tumor suppressor genes involved in hereditary multiple exostoses,45heterodimerize and are critical in the synthesis and display of cell surface heparan sulfate glycosaminoglycans (GAGs).46,47EXT1 is expressed in both MM and normal PCs.EXT2L was overexpressed in MM, suggesting that a functional glycosyltransferase could be created in MM. It is of note that syndecan-1 (CD138/SDC1), a transmembrane heparan sulfate proteoglycan, is abundantly expressed on MM cells and, when shed into the serum, is a negative prognostic factor.48 Thus, abnormal GAG-modified SDC1 may be important in MM biology. The link of SDC1 to MM biology is furthered by the recent association of SDC1 in the signaling cascade induced by the WNT proto-oncogene products. Alexander et al49 showed that syndecan-1 (SDC1) is required for Wnt-1–induced mammary tumorigenesis. We observed significant down-regulation of WNT10B in primary MM cases. It is also of note that the WNT5A gene and the FRZB gene, which codes for a decoy WNT receptor,50,51 were also marginally up-regulated in newly diagnosed MM (J.S., unpublished data, May 2001). Given that the WNTs represent a novel class of B-cell regulators,52 53 deregulation of the expression of these growth factors (WNT5A, WNT10B) and their receptors (eg, FRZB) and gene products that modulate receptor signaling (eg, SDC1), may be important in the genesis of MM.
In addition to identifying genes that were statistically different between the group of normal PCs and MM PCs, we also identified genes, like FGFR3 and CCND1, that demonstrate highly elevated “spiked” expression in subsets of MMs. Patients with elevated expression of these genes can have significant distribution differences among the 4 gene expression cluster subgroups. For example,FGFR3 spikes are found in MM1 and MM2, whereas spikes ofIFI27 are more likely to be found in MM3 and MM4. Highly elevated expression of the interferon-induced gene, IFI27, may be indicative of a viral infection, either systemic or specifically within the PCs from these patients, as correlation analysis has shown that IFI27 spikes are significantly linked (Pearson correlation coefficient values of .77 to .60) to elevated expression of 14 interferon-induced genes, including MX1, MX2,OAS1, OAS2, IFIT1, IFIT4,PLSCR1, and STAT1 (J.S., unpublished data, May 2001). More recent analysis of a large population of MM patients (n = 280), indicated that nearly 25% of all patients had spikes of the IFI27 gene, thus including a large percentage of patients (J.S., unpublished data, May 2001). Studies are now ongoing that are investigating (1) whether or not the patients showing the IFI27 spike who cluster in the MM4 subgroup are more likely to have a poor clinical course and (2) to identify the suspected viral infection causing the up-regulation of this class of genes. Thus, spiked gene expression may also be used in the development of clinically relevant prognostic groups.
Finally, the 100% coincidence of spiked FGFR3 orCCND1 gene expression with the presence of the t(4;14)(p14;q32) or t(11;14)(q13;q32) translocations as well as the strong correlations between protein expression and gene expression represent important validations of the accuracy of gene expression profiling and suggests that gene expression profiling may eventually supplant the labor intensive and expensive clinical laboratory procedures, such as cell surface marker immunophenotyping and molecular and cellular cytogenetics.
Because cancer is thought to arise from permanent alterations in gene expression, our comparison of global gene expression patterns in normal and malignant PCs provides a snapshot of the genetic abnormalities that create the malignant MM phenotype. Many of the genes known to be involved in myeloma genesis, for example, CCND1, FGFR3,MYC, HGF, and MVP, were identified by high-density oligonucleotide DNA microarray comparison of normal and malignant PCs. Importantly, an abundance of heretofore unrecognized classes of genes have been discovered that may be intimately involved in the malignant transformation of PCs and should provide a new framework for studying MM molecular and cellular biology. Similar to investigations in leukemia54 and lymphoma,55gene expression profiling is anticipated to result in the identification of distinct and prognostically relevant clinical subgroups of MM. Recognition of new therapeutic targets, for example, farnesyltransferase and proteasome components, may lead to a rational design of tumor-specific therapies.
We would like to thank members of the Lambert Laboratory, Jena Derrick, Ailian Li, Kelly McCastlain, Ruston Smith, Elizabeth Williamson, Yan Xiao, and Hongwei Xu for technical assistance without which this project would not have been possible. We thank the MIRT staff, especially Clyde Bailey and Randell Terry for data management; Joth Jacobson and Trey Spencer for statistical support; and P. L. Bergsagel for advice on RT-PCR experimental design; and Paula Card-Higginson for technical writing and editorial assistance.
Supported by private funding from Donna D. and Donald M. Lambert and grant no. CA55819 from the National Cancer Institute, Bethesda, MD.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John D. Shaughnessy Jr, Donna D. and Donald M. Lambert Laboratory of Myeloma Genetics, University of Arkansas for Medical Sciences, 4301 W Markham St, Slot 776, Little Rock, AR 72205; e-mail: shaughnessyjohn@uams.edu.

![Fig. 3. Spiked gene expression corresponds to protein expression in MM. / (A) GeneChip HuGeneFL analysis of MS4A2 (CD20)gene expression. The normalized AD value of fluorescence intensities of streptavidin-PE–stained biotinylated cRNA as hybridized to 2 independent probe sets (accession numbers M27394 [blue] andX12530 [red]) located in different regions of the MS4A2gene is on the vertical axis and samples are on the horizontal axis. Note relatively low expression in 31 normal PCs (NPCs) and spiked expression in 5 of 74 MM samples (MM PCs) Also note similarity in expression levels detected by the 2 different probe sets. (B) Immunohistochemistry for CD20 expression on clonal MM PCs: (i) bone marrow biopsy section showing asynchronous type MM cells (hematoxylin and eosin; original magnification × 500); (ii) CD20+ MM cells (original magnification × 100; inset, original magnification × 500); (iii) biopsy from a patient with mixed asynchronous and Marschalko-type MM cells (hematoxylin and eosin; original magnification × 500); and (iv) CD20+ single lymphocyte and CD20− MM cells (original magnification × 200). CD20 immunohistochemistry was examined without knowledge of clinical history or gene expression findings.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/5/10.1182_blood.v99.5.1745/5/m_h80522227003.jpeg?Expires=1765894302&Signature=osXrxgVcs-z64M5HcUdbcKrvabjHeQM40r-~gFXbJu~y~L3eZZcZ8agl1FY-l5XyAkvtH7BGNMsrSieUdlGw7PqQI-jWDymny4Cz6cSK8kXNLPrV3wbtZ5~0NwHluVldRjRN3TBL1n9e1ScSYANIKmkXvXgKXW-YMWIpMh7cgEAOyucZzSGg~~azfHPaGaCzxKLBQnFRszE9kvBDkLO1giIASVLUbxod~793Ttr3QgiguqV~N96xqmWArCGraR-z0GfpObR001vKlohM0ase~14BORYbp4e0zP1oTID3ftSirljXOKD2S7~i3egyazLSte7ZC-it80aVcmxN8WXL3w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal