Immunoglobulin (Ig) and T-cell receptor (TCR) gene rearrangements are excellent patient-specific polymerase chain reaction (PCR) targets for detection of minimal residual disease (MRD) in acute lymphoblastic leukemia (ALL), but they might be unstable during the disease course. Therefore, we performed detailed molecular studies in 96 childhood precursor-B–ALL at diagnosis and at relapse (n = 91) or at presumably secondary acute myeloid leukemia (n = 5). Clonal Ig and TCR targets for MRD detection were identified in 94 patients, with 71% of these targets being preserved at relapse. The best stability was found for IGK-Kde rearrangements (90%), followed byTCRG (75%), IGH (64%), and incompleteTCRD rearrangements (63%). Combined Southern blot and PCR data for IGH, IGK-Kde, and TCRDgenes showed significant differences in stability at relapse between monoclonal and oligoclonal rearrangements: 89% versus 40%, respectively. In 38% of patients all MRD-PCR targets were preserved at relapse, and in 40% most of the targets (≥ 50%) were preserved. In 22% of patients most targets (10 cases) or all targets (10 cases) were lost at relapse. The latter 10 cases included 4 patients with secondary acute myeloid leukemia with germline Ig/TCR genes. In 5 other patients additional analyses proved the clonal relationship between both disease stages. Finally, in 1 patient all Ig/TCR gene rearrangements were completely different between diagnosis and relapse, which is suggestive of secondary ALL. Based on the presented data, we propose stepwise strategies for selection of stable PCR targets for MRD monitoring, which should enable successful detection of relapse in most (95%) of childhood precursor-B–ALL.
Introduction
Several large prospective studies have clearly demonstrated the high prognostic value of minimal residual disease (MRD) monitoring in childhood acute lymphoblastic leukemia (ALL).1-4 Based on the sensitive measurement of early response to cytotoxic treatment, it is currently possible to identify not only patients at high risk for relapse but also a group of low-risk patients with an excellent relapse-free survival of more than 95%.4 Hence, MRD information provides a new definition of remission in childhood ALL, which justifies incorporation of MRD data in current treatment protocols to refine risk assignment.5
Most MRD studies in pediatric precursor-B–ALL applied immunoglobulin (Ig) and/or T-cell receptor (TCR) gene rearrangements as polymerase chain reaction (PCR) targets for MRD detection. They can easily be identified in most patients at diagnosis with limited sets of PCR primers.6,7 Moreover, using these molecular targets, sensitivities of 10−4 to 10−6 (1 malignant cell within 104 to 106 normal cells) can be obtained routinely.7 With the advent of novel real-time quantitative (RQ) PCR techniques, Ig/TCR-based MRD techniques are now also quantitative.8-11 However, it is also known that Ig and TCR gene rearrangements might change during the disease course, owing to secondary rearrangement processes mediated via ongoing activity of the V(D)J recombinase enzyme system (reviewed by Szczepański et al12). This might lead to loss of the PCR target and consequently to false-negative MRD results. Such changes were most frequently described for Ig heavy chain genes (IGH) and to lesser extent for TCR genes and were found particularly in cases of precursor-B–ALL that already contained subclones at diagnosis.13-18
Although the presence of clonal evolution phenomena is widely acknowledged, its actual impact on the effectiveness of MRD monitoring has not been defined. So far, studies assessing the stability of Ig and TCR gene rearrangements at diagnosis and relapse of ALL either did not compare junctional region sequences or were limited to particular gene loci.13-18 Therefore, we studied the stability of the currently used Ig/TCR rearrangements (IGH, Igκ light chain[IGK], TCRγ [TCRG], and TCRδ[TCRD] gene rearrangements) in a large series of 96 childhood precursor-B–ALL patients. This information is essential for reliable selection of MRD-PCR targets with minimal chance of false-negative results.
Patients, materials, and methods
Patients
Bone marrow or peripheral blood samples from 96 childhood precursor-B–ALL patients were obtained at initial diagnosis and at relapse (91 patients) or at presumably secondary acute myeloid leukemia (AML) (5 patients). The age distribution ranged from 1 month to 183 months. Eight children (8%) were infants (age < 1 year). The diagnosis of precursor-B–ALL was made according to French-American-British and standard immunophenotypic criteria.19-21 Immunologic marker analysis of the precursor-B–ALL revealed that 6 (6%) were pro-B–ALL, 59 (61%) were common ALL, and 31 (32%) were pre-B–ALL. Seven patients were studied at 2 subsequent leukemia relapses. Cell samples of 52 patients were obtained from the cell bank of the Dutch Childhood Leukemia Study Group.
Comparative immunophenotypic analysis revealed intralineage switches in 21% (18 of 86) of precursor-B–ALL patients with available detailed immunophenotypic data at relapse, which is slightly higher than reported previously.22
The rationale, methodology, and pitfalls of the stepwise molecular comparison of the Ig/TCR gene rearrangements between diagnosis and relapse of precursor-B–ALL were previously exemplified in the case report of patient 5498, also included in these series.23 A small subgroup of patients (n = 21) was studied before by comparative Southern blotting and PCR analysis of κ-deleting element (Kde) rearrangements at diagnosis and relapse.24
Comparative Southern blot analysis
Mononuclear cells were separated from bone marrow or peripheral blood samples by Ficoll-Paque centrifugation (density 1.077 g/cm3; Pharmacia, Uppsala, Sweden). DNA was isolated from frozen mononuclear cells, digested with the BglII enzyme, and blotted to nylon membranes as described previously.25IGH and IGK gene configurations were analyzed with the IGHJ6, IGKJ5, IGKC, and IGKDE probes (DAKO, Carpinteria, CA).26,27 The configuration of the TCRD genes was analyzed with the TCRDJ1 probe (DAKO).28 The diagnosis and relapse samples of the 96 patients were always run in parallel lanes (Figure 1), except for 6 patients who were exclusively analyzed at diagnosis because of insufficient amounts of DNA from relapse samples. The Southern blot configuration of the Ig and/or TCR genes in 73 patients at diagnosis and in 30 patients at relapse has been reported previously.6 15
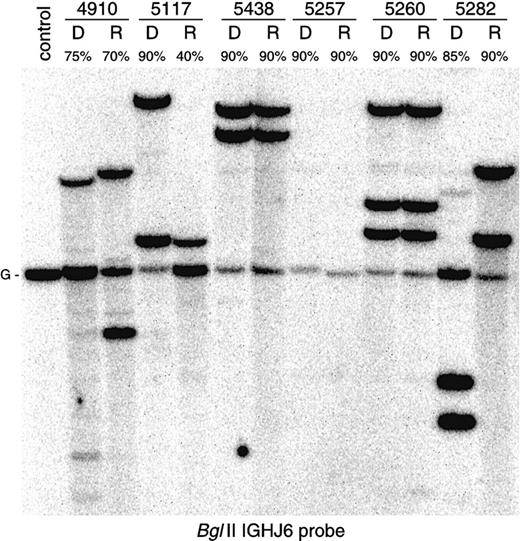
Comparative Southern blot analysis of
IGH gene configuration in 6 precursor-B–ALL patients. Monoclonal patients 5438 and 5260 (patient with trisomy 14) show identical gene rearrangements at diagnosis and at relapse. In monoclonal patient 5117, one allele at relapse is deleted and another one is preserved. Patient 5257 demonstrates biallelic IGHdeletion at both disease stages. Oligoclonal patient 4910 and monoclonal patient 5282 (the upper weak band in the diagnosis lane is derived from previous hybridization) show completely changedIGH gene rearrangement patterns. While sequence analysis has proven a clonal relationship between diagnosis and relapse in patient 4910, patient 5282 most likely represents a secondary ALL.
Comparative Southern blot analysis of
IGH gene configuration in 6 precursor-B–ALL patients. Monoclonal patients 5438 and 5260 (patient with trisomy 14) show identical gene rearrangements at diagnosis and at relapse. In monoclonal patient 5117, one allele at relapse is deleted and another one is preserved. Patient 5257 demonstrates biallelic IGHdeletion at both disease stages. Oligoclonal patient 4910 and monoclonal patient 5282 (the upper weak band in the diagnosis lane is derived from previous hybridization) show completely changedIGH gene rearrangement patterns. While sequence analysis has proven a clonal relationship between diagnosis and relapse in patient 4910, patient 5282 most likely represents a secondary ALL.
PCR amplification and comparative heteroduplex analysis of PCR products
Independent of Southern blotting, PCR analysis could be performed on both diagnosis and relapse samples in 89 patients, essentially as described previously.7,29 Four patients with a secondary AML and 3 precursor-B–ALL patients with insufficient remaining DNA at diagnosis were not analyzed. In each 50 μL PCR reaction, 50 ng DNA sample, 6.3 pM of the 5′ and 3′ oligonucleotide primers, and 0.5 U AmpliTaq Gold polymerase (PE Biosystems, Foster City, CA) were used. The sequences of the 25 oligonucleotide primer pairs used for amplification of IGH (5VH family–specific framework 1 primers and 7 DH family–specific primers with consensus JH primer),IGK-Kde (4 Vκ family–specific primers and intron recombination signal sequence [RSS] primer with Kde primer), TCRG (6 Vγ-Jγ primer combinations most frequently used in precursor-B–ALL), and TCRD gene rearrangements (Vδ2-Dδ3 and Dδ2-Dδ3 primer pairs) were previously published.7,30,31 PCR conditions were as follows: initial denaturation for 10 minutes at 94°C, followed by 40 cycles of 45 seconds at 92°C, 90 seconds at 60°C, and 2 minutes at 72°C using a Perkin-Elmer 480 thermal cycler (PE Biosystems). After the last cycle an additional extension step of 10 minutes at 72°C was performed. Appropriate positive and negative controls were included in all experiments.7
For heteroduplex analysis, the PCR products were denatured at 94°C for 5 minutes after the final cycle of amplification and subsequently cooled to 4°C for 60 minutes to induce duplex formation.32 Afterward the duplexes were immediately loaded on 6% nondenaturing polyacrylamide gels in 0.5 × Tris-borate-EDTA buffer, run at room temperature, and visualized by ethidium bromide staining.32
Relapse samples were at first analyzed with those primer combinations, which showed clonal PCR products at diagnosis. When the clonal PCR product was also found at relapse, its identity was subsequently compared with the PCR product found at diagnosis by means of mixed heteroduplex analysis, ie mixing of the diagnosis and relapse PCR products followed by heteroduplex analysis (Figure2).23 When clonal PCR products found at diagnosis were undetectable at relapse, the relapse sample was then analyzed with additional primer combinations for the involved gene loci.
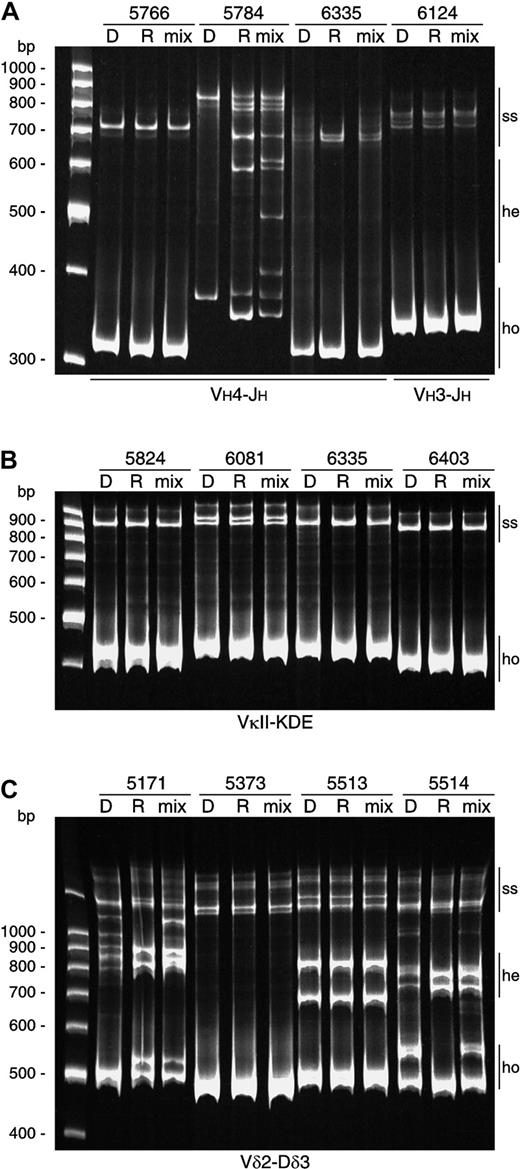
Examples of comparative heteroduplex PCR analysis.
(A) Comparative heteroduplex analysis of IGH gene rearrangements. Monoclonal homoduplexes (ho) in patients 5766, 6335, and 6124 found at diagnosis and at relapse were of the same size. Mixing of the PCR products of these disease phases followed by heteroduplex PCR analysis demonstrated no heteroduplex (he) formation, proving that these gene rearrangements had identical junctional regions. In patient 5784, monoclonal homoduplexes found at diagnosis and at relapse slightly differed in size. Mixing of the VH4-JH PCR products followed by heteroduplex PCR analysis demonstrated clear heteroduplex formation, proving that these VH4-JHgene rearrangements had different junctional regions; (ss) indicates remaining single-strand fragments. (B) Comparative heteroduplex analysis of Kde rearrangements showed completely identical rearrangements at diagnosis and at relapse. (C) Comparative heteroduplex analysis of Vδ2-Dδ3 gene rearrangements. Patients 5373 and 5513 with monoallelic and biallelic rearrangements, respectively, had stable Vδ2-Dδ3 rearrangements. In contrast, Vδ2-Dδ3 joinings at diagnosis in patients 5171 and 5514 are oligoclonal, while in both cases 2 monoclonal Vδ2-Dδ3 rearrangements were found at relapse.
Examples of comparative heteroduplex PCR analysis.
(A) Comparative heteroduplex analysis of IGH gene rearrangements. Monoclonal homoduplexes (ho) in patients 5766, 6335, and 6124 found at diagnosis and at relapse were of the same size. Mixing of the PCR products of these disease phases followed by heteroduplex PCR analysis demonstrated no heteroduplex (he) formation, proving that these gene rearrangements had identical junctional regions. In patient 5784, monoclonal homoduplexes found at diagnosis and at relapse slightly differed in size. Mixing of the VH4-JH PCR products followed by heteroduplex PCR analysis demonstrated clear heteroduplex formation, proving that these VH4-JHgene rearrangements had different junctional regions; (ss) indicates remaining single-strand fragments. (B) Comparative heteroduplex analysis of Kde rearrangements showed completely identical rearrangements at diagnosis and at relapse. (C) Comparative heteroduplex analysis of Vδ2-Dδ3 gene rearrangements. Patients 5373 and 5513 with monoallelic and biallelic rearrangements, respectively, had stable Vδ2-Dδ3 rearrangements. In contrast, Vδ2-Dδ3 joinings at diagnosis in patients 5171 and 5514 are oligoclonal, while in both cases 2 monoclonal Vδ2-Dδ3 rearrangements were found at relapse.
Sequence analysis of Ig/TCR gene rearrangements
Clonal PCR products discordant between diagnosis and relapse of precursor-B–ALL as found by mixed heteroduplex analysis were directly sequenced. Sequencing was performed using the dye-terminator cycle sequencing kit with AmpliTaq DNA polymerase FS on an ABI 377 sequencer (PE Biosystems) as previously described.31 VH, DH, and JH segments were identified using DNAPLOT software (W. Müller, H.-H. Althaus, University of Cologne, Germany) by searching for homology with all known human germline VH, DH, and JH sequences obtained from the VBASE directory of human Ig genes (http://www.mrc-cpe.cam.ac.uk/imt-doc/).33Vγ and Jγ gene segments were identified by comparison to germlineTCRG sequences as previously described.34
Statistical analysis
Statistical analysis using the χ2 test on a 2 × 2 table was performed to compare the frequencies of particular Ig/TCR gene rearrangements between different precursor-B–ALL patient subgroups. Pearson correlation coefficient was calculated to test an association between variables. P values less than or equal to .05 were considered statistically significant.
Results
Southern blot configuration of Ig and TCR genes in relapsed ALL patients
The configuration of IGH, IGK, andTCRD genes was established with multiple Southern blot probes. This concerned all 96 patients at diagnosis of precursor-B–ALL and 91 patients at subsequent relapse or secondary AML (Figure 1). This gave us the unique opportunity to address the question of whether there are any differences in Ig/TCR gene configuration between the patients who relapsed compared with the total childhood precursor-B–ALL group. This comparison is summarized in Table 1, which shows that the Ig/TCR gene rearrangement patterns at diagnosis and at relapse in patients included in this study are largely comparable to each other and to previously published data derived from large series of random childhood precursor-B–ALL cases at initial diagnosis.6 27 Only 2 immunogenotypic features were more prevalent in ALL at relapse. The TCRD gene configuration at relapse was characterized by significantly less frequent Vδ2-Dδ3 and Dδ2-Dδ3 rearrangements and more frequent TCRDdeletions (P < .05), which reflects ongoing deletional rearrangements. Secondly, the frequency of IGH andTCRD oligoclonality at relapse was slightly less frequent, but this difference was only significant for TCRD(P < .05).
Comparison of Ig and TCR gene rearrangement patterns based on Southern blotting
| . | Random precursor B–ALL at diagnosis* . | Relapsed precursor-B–ALL at diagnosis† . | Precursor-B–ALL at relapse . |
|---|---|---|---|
| IGH | |||
| Germline | 0 | 0 | 0 |
| R/G | 1% (1/97) | 2% (2/92) | 5% (4/85) |
| R/R | 88% (85/97) | 88% (81/92) | 85% (72/85) |
| R/D | 8% (8/97) | 7% (6/92) | 7% (6/85) |
| D/G | 2% (2/97) | 1% (1/92) | 1% (1/85) |
| D/D | 1% (1/97) | 1% (1/92) | 2% (2/85) |
| Oligoclonal | 36% (35/97) | 42% (38/92)‡ | 28% (24/85)1-153 |
| IGK-Kde | |||
| Germline | 50% (55/111) | 51% (47/92) | 49% (42/86) |
| R/G | 28% (31/111) | 21% (19/92) | 23% (20/86) |
| R/R | 23% (25/111) | 28% (26/92) | 28% (24/86) |
| Oligoclonal | Not determined | 3% (3/92) | 1% (1/86) |
| TCRG | |||
| Germline | 41% (79/192) | 38% (35/91) | 41% (37/91) |
| R/G or R/R | 59% (113/192) | 62% (56/91) | 59% (54/91) |
| TCRD | |||
| Germline | 11% (22/202) | 8% (7/91) | 12% (10/85) |
| R/G | 15% (30/202) | 11% (10/91) | 8% (7/85) |
| R/R | 23% (47/202) | 24% (22/91) | 12% (10/85)1-155 |
| R/D | 17% (35/202) | 20% (18/91) | 22% (19/85) |
| D/G | 5% (11/202) | 4% (4/91) | 5% (4/85) |
| D/D | 28% (57/202) | 33% (30/91) | 41% (35/85)1-155 |
| Oligoclonal | 21% (13/62) | 26% (24/91) | 12% (10/85)1-155 |
| . | Random precursor B–ALL at diagnosis* . | Relapsed precursor-B–ALL at diagnosis† . | Precursor-B–ALL at relapse . |
|---|---|---|---|
| IGH | |||
| Germline | 0 | 0 | 0 |
| R/G | 1% (1/97) | 2% (2/92) | 5% (4/85) |
| R/R | 88% (85/97) | 88% (81/92) | 85% (72/85) |
| R/D | 8% (8/97) | 7% (6/92) | 7% (6/85) |
| D/G | 2% (2/97) | 1% (1/92) | 1% (1/85) |
| D/D | 1% (1/97) | 1% (1/92) | 2% (2/85) |
| Oligoclonal | 36% (35/97) | 42% (38/92)‡ | 28% (24/85)1-153 |
| IGK-Kde | |||
| Germline | 50% (55/111) | 51% (47/92) | 49% (42/86) |
| R/G | 28% (31/111) | 21% (19/92) | 23% (20/86) |
| R/R | 23% (25/111) | 28% (26/92) | 28% (24/86) |
| Oligoclonal | Not determined | 3% (3/92) | 1% (1/86) |
| TCRG | |||
| Germline | 41% (79/192) | 38% (35/91) | 41% (37/91) |
| R/G or R/R | 59% (113/192) | 62% (56/91) | 59% (54/91) |
| TCRD | |||
| Germline | 11% (22/202) | 8% (7/91) | 12% (10/85) |
| R/G | 15% (30/202) | 11% (10/91) | 8% (7/85) |
| R/R | 23% (47/202) | 24% (22/91) | 12% (10/85)1-155 |
| R/D | 17% (35/202) | 20% (18/91) | 22% (19/85) |
| D/G | 5% (11/202) | 4% (4/91) | 5% (4/85) |
| D/D | 28% (57/202) | 33% (30/91) | 41% (35/85)1-155 |
| Oligoclonal | 21% (13/62) | 26% (24/91) | 12% (10/85)1-155 |
The frequencies of particular Ig/TCR gene rearrangements in the random precursor-B–ALL group at diagnosis are derived from our previous studies.6,27 46
Including one patient with phenotypic switch to AML at relapse.
The frequency of IGH oligoclonality was significantly higher in infant ALL compared with noninfant ALL (75% vs 38%, P < .05).
Not significant.
P < .05.
The configuration of Ig/TCR genes compared between 2 subsequent relapses in the 7 patients analyzed showed evidence for clonal evolution in only 2 cases (concerning 1 or 2 gene rearrangements), while in 5 of the 7 cases we observed some changes in gene rearrangement patterns between diagnosis and first relapse.
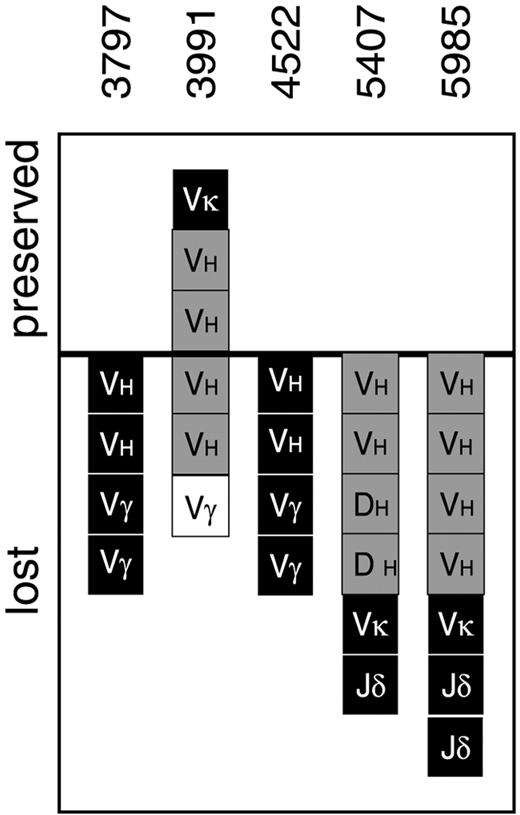
Southern blot analysis in 4 of 5 patients with a presumed secondary AML demonstrated the complete absence of clonal Ig/TCR gene rearrangements (Figure 3), which is in line with the AML diagnosis. However, in one patient (3991) 3 clonal Southern blot bands were preserved in the AML clone, and the sequence identity was confirmed by comparative PCR analysis for 2VH-JH gene rearrangements. Apparently the original ALL clone underwent a phenotypic switch to AML. This patient was included in our further comparative diagnosis-relapse analyses despite the phenotypic shift, while the other 4 patients were excluded.
Ig/TCR gene rearrangement stability in 5 precursor-B–ALL patients subjected to comparative heteroduplex PCR analysis at diagnosis and at presumably secondary AML.
All Ig/TCR gene rearrangements identified at diagnosis in patients 3797, 4522, 5407, and 5985 were lost, which is in line with secondary AML. In contrast, several IGH and IGK gene rearrangements in patient 3991 were preserved, suggesting a phenotypic shift from ALL to AML. Black squares represent monoclonal rearrangements; gray squares, biclonal/oligoclonal rearrangements; white squares, oligoclonal/polyclonal rearrangements. All squares above the line represent stable MRD-PCR targets, while all squares below the line represent Ig/TCR gene rearrangements lost owing to clonal evolution. VH indicates VH-JH gene rearrangements; DH, DH-JH gene rearrangements;Vκ, Vκ-Jκ gene rearrangements; Vγ, Vγ-Jγ gene rearrangements; Jδ, rearrangements to Jδ1-Dδ3 region detected by Southern blot analysis.
Ig/TCR gene rearrangement stability in 5 precursor-B–ALL patients subjected to comparative heteroduplex PCR analysis at diagnosis and at presumably secondary AML.
All Ig/TCR gene rearrangements identified at diagnosis in patients 3797, 4522, 5407, and 5985 were lost, which is in line with secondary AML. In contrast, several IGH and IGK gene rearrangements in patient 3991 were preserved, suggesting a phenotypic shift from ALL to AML. Black squares represent monoclonal rearrangements; gray squares, biclonal/oligoclonal rearrangements; white squares, oligoclonal/polyclonal rearrangements. All squares above the line represent stable MRD-PCR targets, while all squares below the line represent Ig/TCR gene rearrangements lost owing to clonal evolution. VH indicates VH-JH gene rearrangements; DH, DH-JH gene rearrangements;Vκ, Vκ-Jκ gene rearrangements; Vγ, Vγ-Jγ gene rearrangements; Jδ, rearrangements to Jδ1-Dδ3 region detected by Southern blot analysis.
PCR detectability of Ig and TCR gene rearrangements in relapsed precursor-B–ALL patients
A total of 362 clonal PCR products of different Ig/TCR gene rearrangements were identified at diagnosis in 87 (98%) of 89 patients, with an average of 4 targets per patient. In one patient no clonal gene rearrangements were detected by PCR, while Southern blotting showed a single weak rearranged IGH band, identical between the diagnosis and relapse sample. The second patient had an infant ALL and was fully oligoclonal at diagnosis, which precluded identification of clonal Ig/TCR markers for PCR-based MRD monitoring. Generally, IGH oligoclonality at diagnosis was more prevalent in infant ALL patients (6 of 8 cases), compared with the noninfant precursor-B–ALL cases (32 of 84 cases),P < .05.
Stability of particular MRD-PCR targets in monoclonal and oligoclonal precursor-B–ALL patients at relapse
A total of 256 (71%) of 362 clonal Ig/TCR gene rearrangements identified with heteroduplex PCR analysis at diagnosis in 87 patients were preserved at relapse. This concerned 99 (64%) of 155IGH, 54 (90%) of 60 IGK-Kde, 65 (75%) of 87TCRG, and 38 (63%) of 60 TCRD gene rearrangements (Table 2). In 3 additional patients, Southern blot analysis provided conclusive information about stability of gene rearrangement patterns.6 15
Stability of MRD-PCR targets in patients with monoclonal and oligoclonal Ig and TCR gene configuration
| . | Stability of MRD-PCR targets . | Stability in patients (all l ≥ 1) . | ||||
|---|---|---|---|---|---|---|
| Monoclonal . | Oligoclonal . | Total . | Monoclonal . | Oligoclonal . | Total . | |
| IGH | ||||||
| VH-JH | 88% | 47% | 69% | 81%/98% | 37%/73% | 63%/88% |
| (n = 67) | (n = 60) | (n = 127) | ||||
| DH-JH | 57% | 38% | 43% | 67%/67% | 27%/53% | 33%/57% |
| (n = 7) | (n = 21) | (n = 28) | ||||
| All | 85% | 44% | 64% | 76%/98% | 24%/76% | 52%/88% |
| (n = 74) | (n = 81) | (n = 155) | (n = 42) | (n = 33) | (n = 75) | |
| IGK-Kde | ||||||
| Vκ-Kde | 95% | 40% | 91% | 94%/97% | 33%/66% | 88%/94% |
| (n = 39) | (n = 5) | (n = 44) | ||||
| Intron-Kde | 86% | 0% | 87% | 92%/92% | 0% | 85%/85% |
| (n = 15) | (n = 15) | |||||
| All* | 95% | 40% | 90% | 95%/95% | 33%/66% | 88%/93% |
| (n = 55) | (n = 5) | (n = 60) | (n = 37) | (n = 3) | (n = 40) | |
| TCRG | ||||||
| Vγ-Jγ | ND | ND | 75% | ND | ND | 64%/83% |
| (n = 87) | (n = 53) | |||||
| TCRD | ||||||
| Dδ2-Dδ3 | 100% | 14% | 63% | 100% | 14%/14% | 60%/60% |
| (n = 9) | (n = 7) | (n = 16) | ||||
| Vδ2-Dδ3 | 81% | 31% | 63% | 75%/80% | 20%/50% | 59%/71% |
| (n = 27) | (n = 16) | (n = 43) | ||||
| All† | 86% | 26% | 63% | 80%/88% | 14%/36% | 56%/69% |
| (n = 37) | (n = 23) | (n = 60) | (n = 25) | (n = 14) | (n = 39) | |
| . | Stability of MRD-PCR targets . | Stability in patients (all l ≥ 1) . | ||||
|---|---|---|---|---|---|---|
| Monoclonal . | Oligoclonal . | Total . | Monoclonal . | Oligoclonal . | Total . | |
| IGH | ||||||
| VH-JH | 88% | 47% | 69% | 81%/98% | 37%/73% | 63%/88% |
| (n = 67) | (n = 60) | (n = 127) | ||||
| DH-JH | 57% | 38% | 43% | 67%/67% | 27%/53% | 33%/57% |
| (n = 7) | (n = 21) | (n = 28) | ||||
| All | 85% | 44% | 64% | 76%/98% | 24%/76% | 52%/88% |
| (n = 74) | (n = 81) | (n = 155) | (n = 42) | (n = 33) | (n = 75) | |
| IGK-Kde | ||||||
| Vκ-Kde | 95% | 40% | 91% | 94%/97% | 33%/66% | 88%/94% |
| (n = 39) | (n = 5) | (n = 44) | ||||
| Intron-Kde | 86% | 0% | 87% | 92%/92% | 0% | 85%/85% |
| (n = 15) | (n = 15) | |||||
| All* | 95% | 40% | 90% | 95%/95% | 33%/66% | 88%/93% |
| (n = 55) | (n = 5) | (n = 60) | (n = 37) | (n = 3) | (n = 40) | |
| TCRG | ||||||
| Vγ-Jγ | ND | ND | 75% | ND | ND | 64%/83% |
| (n = 87) | (n = 53) | |||||
| TCRD | ||||||
| Dδ2-Dδ3 | 100% | 14% | 63% | 100% | 14%/14% | 60%/60% |
| (n = 9) | (n = 7) | (n = 16) | ||||
| Vδ2-Dδ3 | 81% | 31% | 63% | 75%/80% | 20%/50% | 59%/71% |
| (n = 27) | (n = 16) | (n = 43) | ||||
| All† | 86% | 26% | 63% | 80%/88% | 14%/36% | 56%/69% |
| (n = 37) | (n = 23) | (n = 60) | (n = 25) | (n = 14) | (n = 39) | |
ND indicates not done.
Including one Vκ-intronRSS rearrangement.
Including one Vδ3-Jδ1 rearrangement.
In 36 patients (including patient 4616 studied exclusively by Southern blotting) all MRD-PCR targets identified at diagnosis were preserved at relapse (Figure 4A). In 38 cases (including patients 2665 and 4501 studied exclusively by Southern blotting) at least half of the targets remained stable during the disease course (Figure 4B). In another 10 patients (including patient 3991 with AML at relapse; Figure 3) most MRD-PCR targets were absent at relapse but at least one rearrangement was common for both diagnosis and relapse samples (Figure 4C). Consequently, at least one MRD-PCR target was preserved at relapse in 84 (93%) of 90 patients with available clonal MRD-PCR targets at diagnosis. In the remaining 6 patients all clonal markers found at diagnosis seemed to be lost. However, in 3 of these 6 cases the clonal relationship between diagnosis and relapse was confirmed by the identification of a common DH-JH stem shared by respective VH-JH gene rearrangements. In another 2 cases additional analyses35 36 showed at both disease stages identical DNA sequences of Vκ-Jκ (5771) and chimericMLL-AF4 fusion genes (5978), respectively (data not shown). Finally, in one patient (5282) there was no evidence for a common origin of diagnosis and relapse clones (Figures 1 and 4C). Because all original monoclonal rearrangements were lost in this case at relapse, we believe that this is in fact a secondary ALL.
Ig/TCR gene rearrangement stability patterns in 87 precursor-B–ALL patients subjected to comparative heteroduplex PCR analysis at diagnosis and at relapse.
Black squares represent monoclonal rearrangements; gray squares, biclonal/oligoclonal rearrangements; white squares, oligoclonal/polyclonal rearrangements detectable as weak Southern blot bands. All squares above the upper line represent stable MRD-PCR targets, while all squares below the lower line represent Ig/TCR gene rearrangements lost owing to clonal evolution. VH squares between the 2 lines have common DH-JH stems shared by different IGH rearrangements at diagnosis and at relapse. (A) Thirty-five patients with all PCR-MRD targets identified at diagnosis preserved at relapse; (B) 36 cases with some targets (ranging from one target to half of the targets) lost during disease course; (C) 16 patients with most or all clonal markers absent at relapse. Patient 4790 was not included in our evaluation of MRD-PCR target stability because the detected IGH andTCRG gene rearrangements at diagnosis were oligoclonal.VH indicates VH-JH gene rearrangements;DH, DH-JH gene rearrangements; Vκ, Vκ-Kde gene rearrangements; iκ,intronRSS-Kde gene rearrangements; Vγ, Vγ-Jγ gene rearrangements;Vδ, Vδ2-Dδ3 gene rearrangements; Dδ, Dδ2-Dδ3 gene rearrangements.
Ig/TCR gene rearrangement stability patterns in 87 precursor-B–ALL patients subjected to comparative heteroduplex PCR analysis at diagnosis and at relapse.
Black squares represent monoclonal rearrangements; gray squares, biclonal/oligoclonal rearrangements; white squares, oligoclonal/polyclonal rearrangements detectable as weak Southern blot bands. All squares above the upper line represent stable MRD-PCR targets, while all squares below the lower line represent Ig/TCR gene rearrangements lost owing to clonal evolution. VH squares between the 2 lines have common DH-JH stems shared by different IGH rearrangements at diagnosis and at relapse. (A) Thirty-five patients with all PCR-MRD targets identified at diagnosis preserved at relapse; (B) 36 cases with some targets (ranging from one target to half of the targets) lost during disease course; (C) 16 patients with most or all clonal markers absent at relapse. Patient 4790 was not included in our evaluation of MRD-PCR target stability because the detected IGH andTCRG gene rearrangements at diagnosis were oligoclonal.VH indicates VH-JH gene rearrangements;DH, DH-JH gene rearrangements; Vκ, Vκ-Kde gene rearrangements; iκ,intronRSS-Kde gene rearrangements; Vγ, Vγ-Jγ gene rearrangements;Vδ, Vδ2-Dδ3 gene rearrangements; Dδ, Dδ2-Dδ3 gene rearrangements.
Stability of MRD targets is not related to age, blood cell counts, or remission duration
Stability of MRD-PCR targets did not significantly correlate with age or white blood cell count at diagnosis or with remission duration, ie, time span between diagnosis and relapse.
IGH gene rearrangements
Clonal IGH gene rearrangements were detected by PCR in 75 of the 90 studied childhood precursor-B–ALL patients. In 66 cases (88%) at least one IGH MRD-PCR target was preserved at relapse. In 12 additional patients PCR analysis did not result in identification of clonal IGH rearrangements, while Southern blot data suggested the preservation of at least one target in all 12 patients (fully identical IGH configuration in 9 cases). In 3 patients deletions of JH region were identified by Southern blotting both at diagnosis and at relapse of ALL. The Southern blot results of these 15 patients were not used for calculating the stability of theIGH PCR targets.
The stability of the IGH PCR targets was markedly different between oligoclonal and monoclonal patients, ie, at least one MRD-PCR target was preserved in 76% and 98% of patients, respectively (Table2). This significant difference was even more pronounced at the MRD-PCR target level, with 63 (85%) of 74 monoclonal IGH gene rearrangements being stable compared with only 36 (44%) of 85 oligoclonal rearrangements. Taking into account the type ofIGH gene rearrangement, complete VH-JHrecombinations were characterized by a higher stability compared with incomplete DH-JH rearrangements, with 69% versus 43% of targets preserved, respectively.
IGK deletional rearrangements
A total of 60 IGK deletional rearrangements were detected in 40 childhood precursor-B–ALL patients at diagnosis: 44 Vκ-Kde, 15 intron-Kde, and 1 rarely occurring Vκ-intron RSS recombination. At least one of the rearrangements was preserved in 37 cases (93%). In fact, IGK-Kde recombinations represented the most stable MRD-PCR targets, with 90% of all targets preserved. Most (55 of 60) Kde rearrangements were monoclonal and highly stable (52 targets preserved; 95%), while only 2 of 5 oligoclonalIGK-Kde targets were found at relapse. No significant difference in stability was found between Vκ-Kde and intron-Kde rearrangements.
TCRG gene rearrangements
A total of 87 TCRG gene rearrangements were detected in 53 precursor-B–ALL patients at diagnosis, and in 44 cases (83%) at least one MRD-PCR target was preserved at relapse. Because accurate oligoclonality detection in TCRG locus is rather complex, even by Southern blotting,37 we did not evaluate whether there were any differences in MRD-PCR target stability between monoclonal and oligoclonal patients.
TCRD gene rearrangements
A total of 60 clonal TCRD gene rearrangements (43 Vδ2-Dδ3, 16 Dδ2-Dδ3, and 1 Vδ3-Jδ1) were identified by PCR in 39 precursor-B–ALL patients. At least one of the clonal rearrangements was preserved in 27 (69%) of 39 cases. Once again we observed striking differences in stability between monoclonal and oligoclonal patients, ie, at least one target was preserved in 88% and 36% of patients, respectively (Table 2). Only 26% of oligoclonal targets were preserved compared with 86% of monoclonal targets. Moreover, in 10 patients Southern blot data suggested the presence of a clonally rearranged band corresponding to Vδ2-Dδ3 rearrangements, while heteroduplex PCR analysis of these rearrangements showed oligoclonality or even polyclonality.6
Patterns of clonal evolution in precursor-B–ALL patients with unstable targets
Based on combined Southern blot, PCR, and sequence analysis, it was possible to follow the patterns of clonal evolution leading to disappearance of rearrangements that were originally present at diagnosis.
Clonal evolution in the IGH locus
Owing to clonal evolution phenomena, 62 IGH targets in 36 patients were lost. We could determine the exact patterns of clonal evolution in 8 patients with monoclonal IGH gene rearrangements and in 26 patients with an oligoclonal rearrangement pattern.
In 7 patients (2 monoclonal and 5 oligoclonal) Southern blot rearrangement patterns between diagnosis and relapse were identical, while PCR analyses showed disappearance of a single rearrangement. This might be explained by the disappearance of minor subclones undetectable by Southern blotting. In 3 patients with monoclonal IGHconfiguration, one of the rearrangements was changed both in Southern blotting and PCR, while sequence comparison showed VHreplacement with a preserved VH-N-DH-N-JH junction. In another 2 monoclonal patients we observed clonal “regression” of one of the rearrangements to germline. Finally, in a single patient (5282) both monoclonal IGH rearrangements were replaced by 2 new unrelated rearrangements; this patient was suspected of having developed a secondary ALL (Figure 4C).
In 11 oligoclonal patients, the IGH configuration at relapse was monoclonal, which is suggestive of clonal selection during the treatment. In another 10 oligoclonal patients, IGH was also oligoclonal at relapse, with several rearrangements lost and with emergence of new (sub)clones. Sequence comparison was fully completed in 12 oligoclonal patients with changes at relapse, indicating ongoingVH to DH-JH joinings with preservation of common DH-JH stems in 5 patients and possibly secondary rearrangements in 5 other cases. In the remaining 2 oligoclonal patients, the IGH sequences were unrelated and suggestive of the development of diagnosis and relapse clones from a common clonal progenitor via independent secondary rearrangements.
Clonal evolution in the TCRG locus
Clonal evolution of TCRG gene rearrangements was observed in 19 patients, leading to loss of 22 MRD-PCR targets. In 9 patients this concerned “regression” of clonal rearrangements most probably to germline configuration. In 5 patients the new rearrangements at relapse contained upstream Vγ and downstream Jγ segments as compared with the Vγ-Jγ rearrangements at diagnosis, which is suggestive of ongoing recombination with Vγ-Jγ replacement. In the remaining 5 patients the sequence comparison of Vγ-Jγ rearrangements at diagnosis and at relapse excluded secondary rearrangements and indicated the emergence of a clone related to the initial (pre)leukemic clone but different from the predominant clone at diagnosis.
Clonal evolution in the TCRD locus
Clonal evolution in the TCRD locus resulted in the loss of 22 MRD-PCR targets in 17 patients (5 monoclonal and 12 oligoclonal cases). In 6 patients (including 4 patients with monoclonal Vδ2-Dδ3 rearrangements) ongoing deletions were observed. In contrast, in 8 patients (including 1 patient with monoclonal Vδ2-Dδ3) the rearrangement pattern “regressed” to germline configuration. Finally, in the remaining 3 cases new rearrangements were found at relapse, including 1 Vδ2-Dδ3 joining and 2 unidentified rearrangements to the Dδ3-Jδ1 region. Interestingly, 5 of 10 cases with oligoclonal/polyclonal Vδ2-Dδ3 rearrangements at diagnosis had a monoclonal TCRD configuration at relapse with a single Vδ2-Dδ3 joining. In the remaining 5 patients, ongoingTCRD deletion was assumed in 2 cases, 2 patients demonstrated “regression” to germline configuration, and only a single patient preserved the oligo/polyclonal Vδ2-Dδ3 rearrangement pattern at relapse.
Discussion
Our comparative Southern blot, PCR, and sequencing analyses of childhood precursor-B–ALL at diagnosis and relapse have provided detailed insight in the stability and changes of Ig and TCR gene rearrangements during the disease course. This information is essential for reliable application of Ig/TCR gene rearrangements as MRD-PCR targets in childhood ALL. However, one should be cautious with extrapolating these data to adolescent or adult precursor-B–ALL patients, because the immunogenotype of adult precursor-B–ALL has more immature features.29
The Ig/TCR gene rearrangement patterns at diagnosis in relapsed patients appeared to be comparable to those in a random series of newly diagnosed pediatric precursor-B–ALL, implying that the various Ig/TCR gene characteristics at diagnosis have no prognostic value. This is in contrast to the previously reported strong predictive value of the Vδ2-Dδ3 gene rearrangement or the overall clonal diversity.38 39 The overall Ig/TCR gene configuration patterns at relapse were largely comparable to those at diagnosis but were characterized by less oligoclonality and more frequentTCRD gene deletions. These 2 differences fit with the concept of ongoing clonal selection and continuing rearrangements.
The detailed molecular analyses proved the clonal relationship between diagnosis and relapse in 88 of 89 childhood precursor-B–ALL with identified MRD-PCR markers at diagnosis. In only one patient (5282) were the Ig/TCR gene rearrangement patterns at diagnosis and relapse completely different with unrelated junctional region sequences (Figures 1 and 4C). Consequently, the presumed ALL relapse in this child (3.5 years after diagnosis) might in fact represent a secondary leukemia. This single patient confirms previous observations that ALL rarely occurs as second malignancy after previous cytotoxic treatment.40 This is in contrast to secondary AML, which affects approximately 4% of children treated for ALL with cytotoxic regimens containing topoisomerase II inhibitors.41 We indeed proved the presence of secondary AML with germline Ig/TCR genes in 4 (4%) of the 96 patients (Figure 3). However, in a fifth patient we demonstrated the clonal relationship between the precursor-B–ALL at diagnosis and the presumed secondary AML, which in fact represented a phenotypic switch.
The comparison of Ig/TCR gene rearrangement patterns between diagnosis and relapse showed marked heterogeneity in the occurrence of clonal evolution phenomena. In 40% of patients all PCR-identified clonal Ig/TCR rearrangements were present at relapse (Figure 4A), and in another 42% of cases at least half of the identified gene rearrangements remained stable at relapse (Figure 4B). Extreme clonal evolution with differential outgrowth of subclones characterized the remaining 18% of cases (Figure 4C), in whom most or even all clonal Ig/TCR gene rearrangements identified at diagnosis were lost during disease course. Interestingly, in contrast to the frequent occurrence of clonal evolution between diagnosis and relapse, we did not observe major clonal instability of Ig/TCR genes between 2 consecutive relapses (7 cases), which is in line with previous observations.14
Previous studies suggested that the risk of changes in Ig/TCR rearrangement patterns increases with time.15,42 We did not find a significant correlation between remission duration and target stability in this extensive study. This is in line with the report that clonal selection processes can already occur in early treatment phases and the reports on clonal identity between diagnosis and very late relapse of precursor-B–ALL.43-45
In this extensive molecular study, we wished to identify the factors associated with the occurrence of clonal evolution and therefore the increased risk of false-negative MRD-PCR results. It is entirely clear from our study that discrimination between monoclonality versus oligoclonality at diagnosis is the most powerful predictor of clonal evolution during the ALL disease course. All other variables, such as age, white blood cell count, and immunophenotype at diagnosis, failed to identify patients prone to clonal evolution of their Ig/TCR gene rearrangements. Monoclonal MRD-PCR targets were characterized by high stability, with 89% of all targets detectable at relapse. In contrast, only 40% of the oligoclonal MRD-PCR targets were preserved at relapse. Therefore, it is probably important to discriminate between monoclonal and oligoclonal Ig/TCR rearrangements, which requires a combined Southern blot and PCR approach. Southern blotting is particularly informative for detection of oligoclonality in IGH andIGK gene rearrangements, whereas heteroduplex PCR analysis in combination with Southern blotting is informative for detection of oligoclonal TCRD gene rearrangements. Southern blotting needs more DNA and is more labor intensive and time consuming than PCR techniques. However, with a single BglII restriction enzyme digestion it is possible to detect oligoclonality in IGH, IGK, and TCRD loci.26-28 Judging clonality solely from the number of PCR products per gene would result in marked underestimation of oligoclonality; eg, at least one third of the oligoclonal IGH targets (29 of 85, including 15 lost MRD-PCR markers) would have been classified as monoclonal.
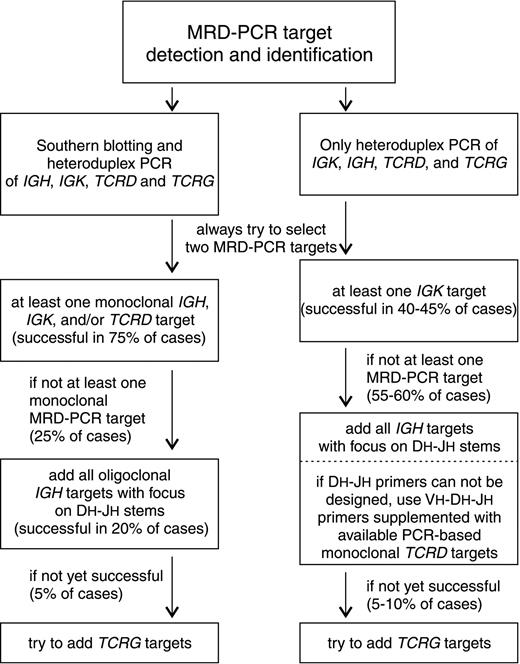
The herein presented detailed comparison of Ig/TCR gene rearrangement patterns provides important information for appropriate selection of PCR targets for MRD monitoring. It is already accepted that preferably 2 MRD-PCR targets should be used per patient. Furthermore, our data show that monoclonal targets should be chosen as first option. As previously suggested, monoclonal Kde rearrangements were characterized by the best stability (95%), owing to their end-stage character.24,30 In addition, approximately 85% of monoclonal IGH and TCRD gene rearrangements remained stable at relapse (Table 2). In monoclonal VH-JHrearrangements, it is particularly attractive to position the patient-specific primers/probes at the VH-DHpart of the junctional region, which is a preferred strategy in current RQ-PCR based strategies.9-11 Identification of preferably 2 monoclonal MRD-PCR targets (IGH, IGK, and/orTCRD) was possible in 67 (77%) of 87 patients. When applying these monoclonal targets, the detection of relapse would have been possible in 65 patients, but false-negative results would have been obtained in 2 patients: one with presumably secondary ALL and an infant case characterized by extensive clonal Ig/TCR evolution. The second choice for target selection should concern oligoclonalIGH gene rearrangements. Although these rearrangements are particularly prone to ongoing and secondary recombination processes, they are the sole MRD-PCR targets in approximately 10% of childhood precursor-B–ALL patients.46 In the case of oligoclonalIGH targets, the patient-specific primers/probes should preferably be positioned at the D-N-J junctions. Moreover, all identified clonal DH-JH stems should be followed because restriction to 2 targets would increase the risk of false-negative MRD results. In our series this approach could have been used in an additional 17 (20%) of 87 patients with available MRD-PCR targets and should have resulted in detection of relapse in 15 cases (false negativity in patients 2308 and 5978 with secondary IGH gene rearrangements). Finally, successful MRD detection in the remaining 3 patients could have been accomplished by usage of TCRG gene rearrangements, which were sole MRD-PCR targets in these patients. One could argue for the preferred usage of TCRG gene rearrangements instead of oligoclonal IGH targets. However, in at least 2 of our patients (4910, 5661) usage of common DH-JHstems would have been superior to Vγ-Jγ targets (Figure 4C). More importantly, TCRG gene rearrangements are generally less sensitive markers in RQ-PCR analyses, owing to their limited combinatorial diversity and the abundance of polyclonal Vγ-Jγ joinings in normal T-cells in postinduction follow-up samples.47 Finally, our study clearly shows that oligoclonal Vδ2-Dδ3 and Dδ2-Dδ3 rearrangements should not be used as MRD-PCR targets, because most of them (about 75%) would be modified through clonal evolution processes. The Southern blot and PCR-based strategy for MRD-PCR target selection (priority order: 2 monoclonal IGH, IGK, and/or TCRDtargets, followed by DH-JH stems of oligoclonalIGH rearrangements, followed by TCRG targets with full exclusion of oligoclonal IGK and TCRDtargets) would enable successful detection of relapse in 83 (95%) of 87 patients with the currently available MRD-PCR targets (Figure5).
Flow diagram.
Flow diagram for the stepwise selection of MRD-PCR targets, dependent on the choice of techniques for detection and identification ofIGH,IGK, TCRD, andTCRG gene rearrangements.
Flow diagram.
Flow diagram for the stepwise selection of MRD-PCR targets, dependent on the choice of techniques for detection and identification ofIGH,IGK, TCRD, andTCRG gene rearrangements.
The above-presented strategy is based on combined Southern blot and PCR analyses for discrimination between monoclonal and oligoclonal Ig/TCR gene configuration. However, many MRD-PCR laboratories do not routinely perform Southern blotting, implying that they will underestimate the occurrence of oligoclonality in IGH (44% of patients in this series), TCRD (36%), and IGK (8%) genes. In an exclusively PCR-based strategy for MRD target selection, Kde rearrangements should be chosen as first option. When applying all available Kde targets, the detection of relapse would have been possible in 37 patients in our series (43%), but false-negative results would have been obtained in 2 patients. The second choice for target selection should concern IGH gene rearrangements. Using all identified IGH gene rearrangements with patient-specific oligonucleotides positioned at the DH-JHstems should have resulted in detection of relapse in 43 cases (49%), with false-negative results in 2 patients. If the design ofDH-JH oligonucleotides is not successful, one might decide to design VH-DH-JH oligonucleotides supplemented with the usage of PCR-based monoclonal TCRD targets. Finally, successful MRD detection in the remaining 3 patients could have been accomplished by usage of TCRG gene rearrangements (Figure5). Similarly to the combined Southern blot/PCR–based approach, the exclusively PCR-based approach would enable successful detection of relapse in 95% of patients. Thus, the lack of Southern blot information for discrimination between monoclonal and oligoclonal PCR targets might be compensated by monitoring of a higher number ofIGH and TCRD targets (Figure 5).
Both described strategies for selection of MRD-PCR targets have their advantages and limitations, which should be carefully weighed in the context of the facilities and experience of each MRD-PCR laboratory. Nevertheless, each strategy would enable successful detection of relapse in 95% of patients. If one assumes that the actual relapse rate in childhood precursor-B–ALL is 25% to 30%, the current Ig/TCR-based MRD-PCR methodology should be “effective” in 97% to 98% of cases with identifiable MRD-PCR targets at diagnosis.
We are grateful to Prof dr R. Benner and Prof dr D. Sońta-Jakimczyk for their continuous support, Mrs F. J. Weerden for assistance in statistical analyses, Mrs W. M. Comans-Bitter and Mr T. van Os for preparation of the figures, and Mrs J. A. Boon for her secretarial support. We thank the Dutch Childhood Leukemia Study Group for kindly providing precursor-B–ALL cell samples. Board members of the Dutch Childhood Leukemia Study Group are P. J. van Dijken, K. Hählen, W. A. Kamps, E. Th. Korthof, F. A. E. Nabben, A. Postma, J. A. Rammeloo, G. A. M. de Vaan, A. J. P. Veerman, and R. S. Weening.
Supported by the Dutch Cancer Society/Koningin Wilhelmina Fonds (grants SNWLK 97-1567 and SNWLK 2000-2268).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jacques J. M. van Dongen, Dept of Immunology, Erasmus University Rotterdam, PO Box 1738, 3000 DR Rotterdam, The Netherlands; e-mail: vandongen@immu.fgg.eur.nl.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal