Abstract
Signal transducer and activator of transcription (STAT) 5b-retinoic acid receptor (RAR) α is the fifth fusion protein identified in acute promyelocytic leukemia (APL). Initially described in a patient with all-trans retinoic acid (ATRA)–unresponsive disease, STAT5b-RARα resulted from an interstitial deletion on chromosome 17. To determine the molecular mechanisms of myeloid leukemogenesis and maturation arrest in STAT5b-RARα+ APL and its unresponsiveness to ATRA, we examined the effect of STAT5b-RARα on the activity of myeloid transcription factors including RARα/retinoid X receptor (RXR) α, STAT3, and STAT5 as well as its molecular interactions with the nuclear receptor corepressor, SMRT, and nuclear receptor coactivator, TRAM-1. STAT5b-RARα bound to retinoic acid response elements (RAREs) both as a homodimer and as a heterodimer with RXRα and inhibited wild-type RARα/RXRα transactivation. Although STAT5b-RARα had no effect on ligand-induced STAT5b activation, it enhanced interleukin 6–induced STAT3-dependent reporter activity, an effect shared by other APL fusion proteins including promyelocytic leukemia-RARα and promyelocytic leukemia zinc finger (PLZF)–RARα. SMRT was released from STAT5b-RARα/SMRT complexes by ATRA at 10−6 M, whereas TRAM-1 became associated with STAT5b-RARα at 10−7 M. The coiled-coil domain of STAT5b was required for formation of STAT5b-RARα homodimers, for the inhibition of RARα/RXRα transcriptional activity, and for stability of the STAT5b-RARα/SMRT complex. Thus, STAT5b-RARα contributes to myeloid maturation arrest by binding to RARE as either a homodimer or as a heterodimer with RXRα resulting in the recruitment of SMRT and inhibition of RARα/RXRα transcriptional activity. In addition, STAT5b-RARα and other APL fusion proteins may contribute to leukemogenesis by interaction with the STAT3 oncogene pathway.
Introduction
Nonrandom chromosomal translocations play a critical role in the pathogenesis of human blood malignancies.1 Five different chromosomal translocations have been reported and characterized so far in acute promyelocytic leukemia (APL), a disease effectively treated by agents that target the resultant chimeric transcription factor.2-4 In the great majority of patients, there is a specific chromosomal translocation t(15;17)(q22;q21), which involves the PML (promyelocytic leukemia) gene located on chromosome 15 and the RARA(retinoic acid receptor α) gene located on chromosome 17.5,6 The wild-type PML is a component of a nuclear structure referred to as PML nuclear body or POD (PML oncogenic domain). Almost all patients with t(15;17) APL respond well to differentiation therapy with all-trans retinoic acid (ATRA).4 Several variant chromosomal translocations occur in a small subset of APLs. Two variant translocations that result in ATRA-responsive APL are t(5;17)(q35;q21) and t(11;17)(q13;q21), which involve the RARα locus and a known gene NPM(nucleophosmin) on chromosome 57 and NuMA(nuclear mitotic apparatus protein) on chromosome 11,8respectively. One additional variant translocation t(11;17)(q23;q21) has been found that fuses the RARα locus with the PLZF(promyelocytic leukemia zinc finger) gene on chromosome 11q23.9 In contrast to patients with APL with t(15;17), t(5;17), and t(11;17), those with t(11;17)(q23;q21) have a poor response to ATRA.3 Very recently, a fifth fusion geneSTAT5b-RARA has been identified in one patient with ATRA-unresponsive APL.10 This fusion gene occurred as a result of an interstitial deletion within chromosome 17 and represents the first stable chromosomal abnormality described in a malignancy that involves a member of the STAT protein family.
Seven signal transducer and activator of transcription (STAT) proteins have been identified in mammalian cells, STAT1, 2, 3, 4, 5a, 5b, and 6; each is tyrosine-phosphorylated by JAKs following the binding of cytokine to its receptor.11 Thus far, more than 40 different polypeptide ligands have been shown to cause STAT protein activation.11,12 On tyrosine phosphorylation, STAT proteins form homodimers or heterodimers through reciprocal intermolecular interactions involving the SH2 domain of one STAT protein binding to the phosphorylated tyrosine of its partner. Dimerization is followed by rapid translocation to the nucleus, binding to target DNA, and induction of gene expression. STAT5 was originally identified in sheep as a prolactin-induced mammary gland transcription factor.13 STAT3 was originally termed acute-phase response factor (APRF) because it was first identified as a transcription factor that bound to interleukin 6 (IL-6)–responsive elements within the promoters of various acute-phase protein genes.14 STAT5a and STAT5b are encoded by 2 highly homologous genes located in close proximity to each other and to STAT3 on mouse chromosome 11 and human chromosome 17.15-17 STAT protein activation, especially STAT5 and STAT3, has been implicated in cell transformation and carcinogenesis. In addition to their constitutive activation in solid tumors, aberrant activation of STAT5 and STAT3 has been reported in a variety of hematopoietic cancers including acute and chronic myelogenous and lymphocytic leukemias and lymphomas.18
Retinoic acid receptor α is a member of a superfamily of nuclear hormone receptors, which affects many physiologic processes including differentiation and growth arrest of various cell types including hematopoietic cells.19 In normal myeloid cells, RARα dimerizes with retinoid X receptor α (RXRα); the dimers bind to retinoic acid response elements (RAREs) located in promoter/enhancer regions of specific genes. Recent models suggests that in the absence of ligand (retinoic acid, RA), RARα/RXRα heterodimers associate at a 1:1 ratio with nuclear receptor transcriptional repressor complex corepressor (CoR) composed of SMRT/NCoR, Sin3, and histone deacetylase (HDAC) resulting in repression of basal transcription.20,21 Physiologic levels of RA induce dissociation of the CoR complex followed by recruitment of the transcriptional activation complex coactivator (CoA), consisting of CBP/p300, P/CAF, SRC-1, TIF2 (GRIP1/SRC-2), and p/CIP (TRAM-1/ACTR/AIB1/RAC3/SRC-3). Binding of the CoA complex results in the activation of gene expression and normal differentiation.22-24 In ATRA-responsive APL with t(15;17), the PML-RARα fusion protein binds RARE as a homodimer and recruits 2 CoR complexes with higher affinity than wild-type RARα.25,26 Complete dissociation of CoR from PML-RARα does not occur at physiologic levels of RA, but rather requires higher levels of ligand achieved during treatment with ATRA.25Several hypotheses have been proposed to explain ATRA unresponsiveness in some variant APL including altered stoichiometry and stability of the CoR-fusion protein interaction26,27 and resistance of the APL-fusion gene products to ATRA-induced proteolysis.28
The molecular bases for leukemogenesis, arrested differentiation, and ATRA unresponsiveness in STAT5b-RARα+ APL are unknown. To address these issues, we examined the effect of STAT5b-RARα on myeloid leukemogenic and differentiation pathways involving RARα/RXRα, STAT3 and STAT5 as well as the interaction between STAT5b-RARα and the nuclear receptor coregulators, CoR and CoA. We determined that STAT5b-RARα binds RARE as either a homodimer or as a heterodimer with RXRα and modulates the transcriptional activities of RARα/RXRα and STAT3 but not STAT5. STAT5b-RARα was insensitive to ATRA-induced proteolysis in transient expression COS-7 and HeLa cells. Finally, the ability of STAT5b-RARα to enhance STAT3 activity is shared by other APL fusion proteins including PML-RARα and PLZF-RARα and may therefore represent an important new pathway contributing to leukemogenesis in APL.
Materials and methods
Cell lines and reagents
COS-7, 293T, HepG2, and HeLa cells were cultured in Dulbecco modified Eagle medium (DMEM; Life Technologies, Grand Island, NY) with 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 U/mL). Human IL-2 and IL-6 were purchased from Roche Molecular Biochemicals (Indianapolis, IN) and R & D Systems (Minneapolis, MN), respectively. ATRA was obtained from Sigma (St Louis, MO) and the ovine prolactin (NIDDK-oPRL-20) was obtained from National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; Bethesda, MD).
Plasmids
The PML-RARα, PLZF–RARα, PLZF, RARα, and RXRα expression vectors were described previously.29-31 The (RARE)3-tk-luciferase reporter5 and the APRE-luciferase reporter construct,32 which has 4 copies of acute phase response elements (APREs) were kindly provided by Dr A. Dejean (Paris, France) and Dr I. Matsumura (Osaka, Japan), respectively. The β-casein luciferase reporter gene is from β-casein gene promoter region (−2300 to +490; a gift from Dr J. Rosen, Houston, TX).33 The human expression vectors for STAT5b, IL-2–receptor (IL-2R) β chain and γ common chain were provided by Dr W. J. Leonard (National Institutes of Health [NIH], Bethesda, MD).16,34-36 The human JAK3 and the Nb2 PRL-R expression vector are from Dr O'Shea (NIH) and Dr L. Yu-Lee (Houston, TX), respectively. GST-TRAM-1 (residues 577-821) in pGEX-5X-2 and GST-SMRT (residues 983-1172) in pGEX-5X-1 both containing the receptor interaction domain were produced by polymerase chain reaction (PCR) with pfu DNA polymerase (Stratagene, La Jolla, CA) using TRAM-1 expression vector (kindly provided by Dr W. W. Chin, Boston, MA)24 and SMRT expression vector20 as template, respectively. The STAT5b-RARα (in pSG5 vector) (Stratagene) expression vectors was constructed by fusing human STAT5b16 and RARα using fusion-PCR technology37 based on the published sequence.10 A similar PCR approach also is applied to construct a series of mutant STAT5b-RARαs, STAT5b-RARα(ΔN), STAT5b-RARα(ΔCC), and STAT5b-RARα(ΔDBD). All plasmid constructs were confirmed by DNA sequencing and by in vitro translation and immunoblotting.
Cell transfections
For transient transfections, COS-7 and 293T cells were grown in 6-well (35-mm diameter) tissue culture plates to 50% to 80% confluence. Twelve hours later, the cells were transiently transfected with the indicated expression vectors and reporter genes by standard calcium phosphate coprecipitation method.38 The amounts of plasmid DNA used per well were 1 μg reporter vector, 1 to 4 μg expression vector, and 1 μg β-galactosidase expression vector (Promega, Madison, WI) as transfection control. For HepG2 and HeLa cell, the GeneJuice transfection reagent (Novagen, Madison, WI) was used according to the manufacturer's instruction. Luciferase activity was measured in a luminometer (Luminoskan Ascent, Labsystems, Franklin, MA), expressed in arbitrary units and normalized according to the internal control. Each point is the mean of at least 3 independent experiments.
In vitro translation
The TNT-coupled rabbit reticulocyte lysate (Promega) system was used for in vitro translated proteins according to the manufacturer's instructions. The relative quantity of in vitro translated proteins was estimated as described.30 31 Briefly, parallel translation reactions were performed in the presence of [α-35S] methionine (NEN, Boston, MA) and the proteins were visualized by autoradiography after separation on a 10% sodium dodecyl sulfate-polyacrylamide gel.
Gel-shift DNA-binding assays
The 293T, COS-7 and HepG2 cell lines were transiently transfected in 6-well plates using 2 to 4 μg plasmids. Forty-eight hours later, cells were either not treated or treated for 30 minutes with cytokines, IL-2 (50 ng/mL), prolactin (500 ng/mL), IL-6 (25 ng/mL), and the whole cell extracts (WCEs) were prepared as reported previously.39 About 20 μg WCE was incubated with duplex oligonucleotides in binding buffer (13 mM Hepes, pH 8.0, 65 mM NaCl, 1 mM DTT, 0.14 mM EDTA, 8% glycerol) and separated on 5% polyacrylamide gels. Duplex oligonucleotide probes included the high-affinity serum-inducible element (hSIE), prolactin response element (PRE) contained within the β-casein promoter,13 and the APRE, an IL-6 response element within the rat α2-macroglobulin promoter.32 For supershift, 1 μg antibody was included in this reaction system when needed. Gel shift assays using in vitro protein were performed as described previously.30 Briefly, in vitro translated proteins were preincubated for 15 minutes at room temperature in the following buffer: 20 mM Hepes, pH 7.4, 50 mM KCl, 1 mM 2-mercaptoethanol, 10% glycerol, 1 μg poly (dI-dC) (Pharmacia), and 100 μg bovine serum albumin (BSA). [γ-32P] adenosine triphosphate (ATP; NEN) end-labeled duplex oligonucleotide probe was added and samples incubated at room temperature for 30 minutes and at 4°C for 30 minutes. Protein-DNA complexes were separated on 6% polyacrylamide gels equilibrated in 0.25 times tris-borate-EDTA buffer (TBE). Gels were dried, exposed to PhosphorImager plates and images developed and quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and ImageQuant software.
In vitro protein-binding assays
The SMRT protein was expressed in Escherichia coliDH5α as a GST fusion product and purified by standard methodology. Twelve microliters of α-35S] methionine-labeled in vitro translated proteins (STAT5b-RARα or STAT5b-RARα [ΔCC]) was incubated with 1 μg GST-SMRT fusion protein conjugated to glutathione Sepharose (Amersham-Pharmacia Biotech, Piscataway, NJ) in binding buffer (20 mM Tris, pH 8.0, 150 mM KCl, 1 mM EDTA, 4 mM MgCl2, 0.2% NP-40, 10% glycerol) at 4°C for 2 hours without or with ATRA (Sigma). Bound proteins were washed 3 times with binding buffer, eluted by boiling in sample buffer, and resolved by 10% SDS-polyacrylamide gel electrophoresis (PAGE). Gels were dried, exposed, and analyzed by PhosphorImager. For in vitro coimmunoprecipitation, in vitro translated SMRT protein was incubated with 35S-labeled proteins, PLZF, RARα, and STAT5b, respectively, in binding buffer for 1 hour at 4°C. Immune complexes were isolated by further incubation with SMRT antibody presorbed on protein A/G (Santa Cruz Biotechnology, Santa Cruz, CA), washed 3 times in binding buffer and analyzed by SDS-PAGE.
Immunoblotting
Whole cell lysates were prepared in lysis buffer (20 mM Hepes, pH 7.9, 420 mM NaCl, 20 mM NaF, 1 mM Na3VO4, 1 mM Na4P2O7, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol [DTT], 20% glycerol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). Equivalent amounts of total cellular protein were electrophoresed on 7.5% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membrane (Millipore). Probing of PVDF membranes with primary antibodies and detection of horseradish peroxidase-conjugated secondary antibodies by enhanced chemiluminescence as directed (Amersham-Pharmacia Biotech). Antibodies used in this study are as follows: antihuman-RARα (C-20) antihuman-STAT5b (N-20), antihuman-STAT5b (C-17), and antihuman-SMRT (N-20), all from Santa Cruz Biotechnology, and antihuman-β-actin (monoclonal; Sigma).
Results
STAT5b-RARα binds RARE as a homodimer and preferentially as a heterodimer with RXRα
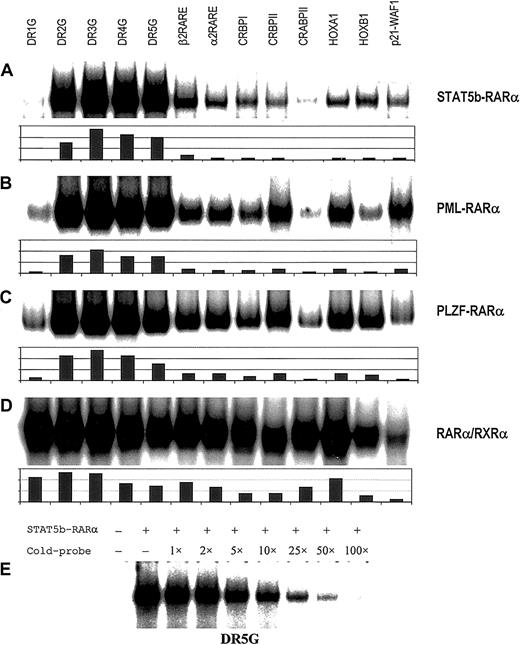
The APL fusion proteins previously identified and characterized contribute to leukemogenesis by binding to RAREs either as homodimers or heterodimers with RXRα, thereby repressing gene transcription essential for myeloid differentiation.3 To determine if STAT5b-RARα is capable of binding to RAREs, in vitro translated STAT5b-RARα protein was examined by gel-shift assay using a series of RARE duplex oligonucleotides. As shown in Figure1A, STAT5b-RARα alone bound to all RAREs tested and this binding could be competitively inhibited by 100-fold excess unlabeled RARE (Figure 1E). The shifted band corresponded to a homodimer of STAT5b-RARα with migration characteristics similar to PML-RARα and PLZF-RARα using RARα/RXRα as a size reference (Figure2A). The STAT5b-RARα homodimer binding preferences overall are similar to PML-RARα and PLZF-RARα (Figure 1A-C).30,31 40 However, a few slight differences in binding preferences were observed: STAT5b-RARα and PLZF-RARα bound to RARE-p21-WAF less efficiently than PML-RARα, and STAT5b-RARα bound to CRBPII and HOXA1 less efficiently than either PLZF-RARα or PML-RARα.
RARE binding by homodimers of APL fusion proteins.
Binding of homodimers of STAT5b-RARα (A), PML-RARα (B), and PLZF-RARα (C) and heterodimers of RARα/RXRα (D) to a series of RAREs. Gel-shift assays were performed with in vitro translated proteins using the following radiolabeled probes: synthetic RARE, DR1, 2, 3, 4 and 5G,40 β2 RARE (DR5T) from the human RARB gene,30 α2 RARE from the human RARA gene,59 the natural enhancer elements from the rat cellular retinal-binding protein type I gene (CRBPI),60 the rat cellular retinal-binding protein type II gene (CRBPII),61 the murine cellular RA-binding protein gene (CRABPII),62HOXA1 gene,63HOXB1gene64 and p21-WAF1gene.65 Equivalent amounts of in vitro translated protein and labeled oligonucleotide were added to each binding reaction. The results of quantitative analysis using ImageQuant software are shown below each autoradiogram. In panel E, radiolabeled DR5G was incubated with in vitro translated STAT5b-RARα and the indicated fold excess unlabeled DR5G followed by gel-shift assay.
RARE binding by homodimers of APL fusion proteins.
Binding of homodimers of STAT5b-RARα (A), PML-RARα (B), and PLZF-RARα (C) and heterodimers of RARα/RXRα (D) to a series of RAREs. Gel-shift assays were performed with in vitro translated proteins using the following radiolabeled probes: synthetic RARE, DR1, 2, 3, 4 and 5G,40 β2 RARE (DR5T) from the human RARB gene,30 α2 RARE from the human RARA gene,59 the natural enhancer elements from the rat cellular retinal-binding protein type I gene (CRBPI),60 the rat cellular retinal-binding protein type II gene (CRBPII),61 the murine cellular RA-binding protein gene (CRABPII),62HOXA1 gene,63HOXB1gene64 and p21-WAF1gene.65 Equivalent amounts of in vitro translated protein and labeled oligonucleotide were added to each binding reaction. The results of quantitative analysis using ImageQuant software are shown below each autoradiogram. In panel E, radiolabeled DR5G was incubated with in vitro translated STAT5b-RARα and the indicated fold excess unlabeled DR5G followed by gel-shift assay.
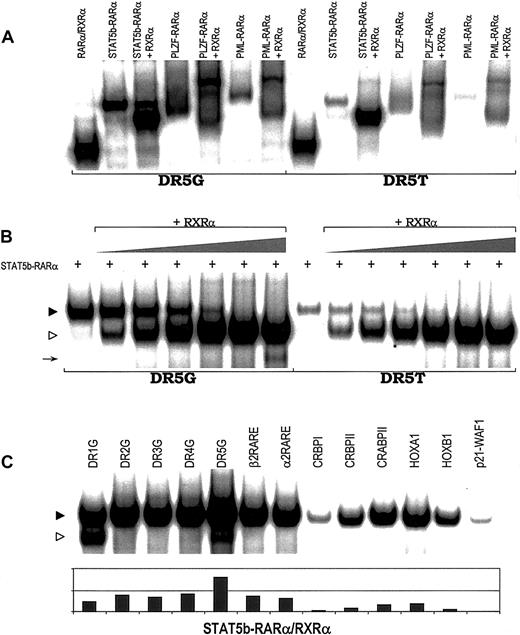
RARE binding by heterodimers of APL fusion proteins.
(A) Binding of heterodimers containing RXRα and STAT5b-RARα, PLZF-RARα, PML-RARα, or wild-type RARα to 2 RAREs, DR5G and DR5T. (B) STAT5b-RARα/RXRα heterodimer formation and RARE binding with increasing concentration of RXRα. In vitro translated STAT5b-RARα protein (2.0 μL) was incubated without or with in vitro translated RXRα protein (0, 0.05, 0.1, 0.2, 0.5, 1.0, and 2.5 μL). Gel-shift assays were performed using 2 RAREs, DR5G and DR5T. The location of the STAT5b-RARα homodimer band is indicated by the filled triangle; the location of the STAT5b-RARα/RXRα heterodimer composed of one molecule of STAT5b-RARα plus one molecule of RXRα is indicated by the unfilled triangle. The arrow indicates the position of the RXRα homodimer binding to DR5G. (C) STAT5b-RARα/RXRα heterodimer can bind a series of RAREs. STAT5b-RARα/RXRα heterodimer is indicated by the filled triangle; RXRα homodimer is indicated by the unfilled triangle.
RARE binding by heterodimers of APL fusion proteins.
(A) Binding of heterodimers containing RXRα and STAT5b-RARα, PLZF-RARα, PML-RARα, or wild-type RARα to 2 RAREs, DR5G and DR5T. (B) STAT5b-RARα/RXRα heterodimer formation and RARE binding with increasing concentration of RXRα. In vitro translated STAT5b-RARα protein (2.0 μL) was incubated without or with in vitro translated RXRα protein (0, 0.05, 0.1, 0.2, 0.5, 1.0, and 2.5 μL). Gel-shift assays were performed using 2 RAREs, DR5G and DR5T. The location of the STAT5b-RARα homodimer band is indicated by the filled triangle; the location of the STAT5b-RARα/RXRα heterodimer composed of one molecule of STAT5b-RARα plus one molecule of RXRα is indicated by the unfilled triangle. The arrow indicates the position of the RXRα homodimer binding to DR5G. (C) STAT5b-RARα/RXRα heterodimer can bind a series of RAREs. STAT5b-RARα/RXRα heterodimer is indicated by the filled triangle; RXRα homodimer is indicated by the unfilled triangle.
Very intriguingly, gel-shift assays containing both STAT5b-RARα and RXRα resulted in the appearance of an additional prominent band (STAT5b-RARα/RXRα heterodimer) with mobility characteristics intermediate between RARα/RXRα heterodimer and STAT5b-RARα homodimer (Figure 2A). Increasing the amount of RXRα while keeping the amount of STAT5b-RARα constant (Figure 2B) resulted in the disappearance of STAT5b-RARα homodimer band and increased the prominence of the STAT5b-RARα/RXRα heterodimer band suggesting that STAT5b-RARα prefers to bind RARE as a heterodimer with RXRα (one molecule of STAT5b-RARα plus one molecule of RXRα) rather than as a homodimer. We and others have demonstrated previously30,31 40 and confirmed in this study (Figure 2A and data not shown) that when PML-RARα or PLZF-RARα binds RARE in combination with RXRα, each does so as a single heterodimer as well as a higher order multimeric complex. In contrast, STAT5b-RARα and RXRα bind RAREs virtually exclusively as a single heterodimer over a wide range of ratios and irrespective of the RARE (Figure 2C).
The STAT5b coiled-coil domain is responsible for STAT5b-RARα homodimer formation and inhibition of RARα/RXRα-mediated transcriptional activity
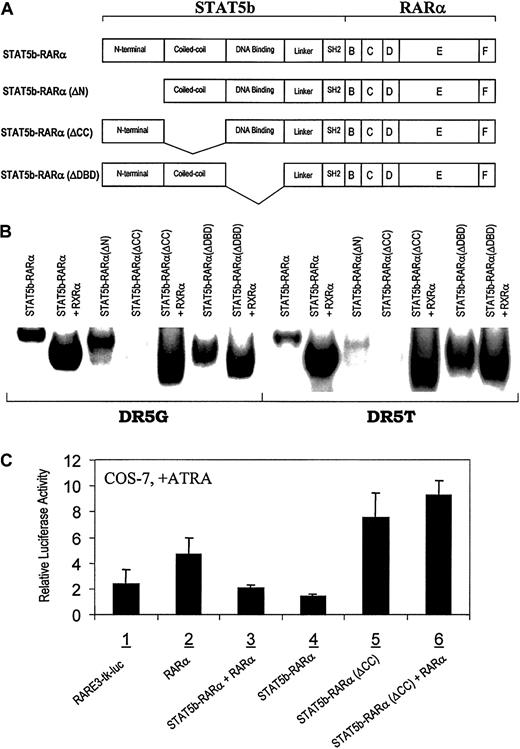
STAT5b-RARα retains 3 complete domains of STAT5b, the N-terminal oligomerization domain, the coiled-coil domain, and the DNA-binding domain, in addition to a truncated SH2 domain (Figure3A). The N-terminal and coiled-coil domains each have been demonstrated to mediate protein-protein interactions.11,41 To investigate whether or not either of these 2 domains or the DNA binding domain is important for the oncogenic functions of STAT5b-RARα, we compared the activities and functions of wild-type STAT5b-RARα with mutants of STAT5b-RARα in which the N-terminal, coiled-coil, or DNA-binding domain was deleted (Figure 3A). In gel-shift assays, STAT5b-RARα(ΔN), STAT5b-RARα(ΔDBD), and STAT5b-RARα(Δlinker and part of SH2) each bound RARE alone as a homodimer or as a heterodimer with RXRα (Figure 3B and data not shown). In contrast, STAT5b-RARα(ΔCC) could not bind RARE as a homodimer, but rather it bound RARE only as a heterodimer with RXRα. These findings indicate that the coiled-coil domain of STAT5b, and not the N-terminal or DNA-binding domains, is important for STAT5b-RARα homodimer formation. These results are reminiscent of those for PML-RARα in which the coiled-coil domain of PML was found to be responsible for the formation of PML-RARα homodimers.40,42 43
Requirement of the coiled-coil domain of STAT5b-RARα for RARE binding and transcription activation.
(A) Schematic illustration of wild-type and mutated STAT5b-RARα constructs. (B) Gel-shift assays with wild-type and mutant STAT5b-RARα using 2 RAREs, DR5G and DR5T. (C) Transactivational activities of wild-type and mutant STAT5b-RARα in COS-7 cells. Following transfection with the indicated constructs, cells were cultured in medium with 10−6 M ATRA for 24 hours. Luciferase activity was normalized for transfection efficiency using a β-galactosidase reporter plasmid. The results presented are the mean ± SD of triplicate wells and are representative of 3 separate experiments.
Requirement of the coiled-coil domain of STAT5b-RARα for RARE binding and transcription activation.
(A) Schematic illustration of wild-type and mutated STAT5b-RARα constructs. (B) Gel-shift assays with wild-type and mutant STAT5b-RARα using 2 RAREs, DR5G and DR5T. (C) Transactivational activities of wild-type and mutant STAT5b-RARα in COS-7 cells. Following transfection with the indicated constructs, cells were cultured in medium with 10−6 M ATRA for 24 hours. Luciferase activity was normalized for transfection efficiency using a β-galactosidase reporter plasmid. The results presented are the mean ± SD of triplicate wells and are representative of 3 separate experiments.
Previously, PML-RARα and PLZF-RARα were demonstrated to have a dominant-negative effect on wild-type RARα/RXRα transcriptional activity.3,5 29 To determine whether or not STAT5b-RARα behaves similarly, we examined the effect of STAT5b-RARα on RA-dependent, RARα/RXRα-mediated transactivation of the RARE3-tk-luc reporter gene construct (Figure 3C). Transfection of the reporter construct alone or with RARα resulted in RA-dependent transactivation. Transfection with STAT5b-RARα alone or with RARα inhibited RA-dependent transactivation. Transfection of STAT5b-RARα(ΔCC) did not inhibit but rather augmented RA-dependent RARα/RXRα-mediated transcriptional activity similar to RARα, indicating that inhibition of RARα/RXRα activity requires the coiled-coil domain of STAT5b-RARα and that in the absence of its coiled-coil domain, STAT5b-RARα functions like wild-type RARα.
Effects of ATRA on the interaction of STAT5b-RARα with CoR SMRT and CoA TRAM-1 and on STAT5b-RARα protein degradation
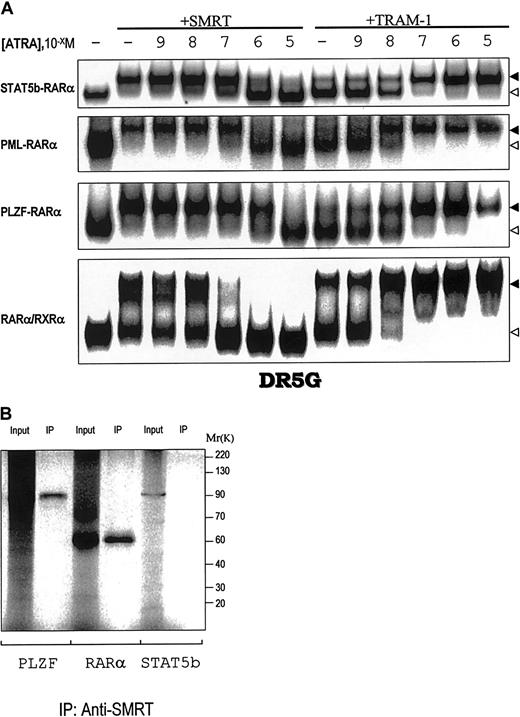
RARα/RXRα and the APL fusion proteins PML-RARα and PLZF-RARα suppress transcription by associating with CoR and CoA depending on the concentration of ATRA.3 To determine the ATRA concentration dependence of the interactions of STAT5b-RARα with CoR and CoA, we examined the composition of complexes containing STAT5b-RARα, GST-SMRT, and GST-TRAM-1 in varying concentrations of ATRA (Figure 4A). SMRT dissociated from STAT5b-RARα at pharmacologic concentrations of ATRA (10−6 M) similar to that required for its dissociation from PML-RARα. This ATRA concentration is one log greater than that required to cause SMRT dissociation from RARα/RXRα (10−7 M) and one log lower than that required for SMRT dissociation from PLZF-RARα (10−5 M). Immunoprecipitation assays (Figure 4B) demonstrated that although SMRT can form a complex with PLZF and RARα, as shown previously,25 it does not bind STAT5b. Complete recruitment of the CoA TRAM-1 to STAT5b-RARα occurred at 10−7 M ATRA similar to that for PLZF-RARα (Figure 4A), but one log greater that that for PML-RARα and RARα/RXRα (10−8 M).
Interactions of STAT5b-RARα, PML-RARα, and PLZF-RARα and wild-type RARα/RXRα with the CoR SMRT and CoA TRAM-1.
(A) The in vitro translated proteins indicated on the left were incubated with or without SMRT or TRAM-1 as indicated and the radiolabeled RARE, DR5G. The location of the complex containing the chimeric or wild-type receptor plus SMRT/TRAM-1 is indicated by the solid triangle; the location of the chimeric or wild-type receptor alone is indicated by the open triangle. (B) SDS-PAGE and autoradiography were performed on in vitro translated and35S-methionine–labeled PLZF, RARα, or STAT5b alone or following incubation with SMRT and immunoprecipitation with SMRT antibody as indicated.
Interactions of STAT5b-RARα, PML-RARα, and PLZF-RARα and wild-type RARα/RXRα with the CoR SMRT and CoA TRAM-1.
(A) The in vitro translated proteins indicated on the left were incubated with or without SMRT or TRAM-1 as indicated and the radiolabeled RARE, DR5G. The location of the complex containing the chimeric or wild-type receptor plus SMRT/TRAM-1 is indicated by the solid triangle; the location of the chimeric or wild-type receptor alone is indicated by the open triangle. (B) SDS-PAGE and autoradiography were performed on in vitro translated and35S-methionine–labeled PLZF, RARα, or STAT5b alone or following incubation with SMRT and immunoprecipitation with SMRT antibody as indicated.
The coiled-coil domain of PML-RARα APL fusion proteins contributes to the stability of the APL fusion protein/SMRT complex.26 To determine if this is the case for STAT5b-RARα, we examined the stability of the binding of in vitro translation proteins STAT5b-RARα and STAT5b-RARα(ΔCC) with GST-SMRT in a GST pull-down assay under varying concentrations of ATRA (Figure5A,B). STAT5b-RARα dissociated from GST-SMRT beginning at 10−6 M ATRA, whereas STAT5b-RARα(ΔCC) dissociated from GST-SMRT beginning at 10−7 M ATRA, one log lower. These findings indicate that the coiled-coil domain plays an important role in the stability of STAT5b/SMRT-RARα complexes.
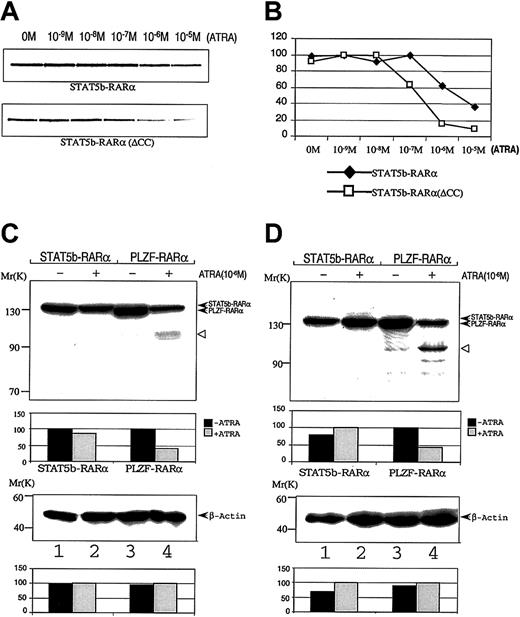
Effect of ATRA on interactions STAT5b-RARα with GST-SMRT and on STAT5b-RARα stability.
Radiolabeled wild-type or mutant STAT5b-RARα was incubated with GST-SMRT in the presence of the indicated concentrations of ATRA. After absorption with glutathione Sepharose, the proteins were separated and analyzed by autoradiography (A) and PhosphorImager analysis (B). COS-7 cells (C) and HeLa cells (D) were transiently transfected with STAT5b-RARα or PLZF-RARα and incubated without (lanes 1 and 3) or with (lanes 2 and 4) 10−6 M ATRA for 24 hours. Cells were lysed; proteins were separated by SDS-PAGE and immunoblotted with RARα antibody (upper panel) as well as β-actin antibody (bottom panel), which was used as loading control. Densitometry analysis is presented below each immunoblot. The results shown are representative of up to 3 separate experiments. The empty triangle to the right of each upper panel indicates presumed proteolytic fragments of PLZF-RARα.
Effect of ATRA on interactions STAT5b-RARα with GST-SMRT and on STAT5b-RARα stability.
Radiolabeled wild-type or mutant STAT5b-RARα was incubated with GST-SMRT in the presence of the indicated concentrations of ATRA. After absorption with glutathione Sepharose, the proteins were separated and analyzed by autoradiography (A) and PhosphorImager analysis (B). COS-7 cells (C) and HeLa cells (D) were transiently transfected with STAT5b-RARα or PLZF-RARα and incubated without (lanes 1 and 3) or with (lanes 2 and 4) 10−6 M ATRA for 24 hours. Cells were lysed; proteins were separated by SDS-PAGE and immunoblotted with RARα antibody (upper panel) as well as β-actin antibody (bottom panel), which was used as loading control. Densitometry analysis is presented below each immunoblot. The results shown are representative of up to 3 separate experiments. The empty triangle to the right of each upper panel indicates presumed proteolytic fragments of PLZF-RARα.
Exposure to ATRA induces intracellular degradation of the PML-RARα and PLZF-RARα fusion proteins.44,45 We examined whether or not ATRA exposure similarly resulted in degradation of STAT5b-RARα. COS-7 and HeLa cells were transiently transfected with expression constructs containing STAT5b-RARα or PLZF-RARα and the cells incubated in 10−6 M ATRA for 24 hours. As previously described,44 PLZF-RARα protein levels sharply decreased in cells incubated with ATRA (Figure 5C,D). In contrast, the level of STAT5b-RARα protein in cells incubated with ATRA was virtually unchanged.
STAT5b-RARα enhances STAT3 transcriptional activity but has no effect on STAT5 transcriptional activity
Aberrant STAT3 activation has been demonstrated to occur in human leukemias and lymphomas,18 to be critical for v-Src transformation,46,47 and alone to be able to transform mouse and rat fibroblasts.41 To examine whether or not STAT5b-RARα modulated STAT3 activity, we used the HepG2 cell line in which the IL-6 receptor signaling pathway is intact together with an APRE-luciferase reporter gene construct.32 IL-6 exposure of HepG2 cells increased APRE-luciferase reporter gene activity 120-fold through the activation of endogenous STAT3 (Figure6A and data not shown). This activation was inhibited 75% by cotransfection of HepG2 cells with STAT5b (Figure6A, lane 2). Cotransfection of HepG2 cells with STAT5b-RARα augmented IL-6–induced reporter-construct activity 8-fold (Figure 6A, lane 3). In contrast, cotransfection with RARα had minimal effect. To assess whether or not the ability of STAT5b-RARα to augment STAT3 transcriptional activity is limited to STAT5b-RARα, we examined other APL fusion proteins in a similar fashion in HepG2 cells (Figure 6A, lanes 5 and 6). In addition to STAT5b-RARα, both PML-RARα and PLZF-RARα enhanced IL-6–mediated STAT3 transcriptional activity 8- to 26-fold.
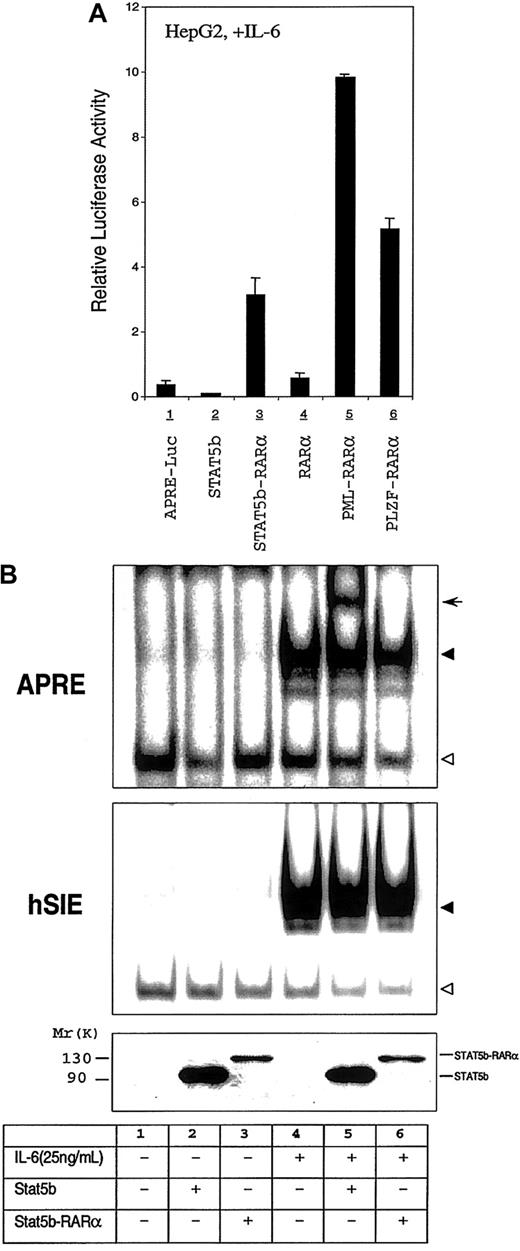
Effect of STAT5b-RARα and other APL fusion proteins on the STAT3 transcriptional and DNA binding activity.
(A) Transactivation activities of STAT5b, STAT5b-RARα, RARα, PML-RARα, and PLZF-RARα in HepG2 cells. HepG2 cells were transiently transfected with 500 ng of APRE-luciferase reporter gene, 500 ng of β-galactosidase expression vector, and 2.0 μg STAT5b, STAT5b-RARα, RARα, PML-RARα, and PLZF-RARα. Transfected cells were stimulated with IL-6 (25 ng/mL) for 24 hours. Luciferase activity was measured and normalized for transfection efficiency using a β-galactosidase reporter construct. Data represent the mean ± SD of 5 separate experiments. The luciferase activity shown in lane 1 is increased 120-fold over the activity of identical cells incubated without IL-6 (not shown). (B) Gel-shift assays with WCEs from HepG2 cells transiently transfected with STAT5b or STAT5b-RARα. Transfected cells were incubated without or with IL-6 (25 ng/mL) for 30 minutes. The APRE (upper panel) and the hSIE (middle panel) were used as duplex oligonucleotide probes in this study. The location of the specific STAT3/DNA complex (upper panel and middle panel) and STAT5b/DNA (upper panel, lane 5) are indicated by the solid triangle and arrow, respectively; the empty triangle indicates the location of a nonspecific band. Levels of protein expression in the transiently transfected cells were determined by immunoblotting with the antibody against the N-terminal region of STAT5b (bottom panel).
Effect of STAT5b-RARα and other APL fusion proteins on the STAT3 transcriptional and DNA binding activity.
(A) Transactivation activities of STAT5b, STAT5b-RARα, RARα, PML-RARα, and PLZF-RARα in HepG2 cells. HepG2 cells were transiently transfected with 500 ng of APRE-luciferase reporter gene, 500 ng of β-galactosidase expression vector, and 2.0 μg STAT5b, STAT5b-RARα, RARα, PML-RARα, and PLZF-RARα. Transfected cells were stimulated with IL-6 (25 ng/mL) for 24 hours. Luciferase activity was measured and normalized for transfection efficiency using a β-galactosidase reporter construct. Data represent the mean ± SD of 5 separate experiments. The luciferase activity shown in lane 1 is increased 120-fold over the activity of identical cells incubated without IL-6 (not shown). (B) Gel-shift assays with WCEs from HepG2 cells transiently transfected with STAT5b or STAT5b-RARα. Transfected cells were incubated without or with IL-6 (25 ng/mL) for 30 minutes. The APRE (upper panel) and the hSIE (middle panel) were used as duplex oligonucleotide probes in this study. The location of the specific STAT3/DNA complex (upper panel and middle panel) and STAT5b/DNA (upper panel, lane 5) are indicated by the solid triangle and arrow, respectively; the empty triangle indicates the location of a nonspecific band. Levels of protein expression in the transiently transfected cells were determined by immunoblotting with the antibody against the N-terminal region of STAT5b (bottom panel).
To begin to explore the mechanism of the enhanced STAT3 transcriptional activity, we assessed STAT3 DNA binding activity and Ser727 phosphorylation status in extracts of transfected versus nontransfected HepG2 cells. Cotransfection of STAT5b or STAT5b-RARα did not affect IL-6–stimulated binding to APRE or hSIE nor did it affect levels of Ser727 phosphorylation (Figure 6B and data not shown).
Gene deletion mouse models48 and studies of dominant negative mutant constructs in cell lines49 support an important role for STAT5b in myeloid cell development. Consequently, the leukemogenic effect of STAT5b-RARα could also be mediated, in part, through its interference with normal STAT5b function. To investigate this possibility, we examined the effect of STAT5b-RARα on STAT5 activity downstream of the prolactin and IL-2Rs following reconstitution of these signaling pathways in COS-7 and 293T cells (Figure 7A and 7C). Transactivation of a reporter construct containing a β-casein promoter was increased following transient transfection of prolactin receptor–reconstituted COS-7 and IL-2R–reconstituted 293T cells with STAT5b. Transient transfection of these cells with STAT5b-RARα, however, did not increase reporter construct activation nor did it inhibit the augmentation seen with transient overexpression of STAT5b. Similar results were obtained in IL-2R–reconstituted COS-7 cells (data not shown). In addition, transient coexpression of the fusion protein did not affect STAT5 DNA binding activity induced by prolactin in prolactin receptor–reconstituted COS-7 cells (Figure 7B) or by IL-2 in IL-2R–reconstituted COS-7 cells (Figure 7D).
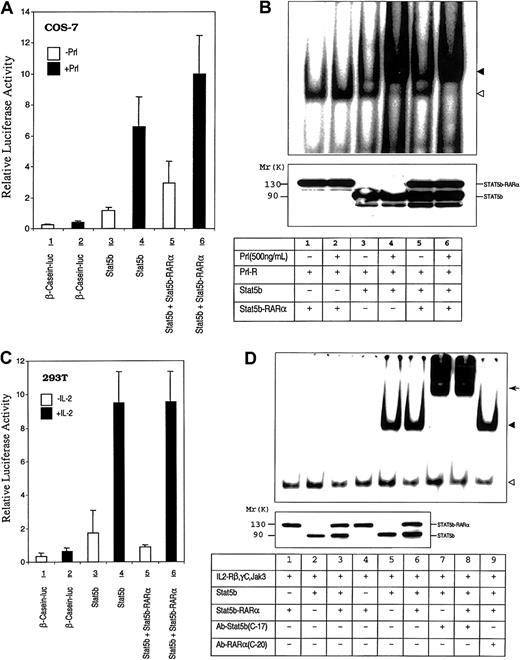
Effect of STAT5b-RARα on STAT5b transcriptional and DNA binding activity.
(A) COS-7 cells were transiently transfected with the human prolactin receptor, a β-casein–luciferase reporter gene construct, and STAT5b with or without STAT5b-RARα. Transfected cells were stimulated without or with ovine prolactin (500 ng/mL) for 24 hours. Luciferase activity was measured and normalized for transfection efficiency using a β-galactosidase reporter construct. Data presented represent the mean ± SD of 3 separate experiments. (B) Gel-shift assays were performed using the PRE and WCEs of transiently transfected COS-7 cells that were incubated without or with prolactin (500 ng/mL) for 30 minutes. The location of the specific STAT5b/DNA complex is indicated with the solid triangle; a nonspecific band is indicated with the empty triangle. The level of protein expression, determined by immunoblotting with antibody against STAT5b, is shown in the bottom panel. (C) 293T cells were transiently transfected with the human β-casein–luciferase reporter gene construct and expression constructs for IL-2Rβ, γC, Jak3, and STAT5b with or without STAT5b-RARα. Transfected cells were incubated without or with IL-2 (50 ng/mL) for 24 hours. Luciferase activity was measured and normalized for transfection efficiency using a β-galactosidase reporter construct. Data presented represent the mean ± SD of 4 separate experiments. (D) Gel-shift assays were performed using the PRE and WCEs from IL-2R reconstituted and transiently transfected COS-7 cells that were incubated without or with IL-2 (50 ng/mL) for 30 minutes. The location of the specific STAT5b/DNA complex is indicated with the solid triangle; a nonspecific band is indicated with the empty triangle. The arrow indicates the position of the supershifted band. Levels of protein expression, determined by immunoblotting with antibody against STAT5b, are shown in the bottom panel for lanes 1 through 6.
Effect of STAT5b-RARα on STAT5b transcriptional and DNA binding activity.
(A) COS-7 cells were transiently transfected with the human prolactin receptor, a β-casein–luciferase reporter gene construct, and STAT5b with or without STAT5b-RARα. Transfected cells were stimulated without or with ovine prolactin (500 ng/mL) for 24 hours. Luciferase activity was measured and normalized for transfection efficiency using a β-galactosidase reporter construct. Data presented represent the mean ± SD of 3 separate experiments. (B) Gel-shift assays were performed using the PRE and WCEs of transiently transfected COS-7 cells that were incubated without or with prolactin (500 ng/mL) for 30 minutes. The location of the specific STAT5b/DNA complex is indicated with the solid triangle; a nonspecific band is indicated with the empty triangle. The level of protein expression, determined by immunoblotting with antibody against STAT5b, is shown in the bottom panel. (C) 293T cells were transiently transfected with the human β-casein–luciferase reporter gene construct and expression constructs for IL-2Rβ, γC, Jak3, and STAT5b with or without STAT5b-RARα. Transfected cells were incubated without or with IL-2 (50 ng/mL) for 24 hours. Luciferase activity was measured and normalized for transfection efficiency using a β-galactosidase reporter construct. Data presented represent the mean ± SD of 4 separate experiments. (D) Gel-shift assays were performed using the PRE and WCEs from IL-2R reconstituted and transiently transfected COS-7 cells that were incubated without or with IL-2 (50 ng/mL) for 30 minutes. The location of the specific STAT5b/DNA complex is indicated with the solid triangle; a nonspecific band is indicated with the empty triangle. The arrow indicates the position of the supershifted band. Levels of protein expression, determined by immunoblotting with antibody against STAT5b, are shown in the bottom panel for lanes 1 through 6.
Discussion
To understand the contribution to the pathogenesis of APL of a chromosomal abnormality resulting in a new RARα-containing fusion protein, it is necessary to evaluate several potential consequences of the abnormality including the effect of the resultant fusion protein on RARα function, the effect of the fusion protein on the function of the normal allele of the fusion partner, and the effect of the reciprocal fusion protein.3
In the studies outlined in this report, we demonstrated that STAT5b-RARα can bind RARE as a homodimer and can recruit SMRT. SMRT remained bound to STAT5b-RARα at physiologic concentrations of ATRA. Because STAT5b by itself did not bind SMRT, these findings suggest that the STAT5b portion of the fusion protein confers an allosteric change in the RARα portion of the fusion protein, similar to the non-RARα portions of other APL fusion proteins, thereby increasing its affinity for SMRT.3 This alone or together with the potential for each homodimer of STAT5b-RARα to bind 2 molecules of SMRT may result in superrepression of gene transcription by RAR/RXR.26,50 In addition to binding RARE as a homodimer, STAT5b-RARα preferentially bound RARE as a heterodimer with RXRα. This feature is unique for STAT5b-RARα among APL fusion proteins and may contribute to its oncogenic potential by sequestering RXRα and other essential transcription cofactors. The effect of the interstitial deletion and creation of the STAT5b-RARα on one allele of STAT5b has the effect of reducing by one half the amount of STAT5b expression. This by itself, however, would not be expected to affect myeloid development because deletion of both alleles of STAT5b in mice did not affect the myeloid lineage51 presumably because of its functional redundancy in myelopoiesis shared with STAT5a. The STAT5b portion of the fusion protein contains a truncated SH2 that includes the phosphotyrosine binding pocket.39 Consequently, STAT5b-RARα might retain the ability to bind activated STAT5a/b and act as a dominant negative. Our findings, however, that STAT5b-RARα did not affect ligand-induced STAT5 transcriptional activity or DNA binding activity do not support this possibility. Finally, because STAT5b-RARα was the result of an interstitial deletion, no reciprocal fusion protein was created.
Recent evidence suggests that dimerization of PML-RARα is critically important for its oncogenic activity including inhibition of RA-mediated myeloid differentiation.26,42,43,50 Deletion of the coiled-coil domain within the PML portion of PML-RARα abrogated dimerization and relieved the inhibitory effects of the fusion protein on RA-induced differentiation.26,40 43Similarly, we have demonstrated that the coiled-coil domain within the STAT5b portion of STAT5b-RARα is required for homodimerization in the presence of RARE, for its inhibitory effect on RARα/RXRα transcriptional activity in COS-7 cells, and for its ability to stably interact with SMRT.
Our results demonstrated that STAT5b-RARα augmented STAT3 transcriptional activity, whereas STAT5b inhibited it. Inhibition of STAT3 by STAT5b may be due to competition for binding to the APRE reporter construct coupled with the failure of STAT5b, relative to STAT3, to recruit the basal transcription machinery to the reporter construct. Alternatively, although STAT5b does not interact with the CoR SMRT, its ability to bind the APRE site and yet reduce transcriptional activity suggests it may bind and recruit another unidentified CoR.
In addition to STAT5b-RARα, we demonstrated that other APL fusion proteins including PML-RARα and PLZF-RARα also had this enhancing effect on STAT3 transcriptional activity. STAT protein activation, especially STAT3, has been implicated in cell models of transformation and carcinogenesis. STAT3 was shown to be constitutively activated in cells transformed by oncoproteins such as v-Src.52,53 In addition, use of dominant-negative STAT3 constructs has shown that STAT3 is essential for fibroblast transformation by v-Src.46,47 Overexpression of a constitutively active form of STAT3 in immortalized rat or mouse fibroblasts induced their transformation and conferred the ability to form tumors in nude mice indicating that STAT3 alone can function as an oncogene.54Aberrant activation of STAT3 also has been demonstrated in various human blood malignancies including leukemia and lymphoma.55-58 In each of these instances cited above, increased STAT3 activation was established in DNA binding assays. In our studies, however, the increased STAT3 transcriptional activity mediated by STAT5b-RARα was not accompanied by either increased STAT3 DNA binding activity or increased Ser727 phosphorylation. This result suggests that STAT5b-RARα and other APL fusion proteins participate in a novel mechanism of leukemogenesis involving enhanced STAT3 transcriptional activity independent of its DNA binding activity and Ser727 phosphorylation status.
Based on its response to RA treatment, APL can be divided into 2 syndromes, RA-responsive APL, in which the RARα fusion gene partner is PML,5 NPM,7 or NuMA,8 and RA-resistant APL, in which the RARα fusion gene partner is PLZF9 or STAT5.10 Although it may be premature to designate STAT5b-RARα+ disease as RA unresponsive judging from only a single case,10 the RA response of the initial case of each of the other 3 APL variants reported has accurately reflected the RA response of subsequent cases within the variant group.3
To gain a molecular understanding of the ATRA unresponsiveness of STAT5b-RARα+ APL, we performed a series of studies intended, in part, to identify differences between STAT5b-RARα and PML-RARα, the most common fusion protein within the ATRA-responsive group, and to highlight similarities between STAT5b-RARα and PLZF-RARα, the sole other protein representative of the ATRA-unresponsive group. RARE-binding studies revealed that although the DNA-binding preferences of STAT5b-RARα homodimers resembled those of PML-RARα homodimers overall, STAT5b-RARα differed from PML-RARα and resembled PLZF-RARα in binding weakly to RARE-p21-WAF. Also, STAT5b-RARα and PLZF-RARα both required 10−7 M ATRA to fully recruit TRAM-1, whereas PML-RARα recruited TRAM-1 in 10−8 M ATRA. Our studies also identified features of STAT5b-RARα unique among APL fusion proteins. STAT5b-RARα differed from both PML-RARα and PLZF-RARα in that it heterodimerized with RXRα almost exclusively as a single heterodimeric complex, whereas PML-RARα and PLZF-RARα heterodimerized with RXRα and formed both single and multimeric complexes.30 40 Furthermore, STAT5b-RARα was insensitive to degradation within cells exposed to ATRA; although the importance of this finding is uncertain because PLZF-RARα is sensitive to ATRA-induced degradation yet results in ATRA-unresponsive disease. As outlined above, the finding that STAT5b can bind to and repress APRE reporter construct activity suggests that it may bind to a CoR distinct from SMRT. This raises the possibility that this CoR may remain bound to STAT5b-RARα in the presence of pharmacologic levels of ATRA and contribute to ATRA unresponsiveness in STAT5b-RARα+ APL.
Supported in part by National Institutes of Health R01 grants CA72261 and CA86430.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David J. Tweardy, Section of Infectious Disease, Department of Medicine, Baylor College of Medicine, 1 Baylor Plaza, BCM 286, Rm N1319, Houston, TX 77030; e-mail: dtweardy@bcm.tmc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal