Abstract
The IRTA1 and IRTA2 genes encode immunoglobulinlike cell surface receptors expressed in B cells and involved in chromosome 1q21 translocations in B-cell malignancy. We have now characterized and comparatively analyzed the structure and expression pattern of the entire family of IRTA genes, which includes 5 members contiguously located on chromosome 1q21. The IRTA messenger RNAs are expressed predominantly in the B-cell lineage within discrete B-cell compartments: IRTA1 is specific to the marginal zone, IRTA2 and IRTA3 are found in the germinal center light zone and in intraepithelial and interfollicular regions, and IRTA4 and IRTA5 are expressed predominantly in the mantle zone. All IRTA genes code for transmembrane receptors that are closely related to Fc receptors in their most amino-terminal extracellular domains and that possess cytoplasmic domains containing ITIM (immunotyrosine inhibition motifs)– and, possibly, ITAM (immunotyrosine activation motifs)–like motifs. These structural features suggest that the IRTA receptors may play a role in regulating activation of normal B cells and possibly in the development of neoplasia.
Introduction
The immunoglobulin (Ig) superfamily encompasses hundreds of distinct molecules and is one of the major classes of surface receptors. Ig superfamily domains consist of sequences of approximately 100 amino acids homologous to Igs that form 2 disulfide-bonded β sheets, each composed of 3 or 4 β strands.1 Members of this class are central to the diverse signaling processes within the immune system and also regulate numerous nonimmunologic functions as diverse as neurite growth2 and myoblast fusion.3
B cells express more than 20 Ig superfamily proteins (IgSPs), which fulfill specific roles as both positive and negative regulators of the humoral response. Many of these molecules deliver activating signals, such as the B-cell receptor (BCR) with its associated IgSPs Igα and Igβ (CD79a and CD79b),4 the CR2-associated coactivating molecule CD19, and CD130, the common β subunit of the interleukin-6 receptor and of several other 2-chain receptors.5 Many of these activating molecules transduce their signal through immunotyrosine activation motifs (ITAMs) present on the receptor or in associated molecules.6 Negatively signaling molecules, which bear cytoplasmic immunotyrosine inhibitory motifs (ITIMs), include the low-affinity Fc receptor FcγRIIB, a member of a family of broadly expressed molecules with diverse cell type–specific functions.7 FcγRIIB recruits the SH2 domain–containing inositol phosphatase SHIP to the BCR membrane locale, thereby dampening BCR-mediated activation and the clonotypic antibody response.8-11 Other ITIM-bearing IgSPs include platelet endothelial cell adhesion molecule (PECAM),12,13CD22,14-16 the products of the chromosome 19q13.4 leukocyte receptor complex17 genesILT218 and LAIR,19,20and CD2-like IgSPs21 signaling lymphocytic activation molecule (SLAM), Ly9, CD48, and CD58. IgSPs in other functional classes serve in intercellular recognition/adhesion (eg, intercellular adhesion molecules 1,22,23 2,23 and 324,25) and as costimulatory molecules for antigen presentation to T cells (CD80 26 and CD86 27).
We recently identified the IRTA1 and IRTA2 genes, 2 new members of the Ig receptor superfamily that are located on chromosome 1q21 and are expressed in B cells.28 In B-cell malignancy carrying 1q21 abnormalities, IRTA2 gene expression is frequently deregulated, while IRTA1 was found to be involved in a chromosomal translocation that fused it to the Ig Cα domain to produce a chimeric IRTA1/Cα fusion protein. Here we describe the identification of 3 additional IRTA genes (IRTA3, IRTA4, IRTA5), which define a subfamily of IgSP genes comprising 5 members all contiguously located on chromosome 1q21. IRTA genes have ITIM- and ITAM-like motifs and show topographically distinct patterns of expression in lymphoid organs, suggesting a role in modulating normal B-cell development and immune responses.
Materials and methods
Complementary DNA library screening and cloning
An oligo(dT)/random-primed spleen complementary DNA (cDNA) library in λgt11 (Clontech, Palo Alto, CA) was screened at high stringency (0.1 × SSC, 68°C) according to established procedures.29 Phage inserts were sequenced using an ABI 373 automated sequencer (Applied Biosystems, Foster City, CA) from template amplified by polymerase chain reaction from phage using flanking primers and treated with ExoI and sALP (USB, Cleveland, OH).30 Plasmid clones were generated by TA cloning into pGEM-T (Promega, Madison, WI). 5′ rapid amplification of complementary DNA ends (RACE) was performed on Marathon spleen cDNA (Clontech) according to the manufacturer's recommendations.
Sequence analyses
Northern blotting
PolyA-enriched RNA was prepared from previously described cell lines using Oligotex beads (Qiagen, Valencia, CA) separated on a 1% agarose 3-(4-morpholino)propanesulfonic acid (MOPS)/formaldehyde gel, blotted downward onto Duralose-UV membranes (Stratagene, La Jolla, CA) in 20 × SSC, cross-linked at 120 000 μJ/cm2, and hybridized to α32P-dCTP–labeled random-primed probes in fomamide-containing solution with 10% dextran sulfate. DNA probes, generated by polymerase chain reaction or restriction enzyme digestion, were purified from 0.8% agarose TAE (tris-acetate ethylenediamine tetraacetic acid [EDTA]) gels using GFX spin columns (Amersham Pharmacia Biotech, Piscataway, NJ) and corresponded to the following regions of the cDNA sequences: IRTA1 (3′ untranslated region [UTR]) nucleotides (nt) 1641 to 2225;IRTA2 (5′ end) nt 1 to 743; IRTA3 (3′ end) nt 2337 to 2970, (5′ end) nt 1 to 745; IRTA4 (5′ end) nt 1 to 750; IRTA5 (3′ end) nt 1271 to 1900, (5′ end) nt 1 to 726. Filters were washed at a final wash of 0.1 × SSC at 65°C, exposed to Kodak XAR film, developed on a Kodak 1000A XOMAT processor, digitized with an AGFA T1200 duoscan, and rendered in Adobe Photoshop 5.5.
In situ hybridization
Digoxygenin-labeled probes encompassing the entire cDNA and 5′ UTR of each IRTA gene were transcribed with T7, SP6, or T3 bacteriophage RNA polymerases, as appropriate, to generate antisense RNA probes from linearized plasmid template. A sense RNA-negative control probe of similar length was made from similarly purified plasmid DNA containing the Bcl6 cDNA. Human tonsil tissue was snap-frozen in dry ice/isopentane, and 12 μm serial cryostat sections were prepared. Sections were not treated with proteinase K but were acetylated and then hybridized at 68°C in 50% formamide-containing solution, washed, probed with an alkaline phosphatase–conjugated antidigoxygenin antibody, and developed, all as described.31
Results
Cloning of IRTA cDNAs and structure of predicted IRTA proteins
Cloning of IRTA1 and IRTA2 has been previously described.28 BLAST search of the GenBank database with the IRTA2 sequence identified an expressed sequence tag (EST) (AA63g02) showing 89% homology toIRTA2 over 250 bases within the extracellular domain encoding region. The insert of that clone (IMAGE: 825650) was used to probe a λgt11 human tonsil cDNA library at high stringency (see “Materials and methods”). Twenty-eight clones were sequenced, which segregated into 4 contigs: IRTA2 (2 clones),IRTA3 (GenBank AF459027) (9 clones), IRTA4(GenBank 459633) (5 clones), and IRTA5 (GenBank AF459634) (12 clones). The 3 IRTA sequences not identical to the probe were identified despite the high stringency of library screening, because each contains regions of about 90% nt sequence identity with the probe, which fell into the IRTA5 contig. The 5′ ends ofIRTA4 and IRTA5 were cloned by 5′ RACE. BLAST searches of GenBank databases and of the Celera human genome sequence failed to identify additional homologous genes, suggesting that there are no outstanding members of the IRTA family.
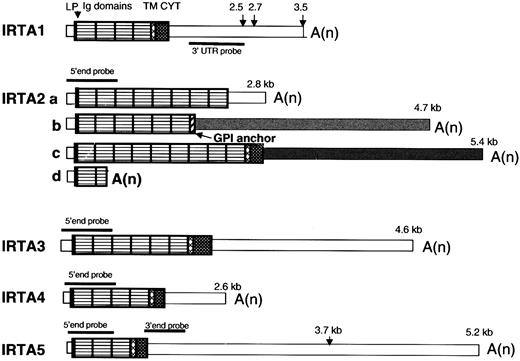
The domain organization of the IRTA RNAs is shown schematically in Figure 1. As previously reported,28IRTA1 undergoes alternative polyadenylation, and IRTA2 undergoes alternative splicing at the last exon to generate 3 protein isoforms that either have no membrane tether (IRTA2a), have a putative glycosyl-phosphatidyl inositol linkage (IRTA2b), or have transmembrane and cytoplasmic domains (IRTA2c). In addition, premature termination within the second Ig domain gives rise to IRTA2d.28 For IRTA3, IRTA4, andIRTA5, only transmembrane-encoding isoforms were found by cDNA cloning and database searches. The 2 bands seen by Northern blot analysis of IRTA5 (see below) are consistent with alternative polyadenylation within the 3′ UTR (at nt 146315 and nt 148601 of the genomic contig AL356276), where multiple ESTs sequences initiate. These bands were seen with both 5′ end and proximal 3′ UTR probes, ruling out the possibility of an unrecognized alternative terminal exon. Whereas predominantly fully processed messenger RNAs (mRNAs) were seen in cultured cell lines (see below), several longer species representing incompletely processed mRNA were seen in RNA from tonsil and spleen, most notably for IRTA5. Recently, based upon sequences of 3 cDNA clones, Xu et al proposed 3 distinct protein isoforms of a putative gene designated SPAP132 (AF319438). The SPAP1a nt sequence is identical to our IRTA4 sequence starting at nt 931 of IRTA4 but is missing the first 5 exons that encode the signal peptide and the first 3 Ig-like domains.
IRTA family mRNAs have a conserved structural organization.
Pattern-filled boxes represent coding domains, and thin open or shaded boxes represent UTRs. The predicted site for signal peptidase cleavage is marked by an arrowhead and was derived according to the SignalIP World Wide Web server.45 The transmembrane domain prediction algorithm is described by Tusnady et al.49 LP indicates leader peptide; Ig, immunoglobulin type; TM, transmembrane; CYT, cytoplasmic domain; A(n), polyA tail; GPI, glycosylphosphatidyl inositol. Lengths of transcripts identified by Northern blot analysis using probes indicated in the figure are shown. The various 3′ UTRs of IRTA2, which result from alternative splicing, are denoted by differential shading.
IRTA family mRNAs have a conserved structural organization.
Pattern-filled boxes represent coding domains, and thin open or shaded boxes represent UTRs. The predicted site for signal peptidase cleavage is marked by an arrowhead and was derived according to the SignalIP World Wide Web server.45 The transmembrane domain prediction algorithm is described by Tusnady et al.49 LP indicates leader peptide; Ig, immunoglobulin type; TM, transmembrane; CYT, cytoplasmic domain; A(n), polyA tail; GPI, glycosylphosphatidyl inositol. Lengths of transcripts identified by Northern blot analysis using probes indicated in the figure are shown. The various 3′ UTRs of IRTA2, which result from alternative splicing, are denoted by differential shading.
IRTA proteins identify a subfamily of IgSP molecules related to Fc receptors
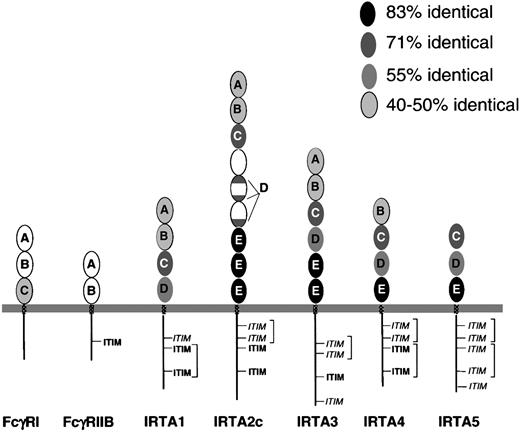
The predicted IRTA proteins each have 3 to 9 extracellular Ig-like domains, followed by transmembrane and cytoplasmic domains (Figure2), while IRTA2 also has secreted and glycosylphosphatidyl inositol–linked isoforms. After signal peptide cleavage, the predicted molecular weights of the unglycosylated polypeptides are 55.7 kd (IRTA1), 104.9 kd (IRTA2c), 78.9 kd (IRTA3), 53.4 kd (IRTA4), and 45.2 kd (IRTA5). The extent of sequence identity of corresponding Ig-like domains varies within the range of 45% to 83% among IRTA family members (Figure 2). Amino acid sequence alignment (Figure 3A) allows 5 Ig-like domain subtypes to be defined based upon homology within the IRTA family (Figure 2A-E). Domain subtypes A and B are similar (37% amino acid similarity) to domains present in the high-affinity Ig receptor FcγRI as well as FcγRII and FcγRIII, which are low-affinity Ig receptors that have only 2 Ig-like domains. Not all of the IRTAs have domains of each subtype. However, a feature common to all of the IRTAs is the presence of domain subtype C, which is 70% preserved among the various IRTA members. Domain C is also 47% identical to the third domain of FcγRI. The adjacent domain, subtype “D,” shows 55% identity among IRTA members and is absent from FcγRI. In IRTA2, however, the “D” consensus sequence is split into 3 parts that are spread over 2 adjacent domains. The most highly conserved domain, domain subtype E, is reiterated with 83% identity in IRTA2c (3 times), IRTA3 (twice), IRTA4 (once), and IRTA5 (once), but it is absent entirely from FcγRI and from IRTA1.
IRTA family proteins have related extracellular and cytoplasmic domains, which can be grouped into domain subtypes.
Conservation and relatedness to Fc receptor family members. Ovals represent extracellular Ig superfamily domains, with IRTA domain subtype indicated by letters “A” through “E.” The extent of conservation of each domain is represented by its degree of shading, as shown in the key. Patterned rectangles represent transmembrane regions. Straight lines (not to scale) represent cytoplasmic domains. ITIM indicates immunotyrosine inhibitory motif; ITIM,immunotyrosine inhibitory motif–like sequence. Potential ITAM pairs are bracketed.
IRTA family proteins have related extracellular and cytoplasmic domains, which can be grouped into domain subtypes.
Conservation and relatedness to Fc receptor family members. Ovals represent extracellular Ig superfamily domains, with IRTA domain subtype indicated by letters “A” through “E.” The extent of conservation of each domain is represented by its degree of shading, as shown in the key. Patterned rectangles represent transmembrane regions. Straight lines (not to scale) represent cytoplasmic domains. ITIM indicates immunotyrosine inhibitory motif; ITIM,immunotyrosine inhibitory motif–like sequence. Potential ITAM pairs are bracketed.
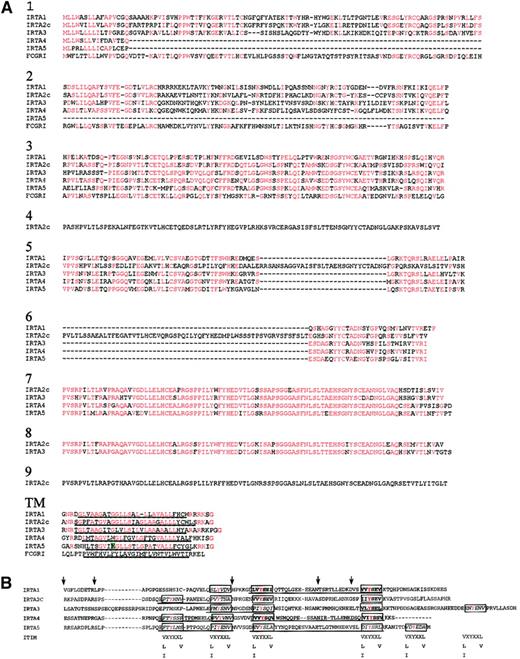
Amino acid sequence homology among IRTA genes and FcγR1 varies by domain.
(A) Extracellular and transmembrane regions. Sequences were aligned using the Clustal 1.8 program46 and formatted by the Boxshade server47 with minor manual readjustments. Residues identical to the consensus (50% or more of sequences) at each position are shown in red. The hydrophobic putative membrane-spanning sequence is underlined. The charged amino acid in the IRTA5 transmembrane region is highlighted in green. (B) Alignment of the cytoplasmic domains of IRTAs 1 through 5. The ITIM consensus sequences are shown below the possible ITIM positions of the IRTA proteins. ITIMs are boxed and bold, ITIM-like sequences are boxed and italicized. ITAM-like sequences are underlined. X denotes any amino acid. Tyrosine residues are colored red. Positions corresponding to exon junctions are marked by arrows.
Amino acid sequence homology among IRTA genes and FcγR1 varies by domain.
(A) Extracellular and transmembrane regions. Sequences were aligned using the Clustal 1.8 program46 and formatted by the Boxshade server47 with minor manual readjustments. Residues identical to the consensus (50% or more of sequences) at each position are shown in red. The hydrophobic putative membrane-spanning sequence is underlined. The charged amino acid in the IRTA5 transmembrane region is highlighted in green. (B) Alignment of the cytoplasmic domains of IRTAs 1 through 5. The ITIM consensus sequences are shown below the possible ITIM positions of the IRTA proteins. ITIMs are boxed and bold, ITIM-like sequences are boxed and italicized. ITAM-like sequences are underlined. X denotes any amino acid. Tyrosine residues are colored red. Positions corresponding to exon junctions are marked by arrows.
The various domains of the IRTA members are encoded by a pattern of exons that is conserved among members of the family: 2 exons encode the signal peptide, 1 exon encodes each Ig domain, and 5 exons encode each cytoplasmic region. IRTA5 is unique among the IRTAs in that it has a charged residue (glutamic acid) in the transmembrane region, suggesting that it may heterodimerize with a protein containing a positively charged amino acid in nearby position, as is the case for many ITAM-bearing proteins, including FcγRIII, and ITAM-bearing polypeptides of the BCR and T-cell receptor complexes.33The cytoplasmic domains of the IRTAs are also similar to each other, with an average of 45% of residues shared among most of the members.
Each cytoplasmic domain contains 3 to 5 tyrosine residues, which suggests the presence of ITIM- or ITAM-like motifs. Based on a consensus sequence for ITIM (V/L/I-X-Y-X-X-L/V),33 7 canonical ITIMs can be found in the IRTA molecules (boxed, bold in Figure 3B), while 14 additional sequences differ from the consensus ITIM at the −2 or +3 position relative to the tyrosine (ITIM-like; boxed, italicized in Figure 3B). Based on the consensus (D/E-X-X-Y-X-X-L/I-X6-8-Y-X-X-L/I), none of the IRTA molecules have canonical ITAM motifs. However, all IRTAs contain ITAM-like sequences (underlined in Figure 3B) that display paired tyrosines preceded by acidic residues at the Y−3 position, although none conform at the Y+3 positions. These ITAM-like sequences also lack other features that Malissen et al have recognized as typical of ITAMs: namely that the 2 tyrosines are encoded by adjacent exons (Figure 3B) (ITAM-like sequences in IRTA1, IRTA4, and IRTA5 span 3 exons) and the presence of intervening introns of type 034 (ie, splice junctions between codons; all IRTA cytotoplasmic domain introns have a pattern of intron types of 1, 2, 1, 1, 0). In conclusion, it appears that all IRTA receptors have credible ITIM sequences, while the presence of ITAMs cannot be firmly postulated based on sequence homology. Taken together, the homology among IRTA members in both cytoplasmic and extracellular regions suggests that they are derived from a single ancestral gene related to a prototype Fc receptor in the Ig-like domains.
IRTA genes are located within a 300 kilobase region on chromosome 1q21
BLAST search of IRTA3, IRTA4, and IRTA5against the human genome revealed that they all lie within a single P1 phage artificial chromosome (PAC) contig (AL356276). All 5 IRTA genes are contained within a 300 kilobase (kb) genomic region (Figure4), and all have a telomere-to-centromere transcriptional orientation. The IRTA locus lies between 2 genes identified at translocation breakpoints in lymphomas: Bcl-9,which was found to be deregulated in a single case of pre–B-cell acute lymphoblastic leukemia35 on the centromeric end, and FcγRIIB, which was found to be up-regulated in 2 cases of follicular lymphoma and in one cell line36 on the telomeric end. Also centromeric to the IRTA locus lies the MUC-1 (EMA) gene, deregulated in 6% of non-Hodgkin lymphomas that have a 1q21 abnormality.36 The IRTA genes are midway between 2 Fc receptor loci, consistent with an evolutionary relationship of the IRTA and the FcR gene families. The IRTA locus also contains a 5 exon pseudogene that is highly homologous to IRTA2; lack of EST sequences derived from this region and the presence of in-frame stop codons in 2 of the exons serve as evidence of its obsolescence. On the telomeric end of the region, the transcriptional start site ofIRTA5 lies 11 kb downstream of the polyadenylation signal of the CD5 antigen–like (scavenger receptor cysteine rich family) gene.
The IRTA locus spans 300 kb in a chromosome segment containing Fc receptors and genes identified through analysis of chromosomal translocations in B-cell malignancy.
The top line shows a portion of chromosome 1q21, indicating the positions of selected genes and their respective genomic sequence accession numbers, according to the working draft sequence of the Human Genome Sequencing Project, as of January 21, 2001. The centromere is to the left. The position of MUC-1 (EMA) is based upon the Celera human genome sequence.48 The second line shows a magnification of the indicated region of line 1, with the IRTA genes depicted as arrows in the direction of transcription under their respective sequence contigs. The third line shows the intron/exon structure of each IRTA gene, with filled boxes indicating coding exons and open boxes indicating noncoding regions of mRNA. The position of the chromosomal breakpoint in the initial cell line analyzed is indicated by an arrow. Scale bars are shown on the left for the lower 2 lines.
The IRTA locus spans 300 kb in a chromosome segment containing Fc receptors and genes identified through analysis of chromosomal translocations in B-cell malignancy.
The top line shows a portion of chromosome 1q21, indicating the positions of selected genes and their respective genomic sequence accession numbers, according to the working draft sequence of the Human Genome Sequencing Project, as of January 21, 2001. The centromere is to the left. The position of MUC-1 (EMA) is based upon the Celera human genome sequence.48 The second line shows a magnification of the indicated region of line 1, with the IRTA genes depicted as arrows in the direction of transcription under their respective sequence contigs. The third line shows the intron/exon structure of each IRTA gene, with filled boxes indicating coding exons and open boxes indicating noncoding regions of mRNA. The position of the chromosomal breakpoint in the initial cell line analyzed is indicated by an arrow. Scale bars are shown on the left for the lower 2 lines.
IRTA genes are expressed in topographically distinct B-cell compartments
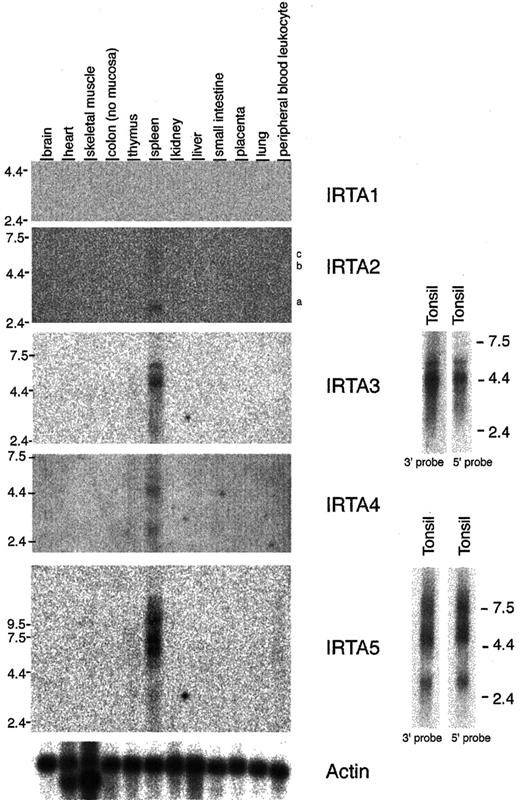
The pattern of expression of the IRTA genes in normal human tissues was examined by Northern blot analysis (Figure5). IRTA2, IRTA3, IRTA4, andIRTA5 are all expressed predominantly in the spleen.IRTA1 was detected in normal tonsil tissue (see below; Figure 6) but is expressed at very low levels in the spleen. Significant heterogeneous signals corresponding to higher molecular weight (incompletely processed) species were seen using probes to IRTA4 and IRTA5in RNA from tissues but not from cell lines (see below; Figure6). Expression of IRTA3, IRTA4, and IRTA5, like IRTA1 andIRTA2,28 is restricted to lymphoid tissue, with predominance in the B-cell–rich spleen over the T-cell–rich thymus.
IRTA family mRNA expression is limited to lymphoid tissues in analysis of normal organs.
Northern blot analysis of IRTA1, IRTA2, IRTA3, IRTA4, andIRTA5 using 2 μg polyA-selected RNA. For IRTA3and IRTA5, both 5′ and 3′ end probes were used. The positions of the probes within the mRNAs are shown in Figure 1. IRTA2 isoforms a, b, and c are indicated.
IRTA family mRNA expression is limited to lymphoid tissues in analysis of normal organs.
Northern blot analysis of IRTA1, IRTA2, IRTA3, IRTA4, andIRTA5 using 2 μg polyA-selected RNA. For IRTA3and IRTA5, both 5′ and 3′ end probes were used. The positions of the probes within the mRNAs are shown in Figure 1. IRTA2 isoforms a, b, and c are indicated.
The pattern of IRTA family mRNA expression in cell lines highlights a correlation of IRTA-2 deregulation with chromosome 1q21 abnormalities.
Northern blot analysis of IRTA2, IRTA3, IRTA4, and IRTA5 using 2 overlapping panels of RNA representative of the major hematopoietic lineages and containing BL cell lines of known 1q21 karyotype, 2 μg polyA RNA per lane; 5′ end probes were used for IRTA2 and IRTA4, and 3′ end probes were used for IRTA3 and IRTA5, as shown in Figure 5. Epstein-Barr virus (EBV) status is indicated on the bottom of the figure. GADPH indicates glyceraldehyde phosphate dehydrogenase.
The pattern of IRTA family mRNA expression in cell lines highlights a correlation of IRTA-2 deregulation with chromosome 1q21 abnormalities.
Northern blot analysis of IRTA2, IRTA3, IRTA4, and IRTA5 using 2 overlapping panels of RNA representative of the major hematopoietic lineages and containing BL cell lines of known 1q21 karyotype, 2 μg polyA RNA per lane; 5′ end probes were used for IRTA2 and IRTA4, and 3′ end probes were used for IRTA3 and IRTA5, as shown in Figure 5. Epstein-Barr virus (EBV) status is indicated on the bottom of the figure. GADPH indicates glyceraldehyde phosphate dehydrogenase.
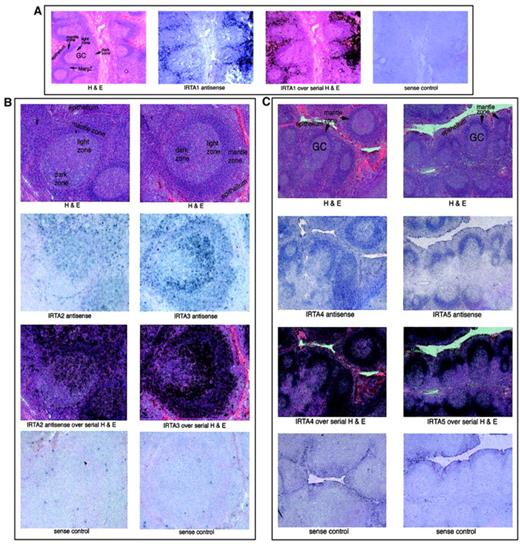
The expression pattern of the IRTA genes within the B-cell compartment was examined by in situ hybridization of hyperplastic human tonsil tissue (Figure 7). IRTA1 is expressed outside of lymphoid follicles in a “marginal zone” pattern and in intraepithelial lymphocytes28 (Figure 7A). This region contains a population of mature/memory B cells that, in lymph nodes of patients with certain infectious conditions (eg, toxoplasmosis and human immunodeficiency virus lymphadenitis), are rich in monocytoid B cells.35
IRTA family mRNAs have gene-specific localization patterns within normal tonsil tissue.
In situ hybridization of hyperplastic human tonsil to IRTA antisense RNA probes, as indicated. For each panel 3 serial sections are shown; the third frame represents an overlay of the antisense RNA signal (with blue changed to black) and the serial section stained with hematoxylin and eosin (H&E). For each set a section was processed in parallel with a sense RNA probe to control for background signal. GC indicates germinal center; MargZ, marginal zone. Original magnification A, × 20; B, × 50; C, × 15.
IRTA family mRNAs have gene-specific localization patterns within normal tonsil tissue.
In situ hybridization of hyperplastic human tonsil to IRTA antisense RNA probes, as indicated. For each panel 3 serial sections are shown; the third frame represents an overlay of the antisense RNA signal (with blue changed to black) and the serial section stained with hematoxylin and eosin (H&E). For each set a section was processed in parallel with a sense RNA probe to control for background signal. GC indicates germinal center; MargZ, marginal zone. Original magnification A, × 20; B, × 50; C, × 15.
IRTA228 and IRTA3 mRNAs were detected in a polarized pattern within the germinal center, with highest expression in the centocyte-rich light zone and little to no signal in the dark zone (Figure 7B). Outside the germinal center, the highest levels of these mRNAs were detected in intraepithelial lymphocytes and in interfollicular regions. These results indicate that transcriptional activation of these genes occurs during the maturation of centroblasts to centrocytes, with maintenance of expression in postgerminal center B cells. A weak signal for IRTA2 andIRTA3 RNA was detected in the follicular mantle zones.
IRTA4 and IRTA5 are expressed in a third, distinct pattern, with highest levels seen within the mantle zones (Figure 7C). IRTA5 mRNA was not detected outside this region, while IRTA4 signal was slightly above background levels outside the mantle zone. Thus, these 2 genes are expressed almost exclusively in naive B cells.
To more broadly assess IRTA gene expression, we performed Northern blot analysis using a panel of cell lines representative of the major hematopoietic lineages (Figure 6). Expression of all IRTA family members is B lineage–restricted, in that no signal was seen in RNA from HeLa (epithelial) or representative erythroid, myeloid, monocytic, or T-cell lines. As shown here and previously,28IRTA1 and IRTA2 are expressed in some lymphoblastoid cell lines that also weakly express IRTA3.Among B-cell lines examined, all IRTAs are widely expressed in Burkitt lymphoma (BL) lines, each having a unique expression profile. The BL and multiple myeloma lines shown in Figure 6 are grouped according to the presence or absence of a cytogenetic abnormality of 1q21, further defined by fluorescence in situ hybridization as trisomy of the locus in 90% of cases.28 For all of the IRTA mRNAs, expression in BL cell lines can be considered a deregulation of gene expression relative to their presumed cell of origin, the centroblast. However, for IRTA2, this deregulation also correlates with the presence of a 1q21 abnormality in the 24 cell lines shown in Figure 6. The levels of IRTA2 are on average 10-fold higher in 1q21 abnormal BLs than in 1q21 normal BLs.28IRTA2is also expressed by one of the 1q21 abnormal multiple myeloma lines. The other IRTA genes' expression levels do not correlate with the presence of a chromosomal defect in the IRTA region.
Discussion
The IRTA genes encode glycoproteins within the Ig superfamily of receptors. Their closest relatives are the Ig-like receptors for the Fc portions of Ig. These Fc receptors trigger various functions of effector cells,7 such as antibody-dependent cellular cytotoxicity and anaphylaxis. The most extensive homology is to FcγRI (CD64), the high-affinity IgG receptor expressed on monocytes, macrophages, and activated neutrophils that plays a role in phagocytosis, antibody-dependent cellular cytotoxicity, and macrophage activation.37 The homology to Fc receptors is through the amino-terminal Ig-like domains, subtypes A, B, and C of the IRTA proteins. These 3 domains are all present in IRTA1, IRTA2, and IRTA3, while IRTA4 and IRTA5 lack 1 or 2 of these domain subtypes. In preliminary studies, we have found that IRTAs 1 and 2 have a low affinity for IgA and IgG, respectively, as evidenced by their ability to bind aggregates of these molecules specifically when expressed by transient transfection on the surface of HEK 293 cells. However, further studies are needed to conclusively establish a physiologic role of these IRTA proteins as Fc receptors.
Recently, using an approach to identify sequences of Fc receptor homologs, Davis et al38 describe the sequences and expression patterns of FcRH1 (IRTA5), FcRH2 (IRTA4), andFcRH3 (IRTA3). Our Northern results are essentially in agreement for IRTA3 and IRTA4, but we find a more restricted expression and transcript size pattern (3 and 5.5 kb, as opposed to multiple transcripts ranging from 1 to 8 kb) forIRTA5.38 This discrepancy may be due to a different probe used for Northern blot analysis; we note that the 3′ UTR of IRTA5 contains sequences (including a sine/alu repeat element) that cross-hybridize to other transcripts. Thus, the inclusion of these sequences in the probe (not described in detail by Davis et al) may have generated a cross-hybridization. Regarding the assignment of ITIM and ITAM motifs, our analysis agrees with the one by Davis et al38 for ITIM, while we have been more conservative in ITAM assignment because we do not detect any bona fide ITAM motifs (see “Results”). However, tyrosine motif identification based on consensus sequences cannot be conclusive and needs functional validation.
Despite the homology to Fc receptors, however, the strong conservation by all of the IRTA members of the carboxy-terminal Ig-like domains, along with the reiteration of domain subtype E up to 3 times within an individual IRTA protein, suggests that the IRTA proteins bind through this region to a common or closely related ligand(s) that may be distinct from Ig. In addition, approximately 40% of the amino acid residues in this domain are similar to a domain of PECAM-1 (CD31), an ITIM-containing molecule that plays lineage- and stage-specific roles in intercellular recognition and adhesion. Thus, despite the similarity to Fc receptors, we cannot rule out other functions for the IRTA proteins, such as a role in intercellular signaling via interaction with other cell-bound ligands or even association with the BCR itself.
The functional consequences of IRTA ligation on the cell surface are under investigation and hinge upon the nature of the cytoplasmic tyrosine motifs. Where ITIMs are present in the IRTA proteins, they may bind a phosphatase, possibly SHP-1,32 to repress B-cell activation. Alternatively, ITIMs could provide activating signals, by analogy to the ITIM-bearing IgSPs PECAM12 and signaling lymphocytic activation molecule,39 which are thought to interact with the physiologically activating phosphatase SHP-2.40 However, based upon primary structure alone, we cannot rule out the possibility that several pairs of tyrosine-containing motifs (Figure 3B) function together as ITAMs to recruit a src family protein kinase to the receptor.
The IRTA mRNAs are expressed on distinct B-cell subsets as defined architecturally within human tonsil tissue. IRTA4 andIRTA5 are expressed in a pattern that corresponds to naive B cells, which suggests a unique role for these molecules in the early stages of B-cell activation or in turnover and migration of this population prior to specific antigenic stimulation. IRTA1 is expressed in the marginal zone and in intraepithelial lymphocytes; this, together with its affinity for IgA, suggests a role in mucosal immunity. IRTA2 and IRTA3 are expressed mainly within the germinal center light zone and in postgerminal center cells, which may imply regulation in the context of T-cell–dependent immune responses. Thus, the IRTA family may provide a means for fine regulation of the immune response at various stages in response to a similar molecule or complex.
Because of their B-cell subtype–specific expression, discovery of the IRTA proteins may have clinical implications in diagnosis and therapy. For instance, the marginal zone/intraepithelial pattern of IRTA1 has been confirmed by immunohistochemistry (not shown), and it is present in the monocytoid B cells of human immunodeficiency virus lymphadenitis as well as in most lymphomas of mucosa-associated lymphoid tissue (G.C., manuscript in preparation). Analogously, the other IRTAs may define lymphoma subtypes of distinct cellular derivation, which may prove to be useful in diagnosis. Finally, as B-cell subtype–specific surface markers, the IRTAs may serve as specific targets in immunotherapy of B-cell lymphomas and nonmalignant disorders, analogous to anti-CD20 antibodies.41 42
Supported in part by a grant from the National Institute of Health (CA-37295 to R.D.-F).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Riccardo Dalla-Favera, Institute of Cancer Genetics, Columbia University, Russ Berrie Science Pavilion, 1150 St Nicholas Ave, Room 303B, New York, NY 10032; e-mail:rd10@columbia.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal