Abstract
Acute promyelocytic leukemia (APL) is characterized by the t(15;17)(q22;q11.2), which results in the PML-RARA fusion gene. In previous studies, we demonstrated that expression of a human PML-RARA complementary DNA in murine granulocyte precursor cells initiated the development of leukemia. However, leukemogenesis by PML-RARA required additional genetic alterations. To identify genetic changes that cooperate with PML-RARA in leukemogenesis, we performed spectral karyotyping analysis of myeloid leukemias from hMRP8-PML-RARA mice (11 cases) and from mice coexpressing PML-RARA and BCL2 (8 cases). Clonal abnormalities were detected in 18 of 19 cases (95%). Recurring numerical abnormalities identified in these murine leukemias included +15 (15 cases, 79%); loss of a sex chromosome (12 cases, 63%); +8 (10 cases, 53%); +10 (9 cases, 47%); +4, +7, or +14 (8 cases each, 42%); +16 (7 cases, 37%); and +6 (5 cases, 26%). In a series of 965 patients with APL, we identified secondary abnormalities in 368 (38%). The most common recurring abnormalities were +8 or partial trisomy of 8q (120 patients, 12.4%) and ider(17) t(15;17) (42 patients, 4.4%). The critical consequence of +8 in human leukemias appears to be the gain of 8q24, which is syntenic to mouse 15. Thus, our results suggest that PML-RARA–initiated murine leukemia is associated with a defined spectrum of genetic changes, and that these secondary mutations recapitulate, in part, the cytogenetic abnormalities found in human APL.
Introduction
Acute promyelocytic leukemia (APL) is a distinct clinicopathologic entity characterized by infiltration of the bone marrow by malignant promyelocytes in association with a hemorrhagic diathesis.1,2 APL cells are exquisitely sensitive to the differentiating effect of all-trans-retinoic acid (ATRA).3 Treatment with ATRA and chemotherapy has resulted in a complete remission (CR) rate of about 95% in APL, with a 5-year disease-free survival of 80% to 90%. The t(15;17)(q22;q11.2-12) was first recognized in APL by Rowley et al in 1977,4 and results in a fusion PML-RARA gene that contains most of thePML coding sequences, and the RARAsequences encoding the DNA-binding and ligand-binding domains.2 5-7
A number of investigators have demonstrated that expression of PML-RARA in early myeloid cells in transgenic mice leads to the development of leukemias with features of human APL, thereby documenting the ability of PML-RARA to initiate leukemogenesis.8-10 In previous studies, we developed a transgenic mouse model for APL, in which the human MRP8 promoter drives expression of PML-RARAin myeloid cells.9 Transgenic mice have impaired neutrophil differentiation early in life, which progresses to overt leukemia in 64% of the mice by the age of 12 months. The murine disease recapitulates features of human APL, including a therapeutic response to ATRA with differentiation of the leukemia cells.
However, several characteristics of the model suggest that expression of PML-RARA alone is not sufficient to cause leukemia. The long latency before disease onset (median of 8.5 months), together with the incomplete penetrance (64% at 1 year), supports the hypothesis that leukemogenesis initiated by PML-RARA requires additional genetic changes. This hypothesis is consistent with studies showing that doubly transgenic mice expressing both PML-RARA and BCL2develop leukemia more rapidly (median time to leukemia of 127 days versus 257 days, P = .00002), with 100% penetrance.11 Although BCL2 cooperated withPML-RARA to markedly expand immature myeloid forms in the bone marrow, the transition to acute leukemia required additional genetic changes. However, the combination of PML-RARA and the molecular signals induced by interleukin 3 (IL-3) were sufficient to fully transform normal myeloid cells into acute leukemia in this mouse model of APL.12 Although experiments of this type define genetic lesions and pathways that can cooperate with PML-RARA, their pathogenic significance is uncertain.
To identify genetic changes that cooperate with PML-RARA in vivo, we performed cytogenetic and spectral karyotyping (SKY) analysis of leukemias arising in PML-RARA andPML-RARA/BCL2 mice. Comparative genomic hybridization (CGH) was used previously to demonstrate the presence of chromosomal gains and losses in this APL model.11 SKY analysis has enabled us to confirm and extend these studies by defining the abnormal clones completely (both numerical and structural abnormalities), by identifying clonal evolution, and by recognizing patterns of abnormalities that occur together. Here, we demonstrate that murine leukemias initiated by PML-RARA have a defined spectrum of genetic changes, with trisomy 15, trisomy 8, monosomy X or Y, and trisomy 10 commonly observed. These secondary mutations are likely to contribute to the development of leukemia in transgenic mice and may provide insights into the mutations that cooperate withPML-RARA in the pathogenesis of human APL.
Materials and methods
Generation of leukemias
The leukemias arose spontaneously in MRP8-PML-RARAtransgenic mice, or in PML-RARA/BCL2 mice (either mice reconstituted with MRP8-PML-RARA/MRP8-BCL2 doubly transgenic bone marrow, or in mice reconstituted with MRP8-PML-RARA bone marrow that had been transduced with a retroviral vector expressing BCL2).9 11 Bone marrow was obtained by flushing buffered saline through mouse long bones. Spleen was suspended in buffered saline and filtered through nylon mesh.
Cytogenetic analysis
Cytogenetic analysis was performed on fresh or cryopreserved spleen or bone marrow cells obtained at the time of development of leukemia. Short-term (24-72 hours) cultures were initiated by incubating 1.0 × 106 cells/mL (70% RPMI 1640, 10% heat-inactivated fetal bovine serum, 10% IL-3–conditioned medium,13 10% IL-6–conditioned medium,14100 U/mL penicillin, 100 μg/mL streptomycin, 10 mM Hepes, pH 7.2-7.3) at 37°C (5% CO2/95% air, humidified atmosphere). Metaphase cells were prepared using standard cytogenetic techniques with the following modifications.15 Following mitotic arrest (Colcemid, 0.05 μg/mL final concentration, 2 hours, Invitrogen Life Technologies, Carlsbad, CA), cells were incubated in hypotonic KCl (0.075 M, 8 minutes, 37°C); after 8 minutes, fixative (2 mL, 3:1, absolute methanol-glacial acetic acid) was added dropwise to the cells in hypotonic solution. The cells were mixed, centrifuged, and resuspended in fresh fixative; the fixative step was repeated 3 to 6 times, and air-dried slides were prepared.
SKY analysis
Spectral karyotyping was performed using the Applied Spectral Imaging (ASI, Carlsbad, CA) SkyPaint kit for mouse chromosomes using the procedure recommended by the manufacturer. Probes were detected using the SkyPaint detection reagents as described by the manufacturer. Analysis was performed using ASI image capturing (SI 2.2) and analysis software (SkyView 1.6.1). A minimum of 10 metaphase cells was analyzed per mouse leukemia. Mouse chromosomes were classified according to the standardized karyotype,16 as refined by Cowell.17
Results
Cytogenetic analysis of murine leukemias initiated byPML-RARA
PML-RARA mice.
The results of cytogenetic and SKY analysis of the murine leukemias are given in Table 1. Of the 11 cases analyzed in PML-RARA transgenic mice, clonal chromosomal abnormalities were identified in 10 (91%). Each of the 10 abnormal cases had numerical abnormalities, whereas structural abnormalities were uncommon and observed in only 2 cases. In 8 cases, a complete karyotype could be defined, whereas a composite karyotype was designated for the remaining 2 abnormal cases (cases 802 and 1717). In 5 cases, only a single abnormal clone was detected; however, 3 cases had multiple, related, abnormal clones providing evidence of clonal evolution.
Cytogenetic analysis of murine leukemias initiated by PML-RARA
| Mouse . | Tissue . | Karyotype . |
|---|---|---|
| PML-RARAmice | ||
| 1111 | BM/spleen | 42,XX,+8,+15[7]/40,XX[2] |
| 909 | Spleen | 39,X,−Y,del(2)(C3H1)[4]/40,idem,+8[2]/40,idem,+16[2]/NCA:41,idem,+8,+16[1]/41,idem,+6,+8[1] |
| 935 | Spleen/LN | 39,X,−Y[7]/40,idem,+16[2]/NCA:40,idem,+15[1]/40,idem,+X[1] |
| 1333 | Spleen | 42,X,−Y,+8,+15,+16[6]/41,idem,−13[3]/NCA:42,idem,+X,−13[1] |
| 1717 | BM/spleen | 44,XY,+8[11],+10[10],+15[8],+17[7][cp11] |
| 1718 | BM/LN | 41,XY,+15[5]/NCA:40,X,+X,−Y[1]/40,XY[4] |
| 1950 | Spleen | 41,XY,+15[9]/40,XY[2] |
| 42 | Spleen | 40,XY[15] |
| 3509 | Spleen | 42,X,−Y,+10,+14,+15[10] |
| 802 | Spleen | 42,X,−X,+6,+7,+14[cp5]/40,XX[4] |
| 1097.1 | Spleen | 50,X,−Y,+1,+4,+8,+10,+12,+14,+15,+der(15)t(14;15)(D3;D1 or D2),+16,+17,+18[10] |
| PML-RARA/BCL2mice | ||
| 2995A4 | Spleen | 41,X,−X,+8,+15[9]/NCA:41,X,−X,−5,+8,−9,+15,+18,+19[1] |
| 1712 | BM | 49,X,−Y,+4,+6,+7,+8,+10,+11,+13,+14,+15,+16[cp5]/40,XY[4] |
| 1713 | Spleen | 42,X,−Y,i(7q),+8,+10,+15[8]/43,idem,+4[2] |
| 65 | Spleen | 45,X,−X,+4,+7,+10,+14,+15,+15[cp11] |
| 66 | Spleen | 45,X,−X,+6,+7,+8,+10,+14,+15[3]/59,X,−X,+1,+4,+5,+6,+7,+8,+9,+10,+10,+11,+11, +12,+13,+15,+15,+16,+17,+18,+18,+19[7]/40,XX[1] |
| 67 | Spleen | 46,XY,+4,+6,+7,+12,+15,+18[6]/47,idem,+16[3] |
| 70 | Spleen | 45,XY,+4,+7,+10,+14,+15[10]/40,XY[1] |
| 1711 | Spleen | 50,XX,+1,+4,+6,+7,+8,+10,+11,+14,+15,+18[1]/49,idem,−X[7]/NCA:49,idem, −X,−X,del(6)(D1F3),+12[1]/40,XX[1] |
| Mouse . | Tissue . | Karyotype . |
|---|---|---|
| PML-RARAmice | ||
| 1111 | BM/spleen | 42,XX,+8,+15[7]/40,XX[2] |
| 909 | Spleen | 39,X,−Y,del(2)(C3H1)[4]/40,idem,+8[2]/40,idem,+16[2]/NCA:41,idem,+8,+16[1]/41,idem,+6,+8[1] |
| 935 | Spleen/LN | 39,X,−Y[7]/40,idem,+16[2]/NCA:40,idem,+15[1]/40,idem,+X[1] |
| 1333 | Spleen | 42,X,−Y,+8,+15,+16[6]/41,idem,−13[3]/NCA:42,idem,+X,−13[1] |
| 1717 | BM/spleen | 44,XY,+8[11],+10[10],+15[8],+17[7][cp11] |
| 1718 | BM/LN | 41,XY,+15[5]/NCA:40,X,+X,−Y[1]/40,XY[4] |
| 1950 | Spleen | 41,XY,+15[9]/40,XY[2] |
| 42 | Spleen | 40,XY[15] |
| 3509 | Spleen | 42,X,−Y,+10,+14,+15[10] |
| 802 | Spleen | 42,X,−X,+6,+7,+14[cp5]/40,XX[4] |
| 1097.1 | Spleen | 50,X,−Y,+1,+4,+8,+10,+12,+14,+15,+der(15)t(14;15)(D3;D1 or D2),+16,+17,+18[10] |
| PML-RARA/BCL2mice | ||
| 2995A4 | Spleen | 41,X,−X,+8,+15[9]/NCA:41,X,−X,−5,+8,−9,+15,+18,+19[1] |
| 1712 | BM | 49,X,−Y,+4,+6,+7,+8,+10,+11,+13,+14,+15,+16[cp5]/40,XY[4] |
| 1713 | Spleen | 42,X,−Y,i(7q),+8,+10,+15[8]/43,idem,+4[2] |
| 65 | Spleen | 45,X,−X,+4,+7,+10,+14,+15,+15[cp11] |
| 66 | Spleen | 45,X,−X,+6,+7,+8,+10,+14,+15[3]/59,X,−X,+1,+4,+5,+6,+7,+8,+9,+10,+10,+11,+11, +12,+13,+15,+15,+16,+17,+18,+18,+19[7]/40,XX[1] |
| 67 | Spleen | 46,XY,+4,+6,+7,+12,+15,+18[6]/47,idem,+16[3] |
| 70 | Spleen | 45,XY,+4,+7,+10,+14,+15[10]/40,XY[1] |
| 1711 | Spleen | 50,XX,+1,+4,+6,+7,+8,+10,+11,+14,+15,+18[1]/49,idem,−X[7]/NCA:49,idem, −X,−X,del(6)(D1F3),+12[1]/40,XX[1] |
BM indicates bone marrow; LN, lymph node; NCA, nonclonal abnormal cell.
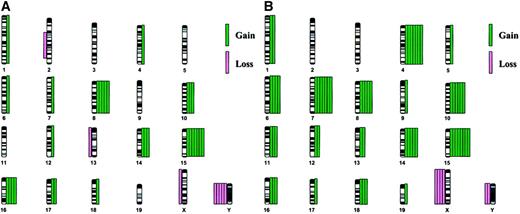
The distribution of chromosome abnormalities is illustrated in Figure1A. The most common abnormality was a gain of chromosome 15, which was observed in 7 (64%) cases; trisomy 15 was the sole abnormality in 2 cases (cases 1718 and 1950, Figure2A-D). Loss of a sex chromosome, an X in females (n = 1) or a Y chromosome in males (n = 5), was noted in 6 cases (55%), and trisomy 8 was observed in 5 (45%) cases. Of note is that trisomy 15 was observed together with trisomy 8 or loss of a sex chromosome in 5 cases.
Chromosomal abnormalities.
The distribution of chromosomal abnormalities in murine leukemias arising in PML-RARA (A, 11 cases) orPML-RARA/BCL2 (B, 8 cases) mice reveals a defined spectrum of numerical abnormalities. Chromosome gain is depicted by green bars on the right of each chromosome, and chromosome loss is depicted by pink bars on the left. Each bar represents a single case.
Chromosomal abnormalities.
The distribution of chromosomal abnormalities in murine leukemias arising in PML-RARA (A, 11 cases) orPML-RARA/BCL2 (B, 8 cases) mice reveals a defined spectrum of numerical abnormalities. Chromosome gain is depicted by green bars on the right of each chromosome, and chromosome loss is depicted by pink bars on the left. Each bar represents a single case.
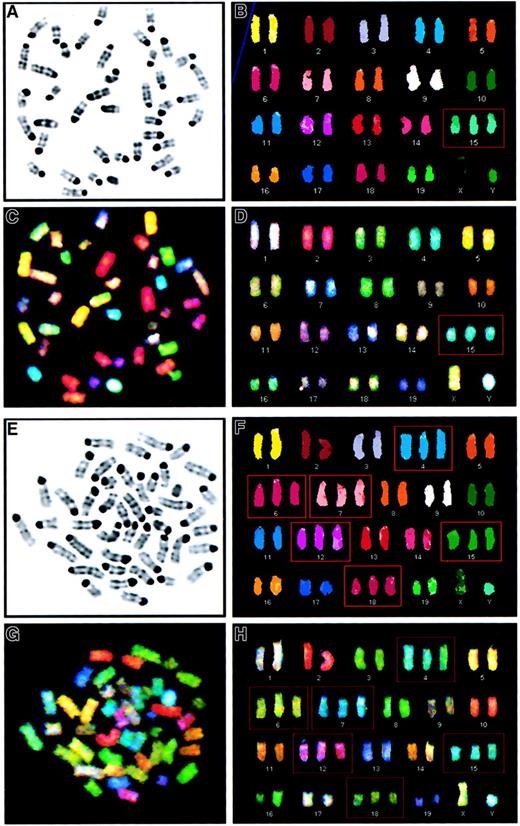
SKY analysis of murine leukemias reveals a pattern of recurring numerical abnormalities.
(A-D). Metaphase cell from the spleen of a PML-RARA mouse with leukemia characterized by trisomy 15 as the sole abnormality (case 1950, 41,XY,+15). (A) Inverted image of the metaphase cell counterstained with 4,6-diamidino-2-phenylindole-dihydrochloride (DAPI). (B) Karyotype of the classified image. (C) Spectral image of the metaphase cell. (D) Karyotype of the spectral image. Trisomy for chromosome 15 is identified by a red box. (E-H). Metaphase cell from the spleen of a PML-RARA/BCL2 mouse with leukemia characterized by a hyperdiploid karyotype with trisomy for chromosomes 4, 6, 7, 12, 15, and 18 (case 67, 46,XY,+4,+6,+7,+12,+15,+18). (E) Inverted image of the DAPI-stained metaphase cell. (F) Karyotype of the classified image. (G) Spectral image of the metaphase cell. (H) Karyotype of the spectral image. Trisomic chromosomes are identified by a red box.
SKY analysis of murine leukemias reveals a pattern of recurring numerical abnormalities.
(A-D). Metaphase cell from the spleen of a PML-RARA mouse with leukemia characterized by trisomy 15 as the sole abnormality (case 1950, 41,XY,+15). (A) Inverted image of the metaphase cell counterstained with 4,6-diamidino-2-phenylindole-dihydrochloride (DAPI). (B) Karyotype of the classified image. (C) Spectral image of the metaphase cell. (D) Karyotype of the spectral image. Trisomy for chromosome 15 is identified by a red box. (E-H). Metaphase cell from the spleen of a PML-RARA/BCL2 mouse with leukemia characterized by a hyperdiploid karyotype with trisomy for chromosomes 4, 6, 7, 12, 15, and 18 (case 67, 46,XY,+4,+6,+7,+12,+15,+18). (E) Inverted image of the DAPI-stained metaphase cell. (F) Karyotype of the classified image. (G) Spectral image of the metaphase cell. (H) Karyotype of the spectral image. Trisomic chromosomes are identified by a red box.
In addition to the frequent involvement of chromosomes 8, 15, and the sex chromosomes, other recurring trisomies observed less often involved chromosomes 10 (3 cases), 14 (3 cases), and 16 (4 cases). With regard to the structural rearrangements observed in 2 cases, an interstitial deletion of chromosome 2 [del(2)(C3H1)] was observed in one case (case 909), and an unbalanced translocation involving chromosomes 14 and 15 [+der(15)t(14;15)(D3;D1 or D2)] was noted in case 1097.1 (Table 1).
Four of the 11 PML-RARA leukemias examined by SKY had been studied previously by CGH.11 In each case, the abnormalities observed with CGH were also seen with SKY. However, in 2 cases, changes present in only some of the metaphase cells were not detected by CGH; this observation was anticipated because SKY allows for the analysis of individual cells.
PML-RARA/BCL2 mice.
Each of the 8 leukemias arising in mice expressing bothPML-RARA and BCL2 had an abnormal karyotype (Table 1) and were characterized by numerical abnormalities. A single case (case 1713) also had a structural abnormality, namely, an isochromosome 7. In 6 cases, a complete karyotype could be defined, whereas a composite karyotype was designated for the remaining 2 abnormal cases (cases 1712 and 65). Multiple clones were detected in 4 cases. Of note is that the karyotypes were more complex, with more abnormalities in each clone, in the PML-RARA/BCL2mice than in the PML-RARA mice. That is, all but 2 cases had 45 or more chromosomes in the abnormal clone(s), as compared to 1 of 11 leukemias arising in PML-RARA mice.
The increased complexity of the karyotypes is illustrated by the distribution of abnormalities in Figure 1B. All 8 cases were characterized by trisomy 15 (100%). Other trisomies that occurred frequently were +4 (7 cases, 88%), +7 (7 cases, 88%), and +10 (6 cases, 75%). Of interest is that every case with +4, also had +7, and 6 of 8 cases had the identical pattern of +4, +7, +10, and +15 (some of these cases had additional abnormalities; Figure 2E-H). Although there are some similarities in the spectrum of abnormalities in leukemias arising in PML-RARA versus PML-RARA/BCL2 mice, there are significant differences. Clonal abnormalities characteristic of both groups include trisomy for chromosomes 8, 10, 14, 15, and 16, and loss of a sex chromosome; however, the frequency of +15 (100% versus 64%), +10 (75% versus 27%), and +8 (63% versus 45%) was greater in the PML-RARA/BCL2 mice. In contrast, recurring trisomy of chromosomes 4, 6, 7, 11, and 18 occurred only in thePML-RARA/BCL2 leukemias, with trisomy 4 and 7 occurring together in 88% of cases.
Only one of the PML-RARA/BCL2 leukemias from the current study had been examined previously with CGH. Although less apparent than in the SKY analysis, the increased karyotypic complexity ofPML-RARA/BCL2 leukemias was also observed with CGH.
Secondary chromosomal abnormalities in human APL
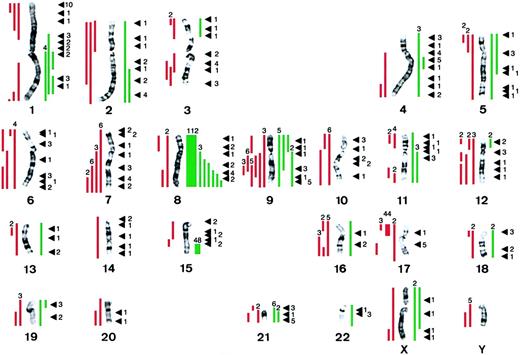
To elucidate the role of secondary chromosomal abnormalities in the pathogenesis of human APL, we accessed Mitelman's compilation in the CGAP database (http://cgap.nci.nih.gov) of 841 cases of APL with the t(15;17) as well as 124 cases ascertained by the Cancer Cytogenetics Laboratory at the University of Chicago (Le Beau et al, unpublished data, August 2001), for a total of 965 cases. Of these, 368 (38%) had numerical or structural abnormalities (or both) in addition to the t(15;17) (Figure 3). In 1986, Heim and Mitelman first reported a high frequency of trisomy 8 in APL,18 and our analysis confirmed this finding. Complete or partial trisomy 8 occurred in 120 or 12.4% of the cases. Rather than having trisomy for an entire chromosome 8, 12 cases had structural rearrangements that led to partial trisomy for 8q. The smallest overlapping region of gain consisted of bands 8q23-24, suggesting that this segment contains the relevant gene(s).
The distribution of secondary chromosomal abnormalities in 965 patients with APL characterized by the t(15;17) reveals a nonrandom pattern with frequent gain of chromosome 8.
Complete chromosomal gain or structural rearrangements leading to partial chromosome gain is depicted by green bars on the right of each chromosome, and complete chromosomal loss or structural rearrangements leading to partial chromosome loss is depicted by red bars on the left. The numbers above the bars indicate the number of patients with the abnormality. Black arrowheads indicate the breakpoints of structural rearrangements; the numbers on the right of each arrowhead indicate the number of patients with a structural abnormality involving this breakpoint.
The distribution of secondary chromosomal abnormalities in 965 patients with APL characterized by the t(15;17) reveals a nonrandom pattern with frequent gain of chromosome 8.
Complete chromosomal gain or structural rearrangements leading to partial chromosome gain is depicted by green bars on the right of each chromosome, and complete chromosomal loss or structural rearrangements leading to partial chromosome loss is depicted by red bars on the left. The numbers above the bars indicate the number of patients with the abnormality. Black arrowheads indicate the breakpoints of structural rearrangements; the numbers on the right of each arrowhead indicate the number of patients with a structural abnormality involving this breakpoint.
An isochromosome of the der(17)t(15;17), which results in loss of 17p, and a gain of 17q11.2-12 and 15q22-qter, was noted in 42 (4.4%) cases. Loss of chromosome 7 or a del(7q) was noted in 17 (1.8%), and −9/del(9q) was identified in 16 cases (1.7%). Other recurring abnormalities observed less commonly (< 1% of cases) were partial trisomy for 1q (7 cases), +9 (5 cases), −10 (6 cases), del(11q) (3 cases), −12 or loss of 12p (7 cases), −17 or loss of 17p (7 cases), and +21 or gain of 21q (8 cases). Loss of a Y chromosome was observed in 5 cases, and loss of an X chromosome occurred in only 1 case. With respect to balanced rearrangements, the most common breakpoints were 1p36 (10 cases, 1.0%), 4q21, 9q34, 17q21, and 21q22 (5 cases each).
Discussion
By using cytogenetic and SKY analysis, we have demonstrated that myeloid leukemias arising in PML-RARA andPML-RARA/BCL2 mice have a defined spectrum of recurring numerical abnormalities. Trisomy 15, trisomy 8, monosomy X or Y, and trisomy 16 are commonly observed in PML-RARA mice, whereas these abnormalities along with +4, +6, +7,+10, +11, +14, and +18 are characteristic of leukemias arising in PML-RARA/BCL2 mice. As described below, these secondary mutations recapitulate, in part, those involved in the pathogenesis of human APL, particularly trisomy 8. Although our data strongly implicate unbalanced secondary mutations, such as numerical abnormalities, as contributing to the pathogenesis of the murine leukemias, we cannot formally exclude the presence of subtle structural rearrangements that are below the limits of detection by SKY analysis.
A number of investigators have demonstrated that expression ofPML-RARA and, in some models, both PML-RARA andRARA-PML fusion genes in myeloid lineage cells result in leukemias with features of human APL after a latency of 6 to 13 months.8-10 However, our understanding of the additional genetic events that contribute to leukemogenesis in these models is poor. Recently, Zimonjic et al19 reported the identification of recurring abnormalities in a cathepsin G-PML-RARA model, particularly in leukemias arising in mice doubly transgenic for both PML-RARA andRARA-PML. These investigators did not define the abnormal clones, precluding a comparison of the karyotypic complexity, and the pattern of abnormalities. Of interest is that they also observed a high frequency of cytogenetic aberrations that led to an unbalanced chromosomal complement, such as the gain or loss of whole chromosomes, or deletions. There are some interesting similarities, as well as notable differences, between their data and ours. For example, Zimonjic and colleagues identified an interstitial deletion of chromosome 2 in 11 of 13 (85%) doubly transgenic mice, and in 1 of 5PML-RARA mice, whereas we detected a del(2) in only 1 of 11PML-RARA mice. These results raise the possibility that RARA-PML expression increases the likelihood of this abnormality. In contrast, trisomy15 occurred more frequently in our series (64% in our study versus 38% in their doubly transgenic mice or 20% in theirPML-RARA mice). Loss of a sex chromosome occurred with equal frequency in both series (55% versus 70% and 40%). Finally, we did not observe loss of chromosome 11, which they detected in 27% of leukemias examined. Importantly, their APL model differs from ours with respect to both genetic background strain (C3H X C57Bl/6 versus FVB/N) as well as the promoter that drives transgene expression (cathepsin G versus MRP8 in our model). Nonetheless, the similarities observed in these 2 studies, such as trisomy 15 and loss of a sex chromosome, suggest that there may be common genetic events in the pathways to leukemia.
The hematologic malignant diseases have provided important model systems for defining the role of chromosomal abnormalities in the pathogenesis of human tumors.20 Primary chromosomal abnormalities are believed to be essential for the initiation and establishment of the neoplasm, whereas secondary chromosomal abnormalities are thought to be involved in tumor progression.21 Our analysis of a large series of human APLs revealed that 38% had secondary abnormalities. A similar frequency (31%-39%) has been reported in earlier studies based on smaller series.22-26 Whether the presence of secondary chromosomal abnormalities alters the outcome in APL is uncertain. Slack et al25 reported the results of a Cancer and Leukemia Group B analysis of 161 APL patients (protocol 8461), and noted that the overall survival did not differ between patients with or without additional cytogenetic abnormalities (27.7 versus 24.4 months, respectively, P = .2825). In an analysis of 198 patients treated in the Medical Research Council 10 trial, Grimwade et al26 observed no difference in CR rate and overall 3-year survival (OS) for patients with the t(15;17) alone (88% CR, 69% OS), as compared to those patients with trisomy 8 (CR 84%, OS 71%) or other secondary abnormalities (CR 90%, OS 66%). In contrast, the results of one study suggested that the presence of additional abnormalities in APL had an adverse effect on prognosis, independent of other prognostic factors.24 A confounding factor is that we do not know the molecular consequences of the common additional chromosomal abnormalities in APL; thus, we cannot determine whether patients without these visible chromosomal abnormalities have sustained mutations that result in the same functional consequences.
The most common secondary abnormality in patients with APL, trisomy 8, was observed in 12.4% of cases in this report, and in 14% to 18% of cases examined in previous reports.22-26 Of note is that the results of a recent study that used fluorescence in situ hybridization of a chromosome 8–specific centromere probe suggested that the frequency of trisomy 8 may approach 30% in newly diagnosed patients with APL.27 The identification of patients with structural rearrangements leading to partial trisomy, rather than complete +8, has led to the proposal that the critical portion is 8q. The smallest overlapping region of gain is 8q23-24.
How do the cytogenetic abnormalities found in human APLs compare with those we have observed in murine leukemias initiated byPML-RARA? Perhaps the most intriguing correlation involves mouse chromosome 15, the most frequently gained chromosome in the murine leukemias. A map of the mouse and human synteny for this chromosome reveals a complex pattern with synteny to 8 different segments from human chromosomes 2, 5, 6, 8, 12, and 22. Of note is that there are 3 regions of synteny to the distal long arm of human chromosome 8, bands q22-24, and this corresponds to the smallest overlapping region of gain, 8q23-24, noted in patients with APL. However, it is also possible that genes encoded by other regions of human chromosome 8 are involved in the pathogenesis of APL. Human chromosome 8 is syntenic to regions of mouse chromosomes 1, 3, 4, 8, 14, 15, and 16. With the exception of murine chromosomes 1 and 3 (syntenic to segments of human 8q13-21), a gain of each of these chromosomes was observed at a high frequency in leukemias (38%-100%) arising in PML-RARA/BCL2 mice.
Trisomy 15 is the most frequent abnormality in murine T-cell lymphomas (40%-90%) arising spontaneously or induced by diverse agents, including ionizing radiation, chemical carcinogens, or tumorigenic viruses; this abnormality has also been identified in B/myeloid leukemias arising in irradiated Eμ-BCL2 transgenic mice.28-32 By analyzing T-cell lymphomas arising in mice with constitutional translocations involving chromosome 15, Silva et al33 demonstrated that trisomy for bands 15D2-3 (syntenic to human 8q23-24) contributes to the development or progression (or both) of these T-cell lymphomas. This region contains theMyc and Pvt1 genes.
At present, we do not know the identity of the relevant gene(s) on human 8q; however, there are 53 fully cloned genes mapped to 8q23-24. A number of these genes, or their family members, have been shown to regulate cell growth, differentiation, or apoptosis. These include the genes encoding interleukin 7, the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase, the E2F5 transcription factor, Nibrin, RAD54B, cyclin E2, and serine/threonine kinase 3 (STK3). Perhaps the best candidate gene is the MYC oncogene, which is overexpressed in a number of human tumors. Gene expression profiling of AMLs characterized by trisomy 8 has demonstrated that the leukemia blasts overexpress genes on chromosome 8, suggesting that gene dosage effects mediate leukemogenesis.34 Of interest is that expression of the MYC gene was not consistently altered in leukemias with trisomy 8.
In the murine models for APL, genetic mutations that suppress apoptosis, for example, coexpression of BCL2, cooperate with PML-RARA in the pathogenesis of APL.11 PML-RARA/BCL2mice had a marked expansion of immature myeloid cells in the bone marrow, perhaps increasing the likelihood that additional leukemogenic mutations would occur. We have demonstrated that murine leukemias initiated by PML-RARA or PML-RARA/BCL2 have a defined spectrum of genetic changes. Nevertheless, the increased karyotypic complexity of the PML-RARA/BCL2 leukemias revealed by SKY suggests that the particular spectrum of mutations may differ depending on the cooperating events. Each cooperating event, such as BCL2 expression, may not only facilitate the acquisition of genetic changes, but also lead to a specific pattern of mutations that complete leukemic transformation.
The pathogenesis of human APL may parallel that of murine APL models. The initial mutation, the t(15;17) and resultant expression of the PML-RARA fusion protein, may initiate leukemogenesis. The acquisition of cooperating genetic mutations that enhance survival, such as BCL2 expression, or impair differentiation, may occur in some patients, and facilitate the development of additional genetic changes that ultimately result in acute leukemia. The results of several studies have suggested that BCL2 is expressed in most APLs; however, the mechanism leading to expression of this gene is unknown.35These results raise the possibility that deregulation of apoptosis, either by down-regulation of proteins that induce apoptosis, or overexpression of proteins that inhibit apoptosis, cooperate with gene dosage effects created by a gain of chromosome 8 in human AMLs, or trisomy 15 in murine AMLs in mediating leukemogenesis.
We thank Dr Kevin Shannon for helpful discussions, the technologists in the Cancer Cytogenetics Laboratory for expert technical assistance, Marjorie Isaacson for data management, and Rafael Espinosa III for assistance in preparing the figures.
Supported by National Institutes of Health (NIH) grants U01 CA84221 (M.M.L, S.C.K) and K08 CA75986 (S.C.K). S.C.K. is a recipient of a Burroughs Wellcome Fund Career Award and is an Edward Mallinckrodt Jr Foundation Scholar.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michelle M. Le Beau, Section of Hematology/Oncology, University of Chicago, 5841 S Maryland Ave, MC2115, Chicago, IL 60637; e-mail: mlebeau@medicine.bsd.uchicago.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal