Abstract
Point mutations were found in the adenosine triphosphate (ATP) binding region of BCR/ABL in 12 of 18 patients with chronic myeloid leukemia (CML) or Ph-positive acute lymphoblastic leukemia (Ph+ ALL) and imatinib resistance (defined as loss of established hematologic response), but they were found in only 1 of 10 patients with CML with imatinib refractoriness (failure to achieve cytogenetic response). In 10 of 10 patients for whom samples were available, the mutation was not detected before the initiation of imatinib therapy. Three mutations (T315I, Y253H, and F317L present in 3, 1, and 1 patients, respectively) have a predicted role in abrogating imatinib binding to BCR/ABL, whereas 3 other mutations (E255K, G250E, and M351T, present in 4, 2, and 2 patients, respectively) do not. Thus we confirm a high frequency of mutations clustered within the ATP-binding region of BCR/ABL in resistant patients. Screening may allow intervention before relapse by identifying emerging mutations with defined impacts on imatinib binding. Certain mutations may respond to higher doses of imatinib, whereas other mutations may mandate switching to another therapeutic strategy.
Introduction
Treatment options are limited for patients with chronic myeloid leukemia (CML) for whom interferon-α therapy has failed or who are in the acute phase of the disease.1 The tyrosine kinase inhibitor imatinib mesylate (Glivic; Novartis Pharmaceuticals, Basel, Switzerland) (formerly STI571) frequently induces significant hematologic and cytogenetic responses in these clinical settings.2,3Imatinib acts as a competitor for adenosine triphosphate (ATP) binding4 and selectively induces apoptosis and blocks proliferation in BCR/ABL–expressing cells.5 A proportion of patients are primarily refractory to imatinib, and some patients, particularly those in the later stages of the disease, acquire clinical resistance after achieving a response.3 In vitro studies of resistance to imatinib indicate various mechanisms of resistance, including overexpression of BCR/ABL because of gene amplification6-8 or increased imatinib efflux mediated by the multidrug resistance P-glycoprotein.9 In a recent report,10 clinical resistance was associated with the reactivation of BCR/ABL tyrosine kinase activity in all patients studied. In 6 of 9 patients, this was attributed to the mutation of a single amino acid (T315I) in the ATP-binding site of BCR/ABL.
We used a reverse transcription–polymerase chain reaction (RT-PCR) strategy to amplify and sequence the ABL kinase domain of BCR/ABL. Twenty-eight patients who showed resistance or cytogenetic refractoriness were selected from patients enrolled in expanded access studies within Australia. Our aim was to determine the frequency and timing of acquired mutations within defined clinical groups and to establish their distribution within the BCR/ABL kinase domain.
Study design
Resistance (n = 18 patients) was defined as loss of complete hematologic remission that had been present for at least 3 months, loss of complete hematologic remission with transformation to accelerated or blastic phase after a period of chronic-phase CML, or relapsed Ph-positive acute lymphoblastic leukemia (Ph+ ALL) after complete hematologic remission had been established. Cytogenetic refractoriness (n = 10 patients) was defined as failure to achieve a major cytogenetic response after at least 6 months of therapy. Three hundred fourteen Australian patients with CML or Ph+ ALL were enrolled in trials of imatinib as the sole therapy. Patients who were eligible and for whom samples were available within the 3-month period of the study were included.
Extraction of RNA from blood, RT, and direct sequencing procedures have been described.11 A long PCR method12 was used to amplify the ABL kinase domain of the BCR/ABL allele with forward primer BCRF (5′- TGACCAACTCGTGTGTGAAACTC) and reverse primer ABLKinaseR (5′-TCCACTTCGTCTGAGATACTGGATT). A second-stage PCR used forward primer ABLkinaseF (5′-CGCAACAAGCCCACTGTCT) and reverse primer ABLkinaseR. The entire kinase domain was sequenced in the forward and reverse directions; this area included 863 bases (GenBank accession number M14752).
Results and discussion
Resistance
Twelve of 18 resistant patients had mutations in the ATP binding region of BCR/ABL (Table 1). In 9 patients for whom samples were available, the mutation was not detected before imatinib administration was initiated, nor was it detected in 4 patients tested at 3 to 9 months—before the onset of resistance in each patient. This might have been because it was not present or it was below the level of detection.
Clinical course and mutation analysis of patients treated with imatinib
| Patient . | Age (y)/sex . | Disease status at start of imatinib . | Duration of imatinib treatment (mo) . | Best response to imatinib . | Response at time of mutation analysis . | Time of mutation analysis* . | Mutation result . | Nucleotide substitution (GenBank accession no. M14752) . |
|---|---|---|---|---|---|---|---|---|
| Resistant | ||||||||
| 1 | 61 F | CP (3rd) | 9 | CCR | LBC | Before study | NM | — |
| CP (3rd) | Before study | NM | — | |||||
| CCR | 3 mo | NM | — | |||||
| CCR | 6 mo | NM | — | |||||
| LBC | 9 mo | T315I | 944C>T | |||||
| 2 | 75 F | MBC | 4.5 | PHR | MBC | Before study | NM | — |
| MBC | 4.5 mo1-154 | T315I | 944C>T | |||||
| 3 | 62 M | LBC | 2 | PHR | LBC | Before study | NM | — |
| LBC | 2 mo1-154 | T315I | 944C>T | |||||
| 4† | 59 F | Rel | 3 | CCR | Rel | Before study | NM | — |
| Rel | 5 mo | Y253H | 757T>C | |||||
| Rel | 5.5 mo | Y253H | 757T>C | |||||
| 5 | 40 M | CP | 8 | CHR | CP | Before study | NM | — |
| CHR | 4 mo | F317L | 951C>G | |||||
| PHR | 7 mo | F317L | 951C>G | |||||
| 6 | 66 M | LBC | 81-153 | CHR | LBC | 8 mo | G250E | 749G>A |
| 7 | 54 F | MBC | 10.5 | CHR | MBC | Before study | NM | — |
| CHR | 6 mo | NM | — | |||||
| CHR | 9 mo | NM | — | |||||
| CHR | 10 mo1-154 | G250E | 749G>A | |||||
| PHR | 10.5 mo1-154 | G250E | 749G>A | |||||
| 8 | 41 M | AP | 6 | CHR | AP | 6 mo1-154 | E255K | 763G>A |
| 9 | 60 M | AP | 5 | CHR | CHR | 3 mo | NM | — |
| PHR | 6 mo1-154 | E255K | 763G>A | |||||
| 10 | 44 M | AP | 4.5 | CHR | AP | Before study | NM | — |
| PHR | 3 mo1-154 | E255K | 763G>A | |||||
| 11† | 60 F | Rel | 3 | CHR | Rel | Before study | NM | — |
| Rel | 3 mo1-154 | E255K | 763G>A | |||||
| 12 | 52 F | CP | 9 | CCR | CP | Before study | NM | — |
| CCR | 6 mo | NM | — | |||||
| CCR | 7 mo | NM | — | |||||
| MCR | 8 mo1-154 | M351T | 1052T>C | |||||
| AP1-155 | 9 mo1-154 | M351T | 1052T>C | |||||
| 13 | 64 M | CP | 10.5 | CHR | AP1-155 | 10 mo | NM | — |
| 14 | 78 F | MBC | 8 | CHR | AP1-155,‡ | 8 mo | NM | — |
| 15 | 47 M | AP‡ | 9 | AP‡ | MBC‡ | 7 mo | NM | — |
| 16 | 56 F | CP | 7 | CHR | AP | 6 mo | NM | — |
| 17 | 37 M | MBC | 7 | PHR | MBC | 4.5 mo | NM | — |
| 18 | 63 M | AP | 7 | PHR | AP | 8 mo | NM | — |
| Refractory | ||||||||
| 19 | 55 F | AP‡ | 6 | AP‡ | AP‡ | 3.5 mo | NM | — |
| 20 | 63 M | AP‡ | 11 | AP‡ | AP‡ | Before study | NM | — |
| AP‡ | 3 mo | NM | — | |||||
| AP‡ | 8 mo1-154 | M351T | 1052T>C | |||||
| AP‡ | 9 mo1-154 | M351T | 1052T>C | |||||
| AP‡ | 10 mo1-154 | M351T | 1052T>C | |||||
| AP‡ | 11 mo1-154 | M351T | 1052T>C | |||||
| 21 | 62 F | AP | 10.5 | CHR | PHR | 9 mo | NM | — |
| 22 | 54 M | AP | 8 | CHR | CHR | 6 mo | NM | — |
| 23 | 61 F | CP | 6 | CHR | PHR | 5 mo | NM | — |
| 24 | 61 F | CP | 10.5 | CHR | PHR | 6 mo | NM | — |
| 25 | 47 F | CP | 10 | CHR | CHR | 6 mo | NM | — |
| 26 | 40 F | CP | 6 | CHR | CHR | 3 mo | NM | — |
| 27 | 60 M | CP | 6 | CHR | CHR | 6 mo | NM | — |
| 28 | 42 M | CP | 7 | CHR | CHR | 6 mo | NM | — |
| Patient . | Age (y)/sex . | Disease status at start of imatinib . | Duration of imatinib treatment (mo) . | Best response to imatinib . | Response at time of mutation analysis . | Time of mutation analysis* . | Mutation result . | Nucleotide substitution (GenBank accession no. M14752) . |
|---|---|---|---|---|---|---|---|---|
| Resistant | ||||||||
| 1 | 61 F | CP (3rd) | 9 | CCR | LBC | Before study | NM | — |
| CP (3rd) | Before study | NM | — | |||||
| CCR | 3 mo | NM | — | |||||
| CCR | 6 mo | NM | — | |||||
| LBC | 9 mo | T315I | 944C>T | |||||
| 2 | 75 F | MBC | 4.5 | PHR | MBC | Before study | NM | — |
| MBC | 4.5 mo1-154 | T315I | 944C>T | |||||
| 3 | 62 M | LBC | 2 | PHR | LBC | Before study | NM | — |
| LBC | 2 mo1-154 | T315I | 944C>T | |||||
| 4† | 59 F | Rel | 3 | CCR | Rel | Before study | NM | — |
| Rel | 5 mo | Y253H | 757T>C | |||||
| Rel | 5.5 mo | Y253H | 757T>C | |||||
| 5 | 40 M | CP | 8 | CHR | CP | Before study | NM | — |
| CHR | 4 mo | F317L | 951C>G | |||||
| PHR | 7 mo | F317L | 951C>G | |||||
| 6 | 66 M | LBC | 81-153 | CHR | LBC | 8 mo | G250E | 749G>A |
| 7 | 54 F | MBC | 10.5 | CHR | MBC | Before study | NM | — |
| CHR | 6 mo | NM | — | |||||
| CHR | 9 mo | NM | — | |||||
| CHR | 10 mo1-154 | G250E | 749G>A | |||||
| PHR | 10.5 mo1-154 | G250E | 749G>A | |||||
| 8 | 41 M | AP | 6 | CHR | AP | 6 mo1-154 | E255K | 763G>A |
| 9 | 60 M | AP | 5 | CHR | CHR | 3 mo | NM | — |
| PHR | 6 mo1-154 | E255K | 763G>A | |||||
| 10 | 44 M | AP | 4.5 | CHR | AP | Before study | NM | — |
| PHR | 3 mo1-154 | E255K | 763G>A | |||||
| 11† | 60 F | Rel | 3 | CHR | Rel | Before study | NM | — |
| Rel | 3 mo1-154 | E255K | 763G>A | |||||
| 12 | 52 F | CP | 9 | CCR | CP | Before study | NM | — |
| CCR | 6 mo | NM | — | |||||
| CCR | 7 mo | NM | — | |||||
| MCR | 8 mo1-154 | M351T | 1052T>C | |||||
| AP1-155 | 9 mo1-154 | M351T | 1052T>C | |||||
| 13 | 64 M | CP | 10.5 | CHR | AP1-155 | 10 mo | NM | — |
| 14 | 78 F | MBC | 8 | CHR | AP1-155,‡ | 8 mo | NM | — |
| 15 | 47 M | AP‡ | 9 | AP‡ | MBC‡ | 7 mo | NM | — |
| 16 | 56 F | CP | 7 | CHR | AP | 6 mo | NM | — |
| 17 | 37 M | MBC | 7 | PHR | MBC | 4.5 mo | NM | — |
| 18 | 63 M | AP | 7 | PHR | AP | 8 mo | NM | — |
| Refractory | ||||||||
| 19 | 55 F | AP‡ | 6 | AP‡ | AP‡ | 3.5 mo | NM | — |
| 20 | 63 M | AP‡ | 11 | AP‡ | AP‡ | Before study | NM | — |
| AP‡ | 3 mo | NM | — | |||||
| AP‡ | 8 mo1-154 | M351T | 1052T>C | |||||
| AP‡ | 9 mo1-154 | M351T | 1052T>C | |||||
| AP‡ | 10 mo1-154 | M351T | 1052T>C | |||||
| AP‡ | 11 mo1-154 | M351T | 1052T>C | |||||
| 21 | 62 F | AP | 10.5 | CHR | PHR | 9 mo | NM | — |
| 22 | 54 M | AP | 8 | CHR | CHR | 6 mo | NM | — |
| 23 | 61 F | CP | 6 | CHR | PHR | 5 mo | NM | — |
| 24 | 61 F | CP | 10.5 | CHR | PHR | 6 mo | NM | — |
| 25 | 47 F | CP | 10 | CHR | CHR | 6 mo | NM | — |
| 26 | 40 F | CP | 6 | CHR | CHR | 3 mo | NM | — |
| 27 | 60 M | CP | 6 | CHR | CHR | 6 mo | NM | — |
| 28 | 42 M | CP | 7 | CHR | CHR | 6 mo | NM | — |
MBC, myeloid blast crisis; LBC, lymphoid blast crisis; Rel, relapsed Ph+ ALL; AP, accelerated phase; CP, chronic phase; PHR, partial hematologic response (blasts in blood + bone marrow < 15%); CHR, complete hematologic response (WBC, < 10.0; platelets, < 450; no blasts, myelocytes, metamyelocytes, < 5%; no promyelocytes, no disease-related symptoms, or extramedullary disease); MCR, major cytogenetic response; CCR, complete cytogenetic response; NM, no mutation detected after sequencing of the kinase region of BCR/ABL in both directions.
Months since treatment initiation.
Patients with Ph+ ALL.
Additional chromosomal abnormalities.
Although the patient was in the study for 8 months, imatinib was alternated monthly with oral arsenic. Therefore, the patient was aministered only 4 months of imatinib during this period.
Double Philadelphia chromosome.
Mixed population of mutant and wild-type BCR/ABL detected by direct sequencing of RT-PCR product.
Mutations were identified that have a predicted or proven role in abrogating imatinib binding to BCR/ABL, as follows: T315I (n = 3), Y253H (n = 1), and F317L (n = 1). The T315I mutation confers resistance in vitro10 and is predicted to disrupt a hydrogen bond between imatinib and BCR/ABL.13 Mutations at amino acids 253 and 317 are predicted to impair binding of imatinib to BCR/ABL by disrupting van der Waals interactions.13Directed mutagenesis of amino acid 253 was shown to activate c-ABL transformation,14 and it induced resistance to imatinib by an increased tendency to autophosphorylate.15 16 Other mutations were also observed—E255K (n = 4), G250E (n = 2), and M351T (n = 1). On prior structural knowledge, these would not have been predicted to disrupt binding between imatinib and the kinase domain. In all patients, acquisition of the mutation was closely linked to the development of resistance. It is likely that mutations at residues adjacent to contact points also led to the disruption of interactions between imatinib and the kinase domain.
The 18 resistant patients could be subdivided by disease stage. Six of 8 patients who had relapses directly into blast crisis/ALL had mutations. In 3 of these patients, the T315I mutation was present, and it has been strongly linked to blast crisis relapse.10 Two of 6 patients who had relapses into the accelerated phase and all 4 patients who had relapses into the chronic phase had mutations.
Cytogenetic refractoriness
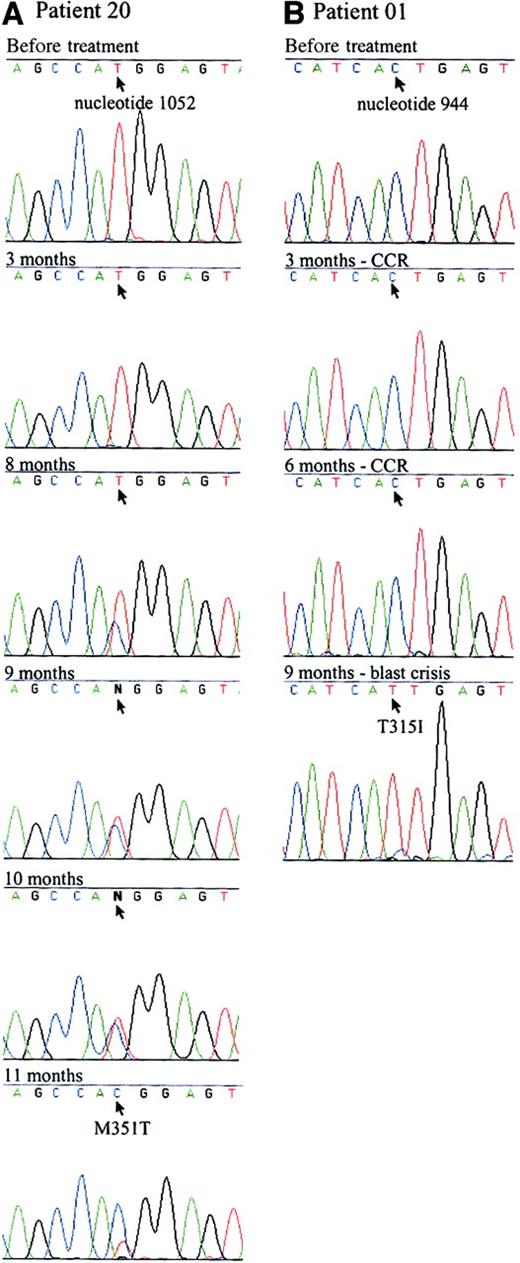
Only 1 of 10 refractory patients had a mutation (Table 1). This mutation was not detected before the administration of imatinib or 3 months after it. The mutant clone emerged at 8 months and persisted in a mixed pattern until it became predominant at 11 months (Figure1). This patient had additional complex chromosomal abnormalities before imatinib therapy that were still present at 6 months. Further disease progression was not evident during the period in which the mutation emerged.
Sequence analysis of 2 patients with point mutations in the ATP binding site.
The 863-bp RT-PCR products of the ABL kinase domain of BCR/ABL were sequenced. Arrows indicate the mutated nucleotide. (A) A point mutation was first detected in patient 20 (Table 1) at the eighth month of imatinib therapy. A mix of mutant and wild-type BCR/ABL was evident from months 8 to 11. The predominant nucleotide switched from wild-type to mutant over the course of 4 months. Sequence Navigator software (Applied Biosystems, Foster City, CA) was unable to distinguish between the wild-type and mutant nucleotides at months 9 and 10, as indicated by N at the mutated site. The patient remains in accelerated phase. (B) Patient 1 (Table 1) had a complete cytogenetic response (CCR) to imatinib therapy but relapsed into blast crisis at 9 months. The T315I mutation was predominant at that time but was not evident when tested before study in chronic phase or in a previous lymphoid blast crisis sample. The patient was refractory to treatment and has since died.
Sequence analysis of 2 patients with point mutations in the ATP binding site.
The 863-bp RT-PCR products of the ABL kinase domain of BCR/ABL were sequenced. Arrows indicate the mutated nucleotide. (A) A point mutation was first detected in patient 20 (Table 1) at the eighth month of imatinib therapy. A mix of mutant and wild-type BCR/ABL was evident from months 8 to 11. The predominant nucleotide switched from wild-type to mutant over the course of 4 months. Sequence Navigator software (Applied Biosystems, Foster City, CA) was unable to distinguish between the wild-type and mutant nucleotides at months 9 and 10, as indicated by N at the mutated site. The patient remains in accelerated phase. (B) Patient 1 (Table 1) had a complete cytogenetic response (CCR) to imatinib therapy but relapsed into blast crisis at 9 months. The T315I mutation was predominant at that time but was not evident when tested before study in chronic phase or in a previous lymphoid blast crisis sample. The patient was refractory to treatment and has since died.
As did Gorre et al,10 we have shown a high frequency of mutations in patients with resistance though our mutations were diverse. This may relate to the greater diversity of patients within the resistance category in our study. Recently, 2 groups reported ABL kinase domain mutations at lower frequency (2 of 44 patients with relapsed or refractory disease).17,18 Specific details were not given to determine how the patients were classified or the exact number of patients in each category. The median duration of therapy, reported in only one of the studies, was 95 days18 compared with 7 months in our study. However, 10 of the patients in our study acquired early resistance (median, 3 months), and we found mutations in 7 of them. None of the 10 patients with refractory disease acquired mutations before 6 months. The differences in the frequency of mutation detection may be attributed to differences in the sensitivities of the techniques, differences in the time point of analysis, or, as Gorre et al19 state, differences between the patient populations in the studies.
We conclude that point mutations within the ATP-binding region of BCR/ABL frequently emerge in patients with CML and Ph+ ALL who show signs of resistant disease. These mutations are likely to partially or totally abrogate imatinib binding to BCR/ABL. Pre-existing mutations or polymorphisms in the BCR/ABL kinase domain, which would explain primary refractoriness, were not detected, but we only studied a limited number of patients. Additional incidences of imatinib refractoriness, particularly in patients who do not demonstrate hematologic responses, must be assessed.
Given the detection of a mixed population of mutant and wild-type BCR/ABL in association with gradual disease progression in some patients (Table 1), regular tests for emerging mutations in patients judged at risk may be warranted. Once a mutation is detected, intervention such as the cessation of imatinib and alternative therapy can be tested. Dose escalation might be of value, depending on the IC50 of the mutation and on whether sufficiently high levels of imatinib could be achieved. The choice of intervention may depend on the impact of the mutation on imatinib binding and kinase activity. Finally, it is important to recognize that the development of point mutations in the ATP-binding region of BCR/ABL kinase is one of a range of mechanisms of drug resistance, including gene amplification and increased expression of P-glycoprotein, that should be considered when designing screening strategies.
We thank Bernadette Miller for invaluable technical support and the trial coordinators and laboratory staff at Royal North Shore Hospital, Royal Melbourne Hospital, Mater Hospital (Brisbane), and Royal Perth Hospital for their excellent assistance. We also thank Arthur Mangos and the Sequencing Centre staff.
Supported in part by grants from Novartis Pharmaceuticals Australia.
T.P.H., A.G., C.A., K.T., and R.H. served as investigators in clinical trials with imatinib. T.P.H. is an adviser to Novartis Pharmaceuticals on clinical issues relating to imatinib. K.P.L. is employed by Novartis Pharmaceuticals.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Susan Branford, Division of Molecular Pathology, Institute of Medical and Veterinary Science, South Australia, 5000, Australia; e-mail: susan.branford@imvs.sa.gov.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal