Key Points
FLT3-TKD/NPM1 double mutation status is associated with superior relapse-free survival compared with NPM1-only–mutated AML.
Allogeneic stem cell transplant in complete responders does not improve outcomes in FLT3-TKD/NPM1 AML patients.
Abstract
Although FLT3 internal tandem duplication (ITD) mutations in acute myeloid leukemia (AML) confer an adverse prognosis, co-occurrence with a nucleophosphomin (NPM1) mutation partially improves response and survival outcomes. In contrast, simultaneous NPM1 and FLT3 tyrosine kinase domain (TKD) mutations were reported to improve response over that of an isolated NPM1 mutation in one as yet unverified report. To validate this, we explored the impact of the co-occurrence of FLT3-TKD and NPM1 mutations on clinical outcomes. Study populations included 21 patients (8%) with FLT3-TKD+NPM1+ mutated, 18 patients (7%) with FLT3-TKD-only–mutated, 117 patients (44%) with NPM1-only–mutated, and 107 patients (41%) with FLT3-ITD+NPM1–mutated AML. Compared with NPM1+-only–mutated AML, FLT3-TKD/NPM1 double mutation status was associated with a significantly superior relapse-free survival (median, not reached vs 18.3 months; P = .03) and a trend toward improved overall survival (OS). The presence of FLT3-TKD/NPM1 double mutation status was an independent positive predictor in multivariable analysis. Allogeneic stem cell transplant did not improve outcomes in the FLT3-TKD/NPM1 cohort. Consistent with historical data, the co-mutation status defined a highly favorable prognostic group characterized by high response rates and prolonged disease-free and OS. These study findings substantiate previous data describing this intriguing paradoxical cooperative effect. Our results emphasize the need for elucidating the mechanistic links between FLT3-TKD and NPM1 in future molecular and murine model studies.
Introduction
Nucleophosphomin (NPM1) is a ubiquitously expressed nucleocytoplasmic shuttling protein that plays an active role in ribosomal protein assembly, chromatin remodeling, and DNA repair, replication, and transcription.1-3 Mutations in the NPM1 gene are noted in ∼35% of acute myeloid leukemia (AML) cases.4 The favorable prognosis conferred by NPM1 mutations was most recently confirmed in a meta-analysis, which demonstrated NPM1 mutations to be associated with higher complete remission (CR) rates and prolonged disease-free and overall survival (OS).5 In regards to FMS-like tyrosine kinase 3 (FLT3), the more common internal tandem duplication (ITD) is associated with short remission durations and high relapse rates after conventional induction therapy,6 whereas the less frequent tyrosine kinase domain (TKD) mutations are of unclear prognostic relevance. The incidence of ITD and TKD mutations in FLT3-mutated AML vary slightly according to age, clinical risk groups, and cytogenetic profile.6 Although both classes result in constitutive activation of the FLT3 receptor, differences in receptor activation likely account for the different clinical and biological features observed for these mutations.7 In vitro studies have demonstrated that FLT3-ITD and FLT3-TKD mutants harbor differing gene expression profiles, especially in regards to STAT5 target gene expression.8
Murine models have shown mutant NPM1 and FLT3-ITD to exhibit a marked and potent molecular synergy toward driving AML pathogenesis.9 Patients with FLT3-ITD and NPM1 mutations have improved response rates and disease-free and OS compared with those who only have the FLT3-ITD mutation, although outcomes are inferior to NPM1-only–mutated AML.5 In a previous study, Bacher et al10 showed that, in contrast to the prognostic impact of FLT3-ITD on NPM1, FLT3-TKD has an additional favorable impact on outcomes in NPM1-mutated AML, suggesting a cooperative positive effect. This favorable effect of FLT3-TKD-NPM1 on OS compared with the overall AML population was recently confirmed by Pappaemanuil et al.11 However, case numbers in this particular group in the Bacher et al10 study were few. We therefore sought to validate this observation in our sample cohort.
Methods
Adult (≥18 years of age) patients newly diagnosed with AML (by standard criteria) who were treated at MD Anderson Cancer Center from January 2009 to January 2017 were evaluated. The study was approved by the institutional internal review board. Informed consent was obtained following institutional guidelines, and patients were treated in accordance with the rules of the Declaration of Helsinki. Molecular mutation analyses on FLT3 and NPM1 mutations, from January 2010 to September 2012, were performed by using a single gene testing approach of polymerase chain reaction amplification followed by Sanger sequencing, using previously described methodology.12 From September 2012 on, NPM1 and FLT3 testing was performed within our panel-based, next-generation sequencing hematologic malignancy platform, as previously described.13 FLT3 AML cases for which NPM1 mutation analysis was not performed, and vice versa, were excluded. Minimal residual disease elimination status, assessed by clearance of NPM1,14 was also collected. The study population was grouped into 4 study cohorts: (1) FLT3-ITD+NPM1+, (2) FLT3-TKD+NPM1+, (3) NPM1 only, and (4) FLT3-TKD only. Patients were treated with a variety of frontline protocols active during this period, and these were broadly classified into: (1) high- or intermediate-intensity idarubicin with cytarabine–based regimens, and (2) low-intensity hypomethylating agent– and low-dose cytarabine–based regimens. Response on hypomethylating agents was classified based on achieving CR or partial response; stable disease was classified as nonresponse. Categorical variables were compared by using the χ2/Fisher’s exact test, and continuous variables were compared by using the Mann-Whitney U test. Response criteria were graded according to international working group criteria.15 OS was measured from time of treatment to date of death or censored at last follow-up date. Relapse-free survival (RFS) was calculated from time of remission to date of death or relapse, or censored at last follow-up date. Response-survival relationships were studied by multivariable Cox regression analysis, and covariates were chosen by stepwise variable selection procedures. Statistical significance was defined as a 2-sided P value of <.05.
Results
A total of 1319 patients were evaluated during this period, and 1282 had an FLT3 mutational analysis available; of these, 239 patients (19%) had ITD and 60 patients (5%) had TKD mutations. NPM1 mutational analysis was available for 1215 patients, and 239 patients (20%) were positive for the mutation. Study populations included 21 patients (8%) with FLT3-TKD+NPM1+ mutated, 18 patients (7%) with FLT3-TKD only mutated, 117 (44%) with NPM1 only mutated, and 107 (41%) with FLT3-ITD+NPM1–mutated AML. Ten cases in the FLT3-ITD+NPM1 cohort had an additional FLT3-TKD mutation, whereas the FLT3-TKD+NPM1+ cohort excluded cases with concurrent FLT3-ITD mutation. Of note, in regards to TKD mutations, all were FLT3-D835 point mutations except one case with a TKD I836 deletion. The median age of the entire cohort was 62 years (range: 17-88 years); 55% of patients were aged ≥60 years. Patient and disease features and differences in baseline characteristics between cohorts are outlined in Table 1.
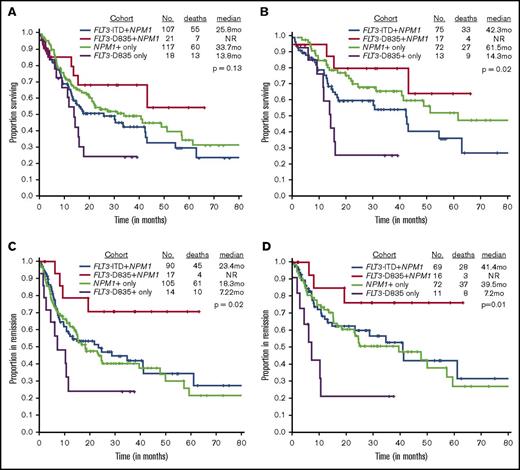
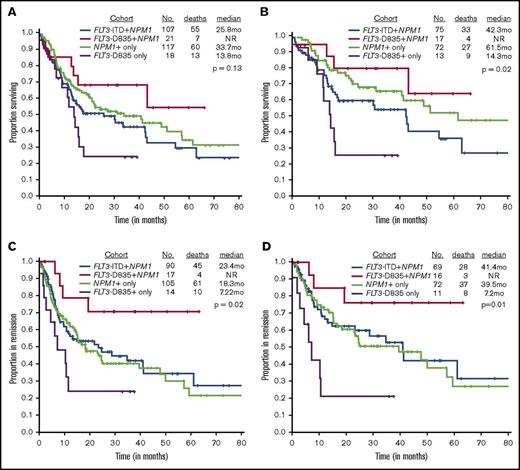
We next evaluated the effect of TKD mutation on outcome. The CR rate did not differ between patients with a TKD domain mutation, whether they had a NPM1 mutation (81%; reference) or not (77%, P > .99), as compared to those with an NPM1 mutation only (90%; P = .27), those with both an ITD and NPM1 mutation (84%; P = .75). The CR rate was similar in patients with FLT3-TKD+NPM1+ and NPM1+ only mutated (81% vs 90%; P = .27). However, there were marked differences in the relapse rates and the median remission durations. Only 24% (4 of 17 patients) of FLT3-TKD+NPM1+ patients relapsed compared with 70% (10 of 14 patients) of TKD+-only cases (P = .01), 58% (61 of 105 patients) of NPM1+-only cases (P = .01), and 50% (45 of 90 patients) ITD+NPM1 mutant cases (P = .06), suggesting that FLT3-TKD+NPM1+ patients are significantly less likely to relapse. Likewise, RFS was not reached (NR) in the FLT3-TKD+NPM1+ mutant patients compared with 18.3 months for NPM1+only, 23.4 months for FLT3-ITD+NPM1+, and 7.2 months for TKD+NPM1 wild-type cases (P = .03).
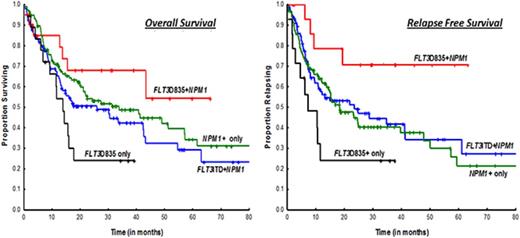
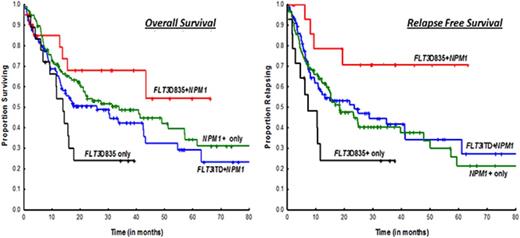
OS was superior in the FLT3-TKD+NPM1+ cohort when compared with the FLT3-TKD+-only cohort (median, NR vs 13.8 months; P = .03) (Figure 1A); there were trends toward improved OS in ITD+NPM1 mutant cases (NR vs 25.8 months; P = .07). Survival was not statistically significant when compared with NPM1+-only mutated AML patients (median, NR vs 33.7 months; P = .29). A similar and more obvious pattern was observed in a subset analysis of patients treated with idarubicin with cytarabine–based regimens (Figure 1B; supplemental Table 1). RFS was significantly superior in FLT3-D835+NPM1+ AML patients compared with all other study cohorts, NPM1+-only mutated patients included (Figure 1C-D). Because transplantation strategies may have influenced outcomes, RFS was evaluated after censoring at the time of transplant and showed a trend toward superior survival in the FLT3-TKD+NPM1+ cohort when compared with NPM1+-only patients (median, NR vs 24.6 months; P = .08; data not shown).
Kaplan-Meier survival curves comparing study cohorts. OS by FLT3 and NPM1 status in (A) all cases and (B) patients treated on intensive cytarabine-based treatment regimens. RFS by FLT3 and NPM1 status in (A) all cases and (B) patients treated on intensive idarubicin and cytarabine–based treatment regimens.
Kaplan-Meier survival curves comparing study cohorts. OS by FLT3 and NPM1 status in (A) all cases and (B) patients treated on intensive cytarabine-based treatment regimens. RFS by FLT3 and NPM1 status in (A) all cases and (B) patients treated on intensive idarubicin and cytarabine–based treatment regimens.
Although numbers were small, we noticed a similar trend among double FLT3 mutants (FLT3-ITD and TKD), where those without NPM1 performed poorly, with survival outcomes equivalent to those with FLT3-TKD only mutations (12.3 vs 13.8 months, P = .71). Furthermore, triple (TKD+ITD+NPM1+)-mutant status was associated with equivalent survival compared with NPM1+-only status (supplemental Figure 2). Eight patients proceeded to transplant after achieving their first CR; there was no difference in pretransplant minimal residual disease status or age in the cohort of patients undergoing transplant versus those treated only with chemotherapy. Importantly, undergoing transplant after CR did not improve RFS (supplemental Figure 1). In multivariable cox regression analysis in NPM1+-mutated AML, FLT3-TKD+ status was an independent favorable prognostic factor for RFS, even after accounting for transplant (supplemental Table 2). However, FLT3-TKD+NPM1+ mutation status did not emerge as a significant predictor of OS on multivariable analysis (supplemental Table 2).
Discussion
Results from our study are consistent with data from previous studies, which have similarly shown FLT3-TKD+NPM1+ mutation status to be associated with superior RFS compared with NPM1+ alone.10 The superior RFS in our study cohort did not translate to an improved OS, which was likely due to the small sample size estimates. More intriguing, however, is the paradoxical cooperative favorable effect on NPM1 conferred by FLT3-TKD, a phenomenon not observed between FLT3-ITD and NPM1 (Figure 1).5 One may surmise, based on the similar response rates observed between FLT3-TKD+NPM1+ and NPM1+-only mutation cohorts, that favorable effects are likely due to durable CRs effected by chemotherapy on an inherently biologically indolent disease. As alluded above, FLT3-ITD and FLT3-TKD mutants show distinct gain-of-function phenotypes with distinct differences in signaling properties and gene expression patterns.8,16 Our results emphasize the need for elucidating the mechanistic links between FLT3-TKD and NPM1 in future molecular and murine model studies.
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported in part by MD Anderson Cancer Center Support Grant CA016672 and award P01 CA049639 from the National Institutes of Health, National Cancer Institute.
Authorship
Contribution: P.B. and S.M.K. designed the study, collected data, and wrote and reviewed the manuscript; S.P. provided data for the study and reviewed the manuscript; and H.K., G.B., T.K., N.D., M.A., F.R., and J.C. provided patients for the study and were involved in writing and reviewing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven M. Kornblau, Department of Leukemia, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Box 448, Houston, TX 77030; e-mail: skornblau@mdanderson.org.