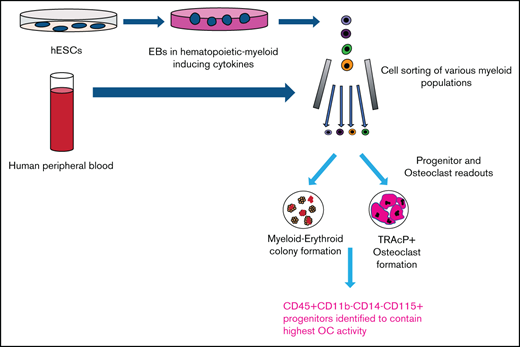
Key Points
Generation, identification, isolation, and expansion of monocyte and osteoclast progenitors from human embryonic stem cells.
CD11b−CD14−CD115+ cells from embryonic stem-derived cultures and peripheral blood mononuclear cells contain monocyte/osteoclast progenitors.
Abstract
Osteoclasts are multinuclear cells of monocytic lineage, with the ability to resorb bone. Studies in mouse have identified bone marrow clonal progenitors able to generate mature osteoclast cells (OCs) in vitro and in vivo. These osteoclast progenitors (OCPs) can also generate macrophages and dendritic cells. Interestingly, cells with equivalent potential can be detected in periphery. In humans, cells with OCP activity have been identified in bone marrow and periphery; however, their characterization has not been as extensive. We have developed reproducible methods to derive, from human pluripotent stem cells, a population containing monocyte progenitors able to generate functional OCs. Within this population, we have identified cells with monocyte and osteoclast progenitor activity based on CD11b and CD14 expression. A population double positive for CD11b and CD14 contains cells with expected osteoclastic potential. However, the double negative (DN) population, containing most of the hematopoietic progenitor activity, also presents a very high osteoclastic potential. These progenitor cells can also be differentiated to macrophage and dendritic cells. Further dissection within the DN population identified cells bearing the phenotype CD15−CD115+ as the population with highest monocytic progenitor and osteoclastic potential. When similar methodology was used to identify OCPs from human peripheral blood, we confirmed a published OCP population with the phenotype CD11b+CD14+. In addition, we identified a second population (CD14−CD11bloCD115+) with high monocytic progenitor activity that was also able to form osteoclast like cells, similar to the 2 populations identified from pluripotent stem cells.
Introduction
Monocytes are a heterogeneous group of cells that play fundamental roles in organogenesis, tissue repair, and immune responses. In addition, their central role on inflammation can define the course of many pathological processes.1-4 Thus, a full understanding of the development and function of this lineage is important to modulate their behavior toward the design of better directed therapies.
Monocytes originate in the bone marrow from a common myelo/erythroid progenitor5 and enter circulation, giving rise to tissue resident macrophages and dendritic cells (DCs) through defined lineage commitment steps.6 The circulating monocyte pool is heterogeneous, consistent with its wide range of potential functions.7,8 Different subpopulations of monocytes have been defined by cell surface markers and function.9 Based on the relative expression of CD14 and CD16, human monocytes have been classified in 3 subsets10 : classical (CD14++CD16−), intermediate (CD14++CD16+), and nonclassical (CD14+CD16++) monocytes.11 Comparison of these subsets to mouse counterparts has shown that the nonclassical subset (CD14+CD16++) has patrolling characteristics similar to that of Gr1− monocytes in mice; and classical and intermediate subsets have inflammatory properties similar to that of Gr1+ mouse monocytes.11,12
Osteoclasts are large multinucleated cells of monocytic origin expressing tartrate-resistant acid phosphatase (TRAcP)13 and form through fusion of committed mononuclear precursors.14,15 Their function is to decalcify bone by laying down an acidic environment16,17 that is degraded by matrix metalloproteinases and cathepsins.18,19 Osteoclasts control bone remodeling, a cellular process by which bone is resorbed (by osteoclasts) and new bone is formed (by osteoblasts).20 This homeostatic process is crucial for the maintenance of bone marrow space, bone growth, repair of microfractures, and maintenance of blood calcium levels.
Until recently, the identity of human osteoclast progenitors has not been well defined. Initial experiments, using mobilized CD34+ hematopoietic precursors in PB, indicated the presence of early progenitors (most likely from bone marrow) able to generate osteoclast like (OCL) cells in vitro.21 Later, osteoclast progenitors were found to be more represented within the CD14+ fraction of PB monocytes, albeit CD11b+ and CD33+, CD61+ fractions also showed the ability to generate bone resorbing TRAcP-positive osteoclasts.22,23 Subsequently, it was shown that the osteoclastic potential was in the CD16− fraction, within the proliferative CD14+CD16− classical monocyte subset.11,24 Recently, a study searching for cells with osteoclastogenic potential was done in human bone marrow aspirates, characterizing cells able to generate osteoclasts, macrophages, and DCs.25 This is consistent with previous work in our laboratory reporting a common monocyte progenitor in murine bone marrow able to give rise, at a clonal level, to macrophages, DCs, and osteoclasts.26-28 Importantly, this myeloid progenitor was able to engraft and form functional osteoclasts that localize to endosteal surfaces when injected into preconditioned mice.
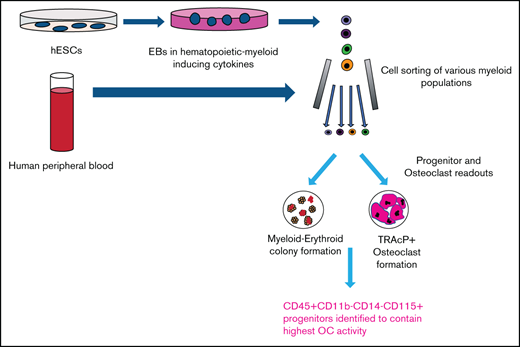
We hypothesize that equivalent human myeloid progenitors could be derived from pluripotent stem cells with the potential in the future to be used as sources of transplantable DCs or osteoclasts able to modulate immune responses or properties of bone marrow microenvironments respectively. Even when mature myeloid cell types such as osteoclasts, macrophages and DCs from human embryonic stem cells (hESCs) have been described, a fine phenotypic dissection of progenitor populations along with comparison with described progenitors from human PB has not been done.29-31 Thus, we designed a sequential approach to define populations with osteoclastogenic potential from early hematopoietic progenitors derived from hESCs. Osteoclast formation was evaluated by the generation of large multinucleated cells expressing TRAcP after induction in media containing MCSF and RANKL.26 The functional identity of osteoclasts was confirmed by their ability to degrade bone matrix as well as gene expression signature. Populations with high osteoclastogenic activity were then tested for their ability to generate macrophages and DCs because this suggests the presence of a common monocyte progenitor. The characteristics of the progenitors generated from hESC cultures were compared with equivalent cells detected in circulating human PB. Surprisingly, this comparison evidenced a monocyte progenitor, previously unidentified in human PB, able to form OCL cells and phenotypically comparable to the one isolated from hESC-derived cells.
Methods
Cell culture
Cells were cultured in a humidified incubator at 37°C and 5% CO2. Basal tissue culture reagents were purchased from Gibco BRL/Invitrogen Life Technologies, unless stated otherwise. Fetal bovine serum (FBS) was from Atlanta Biologicals.
Maintenance of hESCs
The hESC lines, H9 (WA09) and H1 (WA01), were purchased through our institutional stem cell core under our Stem Cell Research Oversight commitee protocol. hESCs were cultured on gelatin-coated dishes with irradiated mouse embryonic feeder cells derived from CF-1 mice.32 The zinc finger nuclease generated ubiquitin mCherry H9 hESC line was a kind gift from Alex Lichtler laboratory at UConn Health.
EB formation and hematopoietic induction
Confluent 6-well dish cultures of hESCs on mouse embryonic fibroblast feeders, were incubated with 1 mg/mL Collagenase IV for 5 to 10 minutes at 37°C. Wells were washed and 1 mL of embryoid body (EB) formation media (KODMEM, 20% FBS, 1% minimum essential medium nonessential amino acids, 1 mM l-glutamine, and 0.1 mM mercaptoethanol), was added per well. Using a p1000 pipette, colonies were detached, aggregates were placed into a 50-mL tube, and allowed to settle by gravity for 10 minutes. The upper supernatant was aspirated leaving the ESC aggregates in about 4 mL of media. Aggregates were plated into Corning Ultra-Low Attachment 6 well plates with 4 mL of EB media/well, placing the equivalent of 3 ESC wells into 2 EB wells.
Hematopoietic induction was done based on a described method.29 EBs were allowed to form overnight and media was switched to EB media containing the following cytokines (EB Cyto6 media): 300 ng/mL hSCF, 300 ng/mL hFLT3L, 50 ng/mL hGCSF, 25 ng/mL hBMP4, 10 ng/mL hIL3, and 10 ng/mL hIL6 (all ISOkine ORF Genetics except for BMP4 from R&D Systems). Full media changes were done at days 5 and 10. Thereafter, half media changes were done every 3 to 4 days by removing half the media, centrifuging at 450g for 5 minutes, suspending the pellet in fresh media, and plating back into the original well.
EB dissociation
For early time points or for dissociation of an entire culture, EBs were incubated at 37°C in 1 mg/mL Collagenase IV for 30 minutes followed by treatment with 0.05% Trypsin-EDTA for 20 minutes. Trypsin was inactivated with serum and cells washed. Older cultures containing visible hematopoietic components (floating and attached) were processed first by collecting floating cells and EB aggregates into tubes and centrifuging at 450g for 5 minutes. The pellet fractions were broken up using gentle trituration. Attached cells were harvested after 5 minutes’ incubation with 0.05% Trypsin-EDTA and then combined with the nondigested fraction. Remaining EB clumps, containing nonhematopoietic components, were filtered out through a 70-μm filter.
Flow cytometry and cell sorting
At each stage of hematopoietic differentiation of hESCs, and for the study of PB progenitors, single-cell suspensions were analyzed by flow cytometry. We used commercial monoclonal antibodies directly conjugated to fluorochromes. All analyses and sorting were done in a BD-FACS ARIA II (BD Biosciences) equipped with 5 lasers, 18 fluorescence detectors, and FACS DIVA software. FACS data were analyzed using BD CellQuest Pro Software. Detailed description of methods to process cells, antibody specificities, and cell-sorting gating strategies can be found in the supplemental Data.
Methylcellulose analysis
Sorted cells were plated at a density of 100 or 4000 cells per well (12-well format) into methylcellulose, Methocult H-4434 (Stem Cell Technologies). After 10 to 14 days of culture, colonies were enumerated, picked for flow cytometry or cytospun, and stained with Wright-Giemsa kit HEMA-3 (Fisher Scientific).
Osteoclast cultures and bone resorption
Osteoclast formation from dissociated EB cultures was done by plating in EB Cyto6 media. The following day, cultures developed floating cells enriched in hematopoietic components. Floating cells were then plated into a new dish at a density of 4000 cells per well in triplicate (96-well format with or without dentine bone slices) in EB media supplemented with osteoclast-inducing cytokines M-CSF and RANKL, both at 100 ng/mL (ISOkine ORF Genetics). Osteoclast formation from total cultures included SCF and FLT3L, both at 100 ng/mL, to aid proliferation of progenitors. After 10 to 14 days of culture, with half media changes every 2 days, cells were processed for osteoclast staining and resorption as described.26-28
Macrophage and dendritic differentiation
For macrophage differentiation, sorted populations were plated in EB media with 100 ng/mL MCSF and for dendritic differentiation plated in EB media with 50 ng/mL IL4 (ISOkine ORF Genetics) and 50 ng/mL GMCSF (R&D Systems). At day 14, cells were analyzed by flow cytometry.
Myeloid cell isolation from human PB
Under the approved institutional review board protocols from the University of Connecticut Medical School (13-119-2), human PB from 23 healthy volunteers was collected into BD Vacutainer tubes containing sodium heparin. Blood was centrifuged at 600g for 10 minutes and plasma removed. An equal volume of 0.5 mM EDTA pH 7.2 was added followed by 3 times the volume of 2% Dextran-500 in phosphate-buffered saline (Sigma 31392). Sedimentation of red blood cells was done at 37°C for 30 minutes and the upper phase containing leukocytes and granulocytes was removed and washed in phosphate-buffered saline. Residual red blood cells were lysed using hypotonic lysis buffer. Cells were processed for flow cytometry or cell sorting. Myeloid populations were evaluated after exclusion of granulocytes and lymphocytes (gating strategy presented in supplemental Figure 3). The study was in accordance with the Declaration of Helsinki.
Human PB progenitor osteoclast differentiation
For osteoclast differentiation, total sorted cells or equal numbers (10 000) were plated into 96-well dishes in 90% MEM-α, 10% FBS, and 1 × Pen-Strep containing MCSF and RANKL (both at 100 ng/mL). Cells were grown for 2 to 3 weeks and were fixed and stained for TRAcP as described.26 For evaluation of myeloid progenitors, cells from sorted populations were plated into Methocult H-4434 at a density of 30 000 for lymphoidneg sorts, 8000 for each of the 4 populations identified by CD11b and CD14 and 1000 for CD115 sorts. After 10 to 14 days, colonies were enumerated.
Results
Development of osteoclast progenitors from hESC
Contrasting methods using stromal feeders or low oxygen chambers to generate osteoclasts,30,31 we modified a published method to differentiate hESCs to hematopoietic cells through EB formation and culture with hematopoietic inducing cytokines.29 EB aggregates were cultured in 6 basic hematopoietic cytokines, and around day 20, clusters of floating cells began to expand rapidly (supplemental Figure 1A). These clusters contained cells expressing CD45 and their representation increased as the cultures proceed for up to 40 days. Cell-surface marker phenotyping was performed and strategies for cell sorting were defined (supplemental Figure 1B).
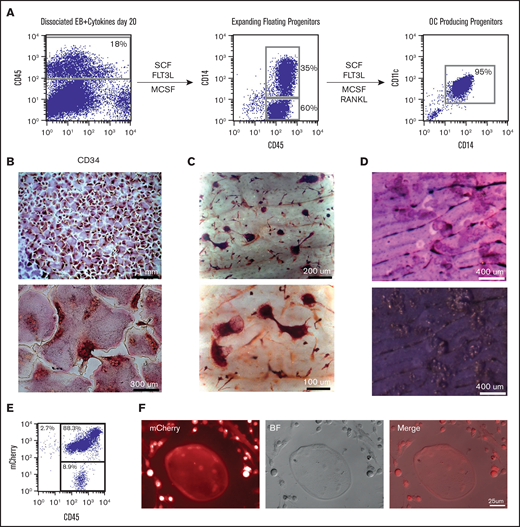
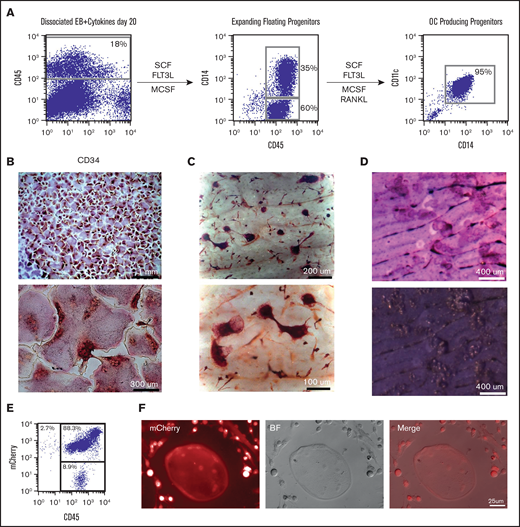
Our initial readout for cells with monocyte progenitor potential has been the evaluation of osteoclastogenesis. Floating hematopoietic components, which were shown to express CD34 and CD45 (Figure 1A), were plated into tissue culture plates and on top of dentine bovine slices in the presence of SCF, FLT3L, and MCSF. These cells expanded and became more homogenous by morphology and cell-surface expression as shown by the upregulation of the monocyte marker CD14. Upon addition of RANKL, cells began to fuse into large multinucleated TRAcP+ cells with the ability to resorb bone, as shown by toluidine staining of the resorption pits (Figure 1B-D). The osteoclastogenic commitment was verified by evaluating the gene expression of key osteoclasts specific genes in the cells obtained along the differentiation progression.33-37 Genes such as Cathepsin K, MMP9, TRAP, and OSCAR were highly expressed in multinucleated TRAcP+ osteoclasts (supplemental Figure 2), whereas cells before the addition of RANKL (termed myeloid progenitors) showed the highest expression for RANK and CSF1R (MCSF receptor or CD115). All of these results indicated that we generated, in a very direct and simple manner, terminally differentiated and functionally active osteoclasts, expressing proper signature genes.
Robust generation of osteoclast progenitors (OCPs) from hESCs and mCherry hESCs. (A) After 20 days of EB differentiation in the 6-cytokine cocktail, EBs were enzymatically dissociated and analyzed for the expression of CD45 and CD34. Cells were plated back into media containing SCF, FLT3L, and MCSF. Expanded floating cells were removed from the dish and analyzed for expression of CD14. Cultures were continued in tissue culture wells with and without dentine bovine slices along with the addition of RANKL. Between 6 and 10 days after RANKL addition, wells or slices were stained for TRAcP and osteoclasts evaluated as multinucleated TRAcP positive cells. (B) TRAcP+ osteoclasts (low and high magnification) grown on tissue culture dishes and (C) on top of dentine slices. (D) Toluidine blue-stained dentine slices after OC removal identifies numerous resorption pits. (E) Zinc finger nuclease-derived ubiquitin-mCherry hESC cell line was evaluated by flow cytometry for mCherry and CD45 after 20 days in EB 6-cytokine cocktail (F) and then switched to osteoclast inducing media which lead to multinucleated mCherry+osteoclasts and mCherry+progenitors.
Robust generation of osteoclast progenitors (OCPs) from hESCs and mCherry hESCs. (A) After 20 days of EB differentiation in the 6-cytokine cocktail, EBs were enzymatically dissociated and analyzed for the expression of CD45 and CD34. Cells were plated back into media containing SCF, FLT3L, and MCSF. Expanded floating cells were removed from the dish and analyzed for expression of CD14. Cultures were continued in tissue culture wells with and without dentine bovine slices along with the addition of RANKL. Between 6 and 10 days after RANKL addition, wells or slices were stained for TRAcP and osteoclasts evaluated as multinucleated TRAcP positive cells. (B) TRAcP+ osteoclasts (low and high magnification) grown on tissue culture dishes and (C) on top of dentine slices. (D) Toluidine blue-stained dentine slices after OC removal identifies numerous resorption pits. (E) Zinc finger nuclease-derived ubiquitin-mCherry hESC cell line was evaluated by flow cytometry for mCherry and CD45 after 20 days in EB 6-cytokine cocktail (F) and then switched to osteoclast inducing media which lead to multinucleated mCherry+osteoclasts and mCherry+progenitors.
Because this culture system could represent a valuable source of cells to be used in transplantation experiments, we wanted to verify this protocol could generate osteoclasts using transgenic hESCs. To this end, we used a mCherry hESC line expressing mCherry under the control of the ubiquitin promoter using zinc finger nuclease technology. mCherry hESCs were able to generate mCherry+CD45+ cells in the same manner as nontransgenic hESCs that, when cultured with MCSF and RANKL, showed the ability to generate mCherry+ progenitors and osteoclasts (Figure 1E-F).
Identification and characterization of osteoclast progenitors
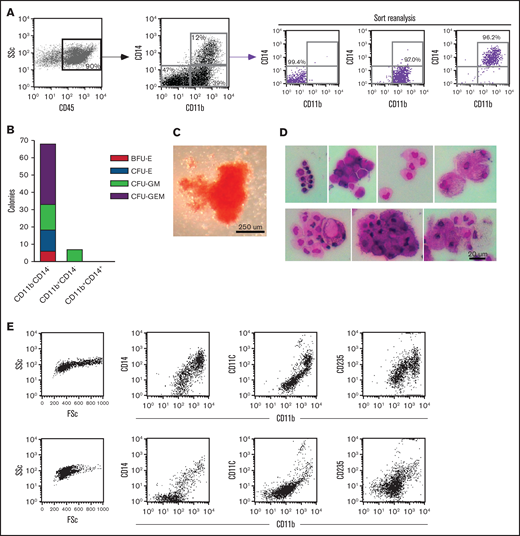
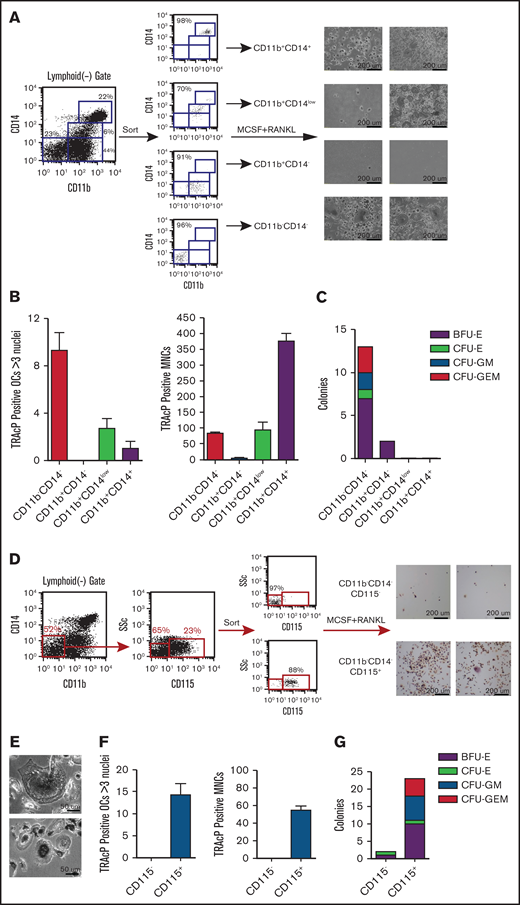
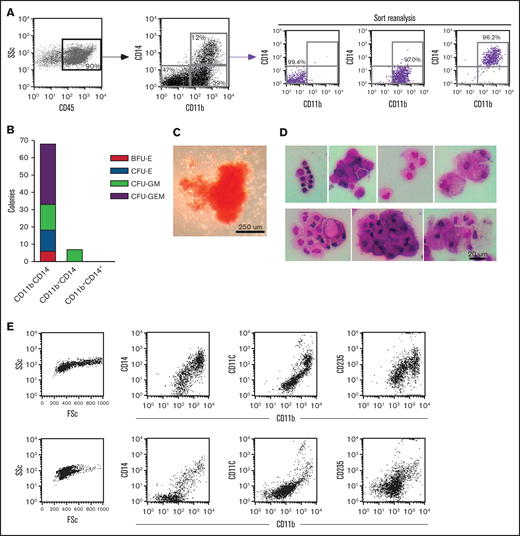
To identify the OC progenitor populations, we initially sorted the hematopoietic cell fractions in the context of CD14 expression. Interestingly, CD14+ and CD14− populations made osteoclasts (OCs) efficiently when plated at equal densities in the presence of MCSF and RANKL (data not shown). Because of these results, we tested the hematopoietic progenitor activity of sorted fractions, dissected by their expression of CD14 and CD11b, using methylcellulose colony formation assays (Figure 2A). Three distinct populations, within the CD45+ gate, were sorted and plated at equal density onto methylcellulose supplemented with myelo/erythroid lineage-inducing cytokines. The CD11bCD14 double negative (DN) population gave rise to the majority of the colonies (Figure 2B-C). On the other hand, the double positive (DP) population failed to generate colonies. Individual methylcellulose colonies obtained from DN sorts were cytospun and stained a heterogeneous population of distinct myeloid subsets including macrophages, foam cells, granulocytes, monocytes, and red blood cells (Figure 2D). Flow cytometric analysis of 2 individual representative colonies was shown to express CD11b, CD11c, CD14, and CD235 at varying levels. These results clearly demonstrated that the CD11bCD14 DN population contains high numbers of early myeloid-erythroid progenitors with the ability to differentiate into mature myeloid-erythroid cell types (Figure 2B-D).
Dissection of myeloid-erythroid progenitor activity in the CD45+ fraction from EB cultures. (A) Staining of EB cultures maintained in the 6-cytokine cocktail with antibodies against CD14 and CD11b evidenced at least 3 discrete population. These were purified using FACS (reanalysis of purity shown) and plated at equal numbers into methylcellulose supplemented with myelo-erythroid lineage-inducing cytokines. (B) Methylcellulose colonies were enumerated at day 10 of culture. (C) Bright-field image of the most primitive colony type, CFU-GEM, from the CD14CD11b DN sort. (D) Individual colonies from the methylcellulose cultures of CD11bCD14 DN sorted cells were cytospun onto glass slides and stained with Wright-Giemsa kit HEMA 3 (Fisher Scientific) (7 colonies shown) and (E) evaluated by flow cytometry for the expression of CD11b, CD11c, CD14, and CD235 (2 representative colonies shown). BFU-E, burst-forming unit-erythroid; CFU- colony forming unit; E, erythrocyte; G, granulocyte; GEM, granulocyte, erythrocyte, and monocyte; GM, granulocyte and monocyte; M, monocyte.
Dissection of myeloid-erythroid progenitor activity in the CD45+ fraction from EB cultures. (A) Staining of EB cultures maintained in the 6-cytokine cocktail with antibodies against CD14 and CD11b evidenced at least 3 discrete population. These were purified using FACS (reanalysis of purity shown) and plated at equal numbers into methylcellulose supplemented with myelo-erythroid lineage-inducing cytokines. (B) Methylcellulose colonies were enumerated at day 10 of culture. (C) Bright-field image of the most primitive colony type, CFU-GEM, from the CD14CD11b DN sort. (D) Individual colonies from the methylcellulose cultures of CD11bCD14 DN sorted cells were cytospun onto glass slides and stained with Wright-Giemsa kit HEMA 3 (Fisher Scientific) (7 colonies shown) and (E) evaluated by flow cytometry for the expression of CD11b, CD11c, CD14, and CD235 (2 representative colonies shown). BFU-E, burst-forming unit-erythroid; CFU- colony forming unit; E, erythrocyte; G, granulocyte; GEM, granulocyte, erythrocyte, and monocyte; GM, granulocyte and monocyte; M, monocyte.
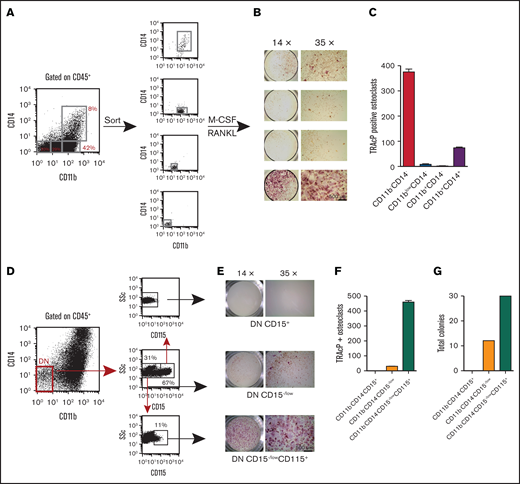
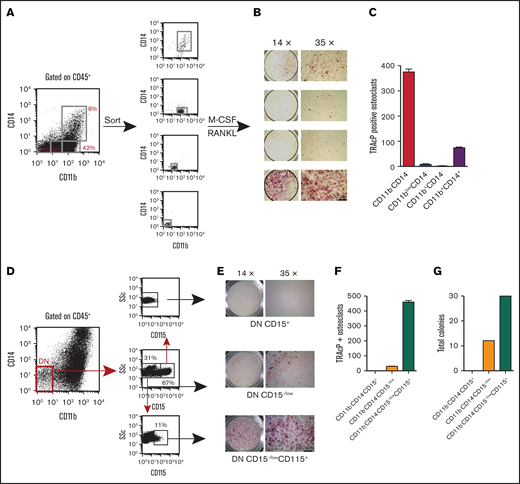
To better evaluate the osteoclastogenic potential within our cell derivatives, we sorted 4 nonoverlapping populations (Figure 3A), cells were plated in MCSF and RANKL, and cultured for 10 days before staining for TRAcP. Figure 3B-C shows the highest OC potential was contained within the CD11bCD14 DN population. The DP population had the ability to make an appreciable number of osteoclasts but not as efficiently as the DN population. In contrast, the other populations had very little to no osteoclast potential.
Evaluation of osteoclast progenitor populations in the CD45+ fraction from EB cultures. (A) Four discrete populations defined by their expression of CD11b and CD14 were sorted to purity and plated in triplicate at equal numbers (4 K). They were then cultured in M-CSF and RANKL for 14 days. (B-C) TRAcP+ cells generated from each population were photographed at ×14 and ×35 magnification and enumerated. Most of the osteoclastogenic activity was contained in the CD11bCD14 DN population. (D) The DN population was further dissected with CD15 and sorted into CD15+ and CD15−/low. The latter population contained a small population expressing CD115. These populations were sorted and plated at equal numbers into (E-F) osteoclast inducing media and (G) myeloid-erythroid methylcellulose for 10 days. TRAcP+ osteoclasts and colonies were enumerated. TRAcP-stained wells were imaged using a LeciaEZ4D stereomicroscope.
Evaluation of osteoclast progenitor populations in the CD45+ fraction from EB cultures. (A) Four discrete populations defined by their expression of CD11b and CD14 were sorted to purity and plated in triplicate at equal numbers (4 K). They were then cultured in M-CSF and RANKL for 14 days. (B-C) TRAcP+ cells generated from each population were photographed at ×14 and ×35 magnification and enumerated. Most of the osteoclastogenic activity was contained in the CD11bCD14 DN population. (D) The DN population was further dissected with CD15 and sorted into CD15+ and CD15−/low. The latter population contained a small population expressing CD115. These populations were sorted and plated at equal numbers into (E-F) osteoclast inducing media and (G) myeloid-erythroid methylcellulose for 10 days. TRAcP+ osteoclasts and colonies were enumerated. TRAcP-stained wells were imaged using a LeciaEZ4D stereomicroscope.
To further define the osteoclastic potential of the DN population, we dissected this progenitor population with CD15, a marker specific to granulocytes. CD15 staining identified a positive and negative fraction. When an antibody to the MCSF receptor (CD115) was included, all the CD115 expression was within the CD15−/low population (Figure 3D). Sorted populations equally plated in osteoclastic or methylcellulose conditions demonstrated that the majority of the osteoclastic activity and myeloid-erythroid progenitor was within this CD15−/lowCD115+ fraction (Figure 3E-G).
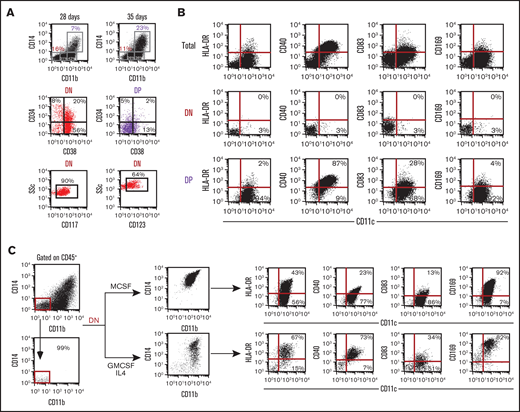
These results confidently demonstrate an early osteoclast progenitor, derived from hESCs through EB hematopoietic induction, with the phenotype: CD45+CD11b− CD14− CD15−/lowCD115+. Interestingly, these cells in culture have the ability to maintain progenitor activity with a slow but steady increase in more committed cells (or DP population) (Figure 4A). Cultures can be grown for more than 60 days while maintaining this colony-forming unit (CFU) and OC progenitor activity. Therefore, we evaluated the DN population for HSC and myeloid progenitor markers and found that 20% of the DN population is CD34+CD38+ compared with 2% of the DP population (Figure 4A). Similarly, myeloid progenitor markers CD117 (c-kit) and CD123 (IL3Ralpha) is highly expressed in the DN population, 90% and 64%, respectively (Figure 4A).
Macrophage and dendritic cell potential of DN cells. (A) Progression of EB cultures, in the 6-cytokine cocktail, from day 28 to 35 shows continued maintenance of the DN population. FACS analysis of the DN and DP population for HSC markers (CD34 and CD38) and myeloid progenitor markers (CD117 and CD123). (B) Day 40 EB cytokine 6 cultures CD11bCD14 DN and DP populations were evaluated for the expression of macrophage and dendritic cell markers CD169, CCR2, HLA-DR, CD40, and CD83 in the context of CD11c. (C) To assess the potential of the DN progenitor population to generate macrophages and DCs, sorted DN cells were expanded in either MCSF or GMCSF + IL4 and then analyzed for upregulation of CD11b and CD14 along with CD169, HLA-DR, CD40, and CD83 in the context of CD11c.
Macrophage and dendritic cell potential of DN cells. (A) Progression of EB cultures, in the 6-cytokine cocktail, from day 28 to 35 shows continued maintenance of the DN population. FACS analysis of the DN and DP population for HSC markers (CD34 and CD38) and myeloid progenitor markers (CD117 and CD123). (B) Day 40 EB cytokine 6 cultures CD11bCD14 DN and DP populations were evaluated for the expression of macrophage and dendritic cell markers CD169, CCR2, HLA-DR, CD40, and CD83 in the context of CD11c. (C) To assess the potential of the DN progenitor population to generate macrophages and DCs, sorted DN cells were expanded in either MCSF or GMCSF + IL4 and then analyzed for upregulation of CD11b and CD14 along with CD169, HLA-DR, CD40, and CD83 in the context of CD11c.
Evaluation of macrophage and DC potentials within the identified progenitor population
Our laboratory has previously identified in mouse a common bone marrow-transplantable progenitor that gives rise to functional osteoclasts, macrophages, and DCs.26,28 With this in mind, we tested the ES cell derivative populations with early myeloid progenitor/osteoclastogenic activity for their ability to form macrophages and DCs. Initially we evaluated the DN and DP populations identified by CD11b and CD14 for the following macrophage and dendritic markers: CD11c, sialoadhesin molecule CD169, major histocompatibility class II or HLA-DR, co-stimulatory molecules CD40, and CD83 (Figure 4B). As expected, our DN population shows no expression of macrophage or DC markers. However, the CD11bCD14 DP population coexpresses CD11c (96%), CD40 (87%), and CD83 (28%). Therefore, the DP population is composed of myeloid lineage committed cell types. Sorted CD11bCD14 DN populations were cultured in conditions favoring the generation of macrophages (MCSF) or DCs (GMCSF+IL4) (Figure 4C). After 14 days in culture, under both conditions, DN cells showed upregulation of CD11b and CD14 (becoming DP) as well as CD11c. Both macrophage and dendritic culture conditions progressed the DN population into cells expressing appropriate and phenotypically different profiles of CD169, HLA-DR, CD40, and CD83 differentiation markers. As expected, costimulatory and major histocompatibility class molecules were more highly expressed in cells placed into DC conditions compared with macrophage conditions.
Early monocyte and osteoclast progenitor potential in human PB correlates with hESC-derived progenitors
Once we confirmed the identity of monocyte progenitors from hESC derivatives, we tested their equivalence to progenitors found in vivo. At the time of these findings, most of the reports on human monocyte/osteoclast progenitors were from PB CD14+ progenitors. Because progenitors identified from hESC derivatives included populations negative for CD14, we determined if we could identify a CD11bCD14 DN cell type with osteoclastogenic potential in human PB in the same fashion as we showed using hESC derivatives (supplemental Figure 3A). Leukocyte suspensions from blood of healthy donors were analyzed by flow cytometry using a gating strategy to analyze live single cells, excluding granulocytes and lymphoid cells (CD3, CD19, and CD56). Finally, within these gates the markers used to dissect the hESC-derived myeloid progenitors were also used: CD11b, CD14, and CD115.
Initial analyses of populations sorted for granulocyte-lymphoid negative cells and plated into myeloid-erythroid methylcellulose (supplemental Figure 3B) showed that these fractions contained all the possible colony types (burst-forming unit-erythroid, colony-forming unit-erythroid, colony-forming unit granulocyte and monocyte, colony-forming unit granulocyte, erythrocyte, and monocyte) indicating that these fractions are enriched in early hematopoietic progenitors. Interestingly, comparison between the results from 2 individuals showed variations, suggesting that the distribution of peripheral early progenitors could be heterogeneous between individual subjects. Further dissection of the negative population using the differential expression of CD11b and CD14, showed a clear enrichment in early progenitor activity (approximately threefold) between the total lymphoid negative with sorted lymphoid negative CD11b−CD14− DN population (supplemental Figure 3C).
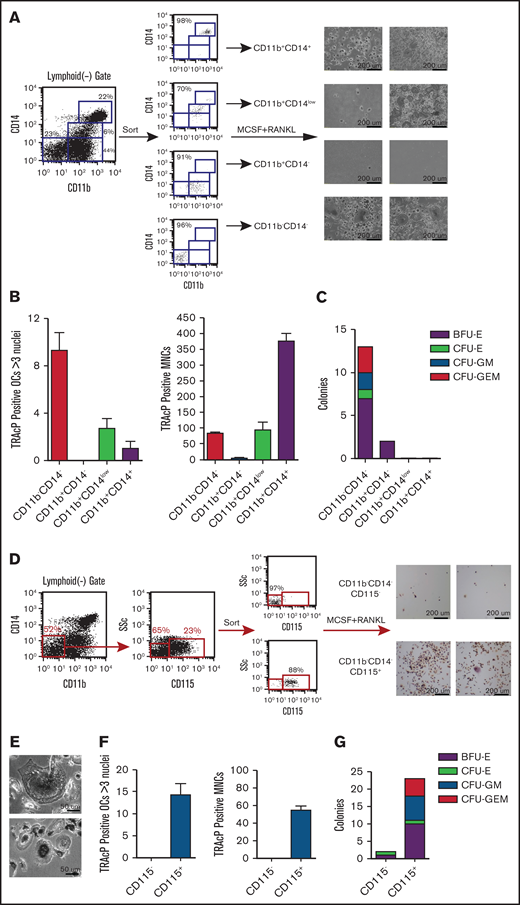
Based on CD14 and CD11b expression, 4 populations were sorted and tested for their ability to generate OCL cells and colony formation in methylcellulose (Figure 5). As expected, the DP populations readily formed osteoclasts after culture in media containing MCSF+RANKL; however, they did not form colonies in methylcellulose. Interestingly the population DN for these markers, with known early hematopoietic potential, also generated OCL when cultured in osteoclastogenic conditions, similar to what we observed in our hESC-derived cultures (Figure 5A-C). Consistent with the observations from methylcellulose cultures, variations were seen in the osteoclast potential of this population between different individuals (2 representative samples shown). Importantly, when plating equal numbers of each of the four populations, multinucleated TRAcP+ OCs were higher in the DN than in the DP fractions (Figure 5B). Because the CD14+ populations had TRAcP+ mononuclear cells that were difficult to tell whether they were OC precursors or macrophages, these cell types were counted separately and were highly represented in the DP population.
Myeloid progenitor potential of human peripheral blood cells. (A) Peripheral blood mononuclear cells (PBMCs) were analyzed in the context of CD11b and CD14 and sorted into 4 distinct populations (FACS plot are shown for 1 patient). Sorted cells were then cultured in the presence of MCSF and RANKL for 2 weeks, stained for TRAcP, and photographed at ×10 magnification (2 representative patients shown). (B) Equal numbers (10 K) from each population were cultured in the presence of MCSF and RANKL and TRAcP+ osteoclasts with 3 or more nuclei and mononuclear cells (MNCs) were counted. (C) Equal numbers of cells from each population plated into methylcellulose and scored at day 10 for BFU-E, CFU-E, CFU-GM, and CFU-GEM. (D) The DN population was dissected further by sorting CD115− and CD115+ cells within the DN gate (2 representative patients shown). 10 K of sorted cells were grown in osteoclastic conditions and stained for TRAcP (photographed at ×35 magnification). (E) Higher magnification shows examples of multinucleated osteoclasts which were enumerated in (F) as well as TRAcP+ MNCs. (G) Equal numbers of sorted cells were grown in myeloid-erythroid methylcellulose for 10 days and then colonies enumerated. BFU-E, burst-forming unit-erythroid; CFU- colony forming unit; E, erythrocyte; G, granulocyte; GEM, granulocyte, erythrocyte, and monocyte; GM, granulocyte and monocyte; M, monocyte.
Myeloid progenitor potential of human peripheral blood cells. (A) Peripheral blood mononuclear cells (PBMCs) were analyzed in the context of CD11b and CD14 and sorted into 4 distinct populations (FACS plot are shown for 1 patient). Sorted cells were then cultured in the presence of MCSF and RANKL for 2 weeks, stained for TRAcP, and photographed at ×10 magnification (2 representative patients shown). (B) Equal numbers (10 K) from each population were cultured in the presence of MCSF and RANKL and TRAcP+ osteoclasts with 3 or more nuclei and mononuclear cells (MNCs) were counted. (C) Equal numbers of cells from each population plated into methylcellulose and scored at day 10 for BFU-E, CFU-E, CFU-GM, and CFU-GEM. (D) The DN population was dissected further by sorting CD115− and CD115+ cells within the DN gate (2 representative patients shown). 10 K of sorted cells were grown in osteoclastic conditions and stained for TRAcP (photographed at ×35 magnification). (E) Higher magnification shows examples of multinucleated osteoclasts which were enumerated in (F) as well as TRAcP+ MNCs. (G) Equal numbers of sorted cells were grown in myeloid-erythroid methylcellulose for 10 days and then colonies enumerated. BFU-E, burst-forming unit-erythroid; CFU- colony forming unit; E, erythrocyte; G, granulocyte; GEM, granulocyte, erythrocyte, and monocyte; GM, granulocyte and monocyte; M, monocyte.
Finally, further fractionation of the DN population using CD115, showed that all of the osteoclastogenic and the majority of progenitor activity was contained in the CD115+ fraction, just as observed in the studies conducted with hESC derivatives (Figure 5D-G). This was true for both TRAcP+ multinucleated OCL cells as well as mononuclear cells (Figure 5F).
All these results combined indicated that in human PB, there are 2 defined populations with osteoclast progenitor activity: a CD14CD11b DP population previously identified in the literature and a newly defined population, negative for CD11b and CD14 that also contains cells with early progenitor activity. Both the DN and DP population correlates to those obtained from hESC-derived progenitors, including further enrichment of osteoclastogenic activity within the DN population using CD115 selection.
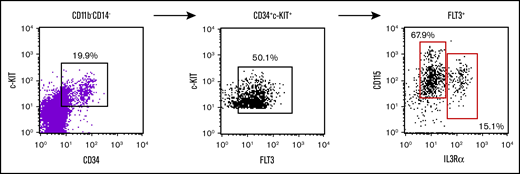
Early myeloid precursors from human bone marrow have been characterized bearing the phenotype CD11b−CD34+CD117+CD135+CD123+ that was able to generate osteoclasts, macrophages, and DCs.25 These unique progenitors are then analogous to murine progenitors identified by others and us. Thus, we analyzed our lymphoid-negative, CD11bCD14 DN population for the expression of these markers and found that there is a variable expression of all of them, including combined positive expression of all the relevant markers (Figure 6). Unfortunately, the abundance of these cells is extremely low, making it challenging to isolate enough cells for functional assays.
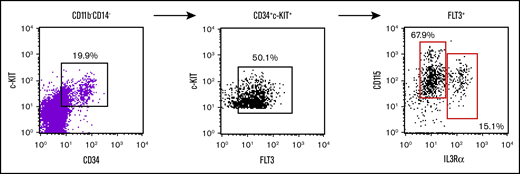
Evaluation of bone marrow myeloid progenitor markers within the human peripheral blood DN CD115 fraction. Flow cytometry analysis of human peripheral blood gated on lymphoid-CD11b−CD14− for c-kit, CD34, FLT3, IL3Ralpha, and CD115. This gating strategy of CD11b−CD14−c-kit+CD34+FLT3+ yields almost all the cells CD115+ which can be separated into 2 populations of IL3Ralpha+ cells.
Evaluation of bone marrow myeloid progenitor markers within the human peripheral blood DN CD115 fraction. Flow cytometry analysis of human peripheral blood gated on lymphoid-CD11b−CD14− for c-kit, CD34, FLT3, IL3Ralpha, and CD115. This gating strategy of CD11b−CD14−c-kit+CD34+FLT3+ yields almost all the cells CD115+ which can be separated into 2 populations of IL3Ralpha+ cells.
Discussion
The ability of hESCs to differentiate into hematopoietic cells has been established in the literature. Hematopoietic progression has been reported using stromal support30,38,39 and EB formation followed by induction with various cocktails of hematopoietic cytokines.8,29 In general, methods using mouse stromal supporting cells result in the generation of heterogeneous populations of derivatives with modest representation of hematopoietic components. In contrast, protocols based on EB formation followed by hematopoietic induction render more defined methods. Myeloid cells (including osteoclasts) have been derived using both methods; however, the reported EB method used a complex 4-step protocol of cytokine induction along with culture in low-oxygen chambers.30,31 In this report, we modified a simple method of EB formation in the presence of 6 basic hematopoietic cytokines. Our method clearly rendered very efficiently hematopoietic cells including cells of the myeloid lineage and also using a mCherry reporter line. We were able to produce large numbers of these progenitors and study myelopoiesis using osteoclast formation as an initial readout. Our initial studies showed that osteoclasts could be generated from a derivative population expressing CD11b and CD14 that gets efficiently expanded upon induction with myelopoietic cytokines. This is consistent with results from studies defining human osteoclast progenitors in PB. However, further dissection of derived hematopoietic progenitors showed clearly that cells with the best osteoclast progenitor potential were contained in the CD11b/CD14 DN fraction, more specifically in cells expressing low levels of CD15 and high levels of CD115. Importantly, these fractions are also capable to generate macrophage and DC-like cells and have the potential to be expanded. The ability of these cultures to robustly generate hematopoietic progenitors is important because these cells are very difficult to obtain from human subjects in high numbers and could serve as a substrate to study human myelopoiesis.
These findings prompted us to search for equivalent progenitor populations in human PB. As expected, we confirmed an osteoclast progenitor population expressing CD14, which we also detected as CD11b+CD14+ DP population. More interestingly, we found a population with osteoclastogenic potential contained in the fractions negative for CD11b and CD14 and with high expression of CD115. We propose that this DN cell population corresponds to a very early monocyte progenitor, more prevalent in the bone marrow, and that the DP population represents a precursor more exclusive in periphery. In fact, this DN population shared markers with the identified osteoclast progenitor population in the bone marrow.25 One interesting observation is that this population seems to be differentially represented in the circulation of different individuals and could represent variable egress of hematopoietic progenitors because of a particular physiological status of the individual.40 In fact, recent literature has suggested that inflammatory signals could dictate the number of circulating early progenitors.41-43 Within our patient cohort, we were unable to identity the cause of such differences when looking at age, race, or sex. Even when very preliminary, the evaluation of these populations could have potential to be a diagnostic marker in many physiopathological processes and would require a large-scale study of control patients first to understand these differences.
Interestingly, the osteoclast progenitor activity of the DN population is superior to the one observed from the DP populations, both in hESC and PB-derived cells. We think that this difference relates to differential ability of these populations to expand under the osteoclastogenic-inducing cytokines. Visually, the DN population proliferates very quickly, creating optimal densities for larger number of fusion events, whereas the DP population, although further along in differentiating toward an OC, has much slower proliferation rate.
Our laboratory has extensively characterized mouse myeloid progenitors from bone marrow and periphery in mouse.26-28 These progenitors are transplantable and can contribute to monocyte cells and progression in different anatomical sites. Future experiments are needed to determine if these human myeloid progenitors isolated from hESCs under these differentiation and sorting techniques could represent a source for a new osteoclast transplantable progenitor and provide a tool to facilitate the study of human monocyte biology.
Acknowledgments
The authors thank the Department of Immunology Department Chair Anthony Vella for his support in this publication as well as Bruce Liang for assistance in obtaining control blood samples under our institutional review board protocol.
This work was funded by grants from the National Institutes of Health National Heart, Lung, and Blood Institute (1RC1HL100569-01) (H.L.A.) and the State of Connecticut Stem Cell Program (06SCC04) (H.L.A.).
Authorship
Contribution: S.H.R. and H.L.A. designed the study, performed experiments, and acquired and analyzed data; both authors were involved in the interpretation of the data, manuscript drafting, and intellectual content; both authors approved the initial manuscript before submission; and S.H.R. was involved in final revisions before submission as well as final revisions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.H.R. is Division of Pediatric Dentistry/Craniofacial Sciences, University of Connecticut School of Dental Medicine, Farmington, CT.
Hector Leonardo Aguila died on 30 March 2020.
Correspondence: Sierra H. Root, Department of Immunology, University of Connecticut School of Medicine, 263 Farmington Ave, Farmington, CT 06030-1319; e-mail: sroot@uchc.edu.
References
Author notes
The full-text version of this article contains a data supplement.