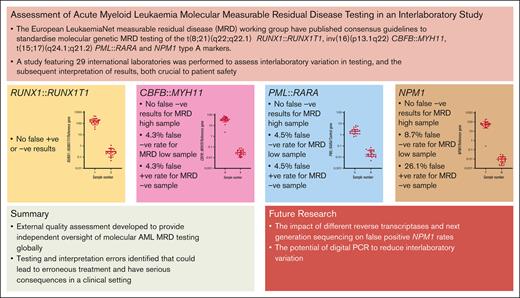
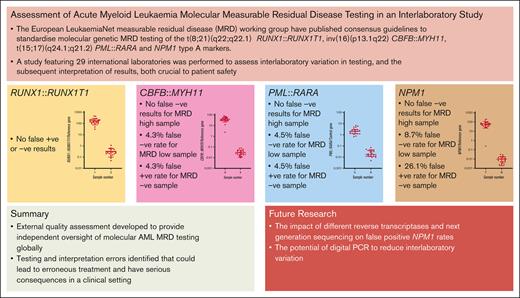
Key Points
External quality assessment developed to provide independent oversight of molecular acute myeloid leukemia MRD testing globally.
Testing and interpretation errors identified that could lead to erroneous treatment and have serious consequences in a clinical setting.
Abstract
The European LeukaemiaNet (ELN) measurable residual disease (MRD) working group has published consensus guidelines to standardize molecular genetic MRD testing of the t(8;21)(q22;q22.1) RUNX1::RUNX1T1, inv(16)(p13.1q22) CBFB::MYH11, t(15;17)(q24.1;q21.2) PML::RARA, and NPM1 type A markers. A study featuring 29 international laboratories was performed to assess interlaboratory variation in testing and the subsequent interpretation of results, both crucial to patient safety. Most participants in this study were able to detect, accurately quantify, and correctly interpret MRD testing results, with a level of proficiency expected from a clinical trial or standard-of-care setting. However, a few testing and interpretive errors were identified that, in a patient setting, would have led to misclassification of patient outcomes and inappropriate treatment pathways being followed. Of note, a high proportion of participants reported false-positive results in the NPM1 marker-negative sample. False-positive results may have clinical consequences, committing patients to unneeded additional chemotherapy and/or transplant with the attendant risk of morbidity and mortality, which therefore highlights the need for ongoing external quality assessment/proficiency testing in this area. Most errors identified in the study were related to the interpretation of results. It was noted that the ELN guidance lacks clarity for certain clinical scenarios and highlights the requirement for urgent revision of the guidelines to elucidate these issues and related educational efforts around the revisions to ensure effective dissemination.
Introduction
Measurable residual disease (MRD) testing is increasingly used and accepted as the standard of care to manage a range of different hematological neoplasms. Its use as a surrogate outcome in clinical trials of new therapies is being explored,1,2 where it has the potential to accelerate drug assessment and approval.
Compared with other diseases, such as chronic myeloid leukemia (CML) or acute lymphoblastic leukemia, where standardized molecular testing is well established, the phenotypic and genetic heterogeneity of acute myeloid leukemia (AML) has limited the use of MRD in this context. However, in recent years, several platforms and markers have started to overcome the initial difficulties and have demonstrated potential to offer accurate and precise MRD assessment in AML.
AML lacks a molecular aberration common to most of the patients, as seen with the BCR::ABL1 rearrangement in CML or immunoglobulin and T-cell receptor rearrangements in lymphoid leukemias. As such, a variety of markers and approaches have had to be developed. Although next-generation sequencing offers the ability to screen for and monitor a wide range of canonical small variants that have been established in AML, its validation and implementation have been hampered by methodological aspects such as the high error rate seen in current sequencing methodologies, higher costs, as well as biological aspects, such as clonal hematopoiesis, of indeterminate potential confounding results.3
Multiple studies have, however, now established that persistent detection of t(8;21)(q22;q22.1) RUNX1::RUNX1T1,4-6 inv(16)(p13.1q22) CBFB::MYH11,4 t(15;17)(q24.1;q21.2) PML::RARA,7,8 and canonical NPM1 exon 11 variants9-12 using sensitive reverse transcriptase quantitative polymerase chain reaction (RT-qPCR)-based approaches are a strong predictor of relapse, and the integration of MRD testing into the patient treatment pathway will improve risk assessment. Between them, these markers are found in around 40% of patients with AML, and they have been validated and standardized as sensitive (10−5−10−7) and stable MRD markers in single laboratory studies.13-18 However, the performance of these assays in interlaboratory studies has yet to be established.
The increasing importance of MRD testing for risk stratification and treatment planning has led to an increase in the number of laboratories performing this testing. As a response to this, the European LeukaemiaNet (ELN) recently evaluated both the technical aspects of flow cytometric and molecular genetic approaches to MRD attesting as well as their clinical application and published a set of consensus guidelines providing recommendations on how and when to perform MRD assessments and how to use the results in clinical practice.3
As has been seen with the application of BCR::ABL1 as an MRD marker in CML,19 harmonizing MRD testing and thus reducing interlaboratory variation between laboratories has a number of benefits, including: (1) ensuring multicenter clinical trials are based on comparable data, (2) allowing the development of clinical guidelines around common MRD milestones ensuring consistent management of patients globally, and (3) facilitating serial/longitudinal MRD assessment of itinerant patients who may present to a number of different caregivers (clinics/practices/laboratories).
Ongoing independent assessment of laboratory testing that is used to diagnose and manage patient treatment is required as part of internationally recognized laboratory accreditation frameworks.20 External quality assessment (EQA)/proficiency testing (PT) provides an excellent tool to establish the performance of an assay and identify factors that are producing results that are out of consensus to promote a reduction of interlaboratory variation. This is especially relevant because most of these assays are based on laboratory-developed tests.
Aims
To establish a viable sample matrix for EQA/PT.
To establish the performance of current molecular MRD testing in AML in an interlaboratory context.
To identify areas for improvement to reduce interlaboratory variation.
Methods
A total of 12 batches of lyophilized cell line-based material were manufactured for this study. These consisted of 3 batches of samples for each marker, all containing 10 × 106 cells: an “MRD-high” sample, an “MRD-low” sample, and an “MRD-negative” sample. The t(8;21)(q22:q22) RUNX1::RUNX1T1-positive samples were manufactured using the KASUMI-1 cell line, which carries a fusion between exon 5 of the RUNX1 gene and exon 2 of the RUNX1T1 gene that is seen in virtually all patients with AML with RUNX1::RUNX1T1.13 The inv(16)(p13q22) CBFB::MYH11–positive samples were manufactured using the ME-1 cell line, which carries the common type A rearrangement variant seen in 88% of inv(16)(p13q22)-positive patients, a fusion between exon 5 of the CBFB gene and exon 12 of the MYH11 gene.13 The t(15;17)(q24.1;q21.2) PML::RARA-positive samples were manufactured using the NB-4 cell line, which carries the bcr 1, L fusion between exon 6 of the PML gene and exon 3 of the RARA gene seen in ∼55% of the t(15;17)(q24.1;q21.2)-positive patients.13 The NPM1-positive samples were manufactured using the OCI-AML3 cell line, which carries the type A variant (NM_002520.7(NPM1):c.860_863dup) seen in ∼75% of patients with AML with an NPM1 variant.21 MRD-positive samples were diluted with HL60 cells to achieve the desired MRD level. MRD-negative samples were manufactured using the HL60 cell line only.
All cell lines were acquired from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany) and tested negative for human immunodeficiency virus I and II, hepatitis B virus, hepatitis C virus, Epstein–Barr virus, human T-lymphotropic virus type I and II, human herpes virus 8 (OCI-AML3 not tested), murine leukemia virus, squirrel monkey retrovirus, and mycoplasma by PCR. Cell lines were grown in RPMI 1640 (Thermo Fisher Scientific, Waltham, MA) medium supplemented with 10% fetal bovine serum (Thermo Fisher Scientific). Predefined dilutions of the cells were prepared and freeze dried for 24 hours in 3 mL glass ampoules to contain 10 × 106 cells. Before distribution, to ensure sample quality and homogeneity, a minimum of 3 selected samples (first, middle, and last samples manufactured) were subjected to RNA extraction, complementary DNA (cDNA) synthesis, and RT-qPCR for the relevant rearrangement/variant following ELN criteria.3 Sample quality was defined as an RNA OD260/280 ratio of between 1.8 and 2.2 and ABL1 levels >10 000 copies per replicate. Replicate samples were required to be within 1.2-fold of each other. The stability of trial samples was ensured by measuring ABL1 (reference gene) levels on a further 3 vials at trial closure.
The samples were shipped at ambient temperature to 29 laboratories in 12 countries. Laboratories currently active in the UK NEQAS for Leucocyte Immunophenotyping’s database were invited to be part of the study on a first-come, first-served basis. Participants were asked to test the blinded samples with their in-house assay, in line with ELN criteria,3 and report the % normalized ratio of the relevant marker to the ABL1 reference gene, alongside additional methodological and technical data, including but not limited to:
Quality of sample/analysis: suitable or not suitable for MRD assessment (e.g., based on an ABL copy number ≥10 000 as recommended by Schuurhuis et al3).
Quantification of target genes, for example, copy number of mutated NPM1 and threshold cycle (Ct) value.
Quantification of reference genes, for example, copy number of ABL1 and Ct value.
Qualitative MRD result (positive vs negative MRD).
Normalized ratio (%) result (target gene or rearrangement copy number /reference gene copy number)∗100.
Based on their result for each sample, participants were also asked to provide a diagnosis based on outcome criteria taken from the ELN recommendations3 (supplemental Data Table), that is,
Complete molecular remission: patients in complete morphological remission with 2 successive MRD-negative samples obtained within an interval of ≥4 weeks at a sensitivity level of at least 1 in 1000.
Molecular persistence at low copy numbers: patients in complete morphological remission with low copy number MRD (<100-200 copies per 104ABL1 copies corresponding to <1%–2% of target to reference gene or allele burden) and a copy number or relative increase <1 log10 between any 2 positive samples collected after the end of treatment.
Molecular progression: molecular progression in patients with molecular persistence at a low copy number with an increase of MRD copy numbers ≥1 log10 between any 2 positive samples.
Molecular relapse: patients with an increase in the MRD level of ≥1 log10 between 2 positive samples in a patient who previously tested negative in technically adequate samples.
To make a clinical interpretation possible, participants were provided with a mock clinical scenario for each sample. The correct clinical interpretation was a classification that was in line with the testing consensus (median) result calculated from all participants’ results.
Standard parametric statistics were used to calculate interlaboratory variation for % normalized ratio results for each sample in Microsoft Excel. Nonparametric statistics were preferred to assign a value to each sample as they are less affected by outliers.
Participant data used in the study have been fully anonymized to allow publication.
Results
A total of 25 laboratories returned results; however, not all laboratories tested all markers, with 23 of them returning results for t(8;21) RUNX1::RUNX1T1, inv(16) CBFB::MYH11, and NPM1 and 22 returning results for t(15;17) PML::RARA.
Participant methods
t(8;21) RUNX1::RUNX1T1
Most participants used RT-qPCR (n = 22), with only a single participant using reverse transcription digital PCR (RT-dPCR). A wide range of protocols were used, with the most popular being the Europe Against Cancer protocol (EAC)14 (n = 12), followed by the Qiagen Ipsogen RUNX1-RUNX1T1 Kit (n = 5) and a modified EAC protocol (n = 3) (supplemental Data Table 1). All participants used ABL1 as a reference gene.
inv(16) CBFB::MYH11
Twenty-one participants used RT-qPCR, with 1 participant using RT-dPCR and 1 using agarose gel electrophoresis that returned a qualitative result only. A wide range of protocols were used, with the most popular being the EAC protocol14 (n = 12), followed by the Qiagen Ipsogen CBFB-MYH11 A Kit (n = 4) (supplemental Data Table 1). Twenty-one participants used ABL1 as a reference gene, and 1 participant used GUSB.
t(15;17) PML::RARA
Twenty-one participants used RT-qPCR (n = 21), with 1 participant using RT-dPCR. A wide range of protocols were used, with the most popular being the EAC protocol14 (n = 11), followed by the Qiagen Ipsogen PML-RARA bcr1 Kit (n = 5) (supplemental Data Table 1). Twenty-one participants used ABL1 as a reference gene, and 1 participant used GUSB.
NPM1
Twenty-two participants used RT-qPCR, with 1 participant using RT-dPCR. A wide range of protocols were used, with the most popular being an in-house assay (n = 11), followed by the Qiagen Ipsogen NPM1 mut A, B, & D MutaQuant Kits (n = 8), Qiagen Ipsogen NPM1 mut A MutaQuant Kits (n = 2), and the Qiagen Ipsogen NPM1 Mutascreen Kit (n = 2) (supplemental Data Table 1). All participants used ABL1 as a reference gene.
Suitability of samples for molecular testing
For all 12 samples issued in the study, >95% of laboratories classified them as suitable for analysis (supplemental Data Table 2), based on the criteria in the consensus recommendations from the ELN MRD Working Party Criteria.3 Median ABL1 copy numbers calculated from all participant results ranged from 88 058 to 482 000 (supplemental Data Table 3). One participant that returned results for all markers, reported all samples in the study as suboptimal using RT-qPCR and the EAC protocol. They used ABL1 as a control gene for the t(8;21) RUNX1::RUNX1T1 and NPM1 markers and reported copy numbers that ranged between 1014 and 2276. The participant used GUSB as a reference gene for the inv(16) CBFB::MYH11 and t(15;17) PML::RARA markers and reported copy numbers between 14 650 and 64 700. Another participant reported sample 6 as suboptimal, reporting a reference gene level of 10 346.
Qualitative and quantitative results
t(8;21) RUNX1::RUNX1T1
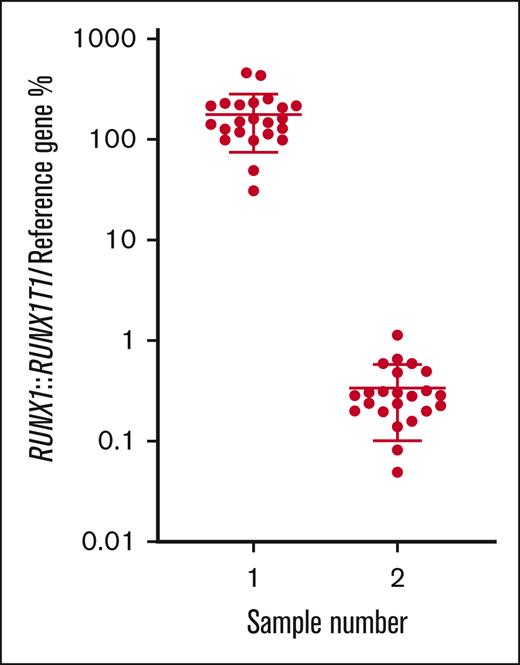
For the t(8;21) RUNX1::RUNX1T1 rearrangement, all participants that returned results (n = 23) classified the MRD-high and MRD-low samples as positive (Table 1) and the MRD-negative sample as negative (Table 2). Calculated from all participants’ results, the mean normalized ratio for the MRD-high sample (001) was 179.0% with a coefficient of variation (CV) of 57.9%. The mean normalized ratio for the MRD-low sample (002) was 0.34% with a CV of 70.2% (Table 1; Figure 1).
% normalized ratio returned by all participants reporting RUNX1::RUNX1T1 MRD levels in samples 1 and 2. Long horizontal line represents average. Short horizontal line represents standard deviation.
% normalized ratio returned by all participants reporting RUNX1::RUNX1T1 MRD levels in samples 1 and 2. Long horizontal line represents average. Short horizontal line represents standard deviation.
inv(16) CBFB::MYH11
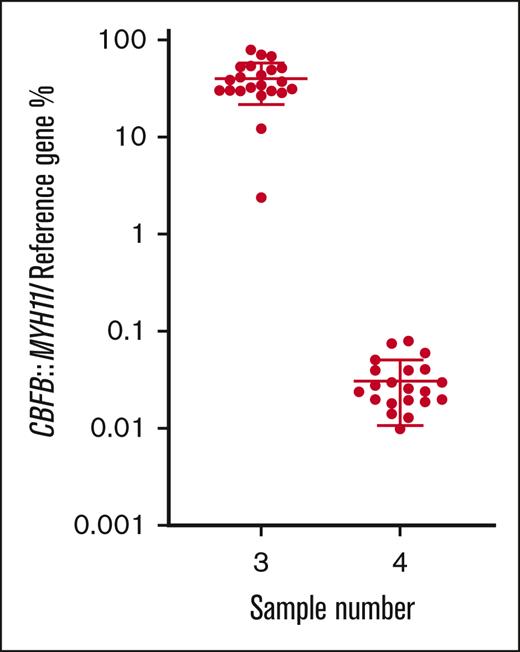
For the inv(16) CBFB::MYH11 rearrangement, all participants that returned results (n = 23) classified the MRD-high sample as positive, 22 of 23 (95.7%) classified the MRD-low sample as positive, and 22 of 23 (95.7%) classified the MRD-negative sample as negative (Table 2). Calculated from all participants’ results, the mean normalized ratio for the MRD-high sample (3) was 39.8% with a CV of 45.6%. The mean normalized ratio for the MRD-low sample (4) was 0.033% with a CV of 60.2% (Table 1; Figure 2).
% normalized ratio returned by all participants reporting CBFB::MYH11 MRD levels in samples 3 and 4. Long horizontal line represents average. Short horizontal line represents standard deviation.
% normalized ratio returned by all participants reporting CBFB::MYH11 MRD levels in samples 3 and 4. Long horizontal line represents average. Short horizontal line represents standard deviation.
t(15;17) PML::RARA
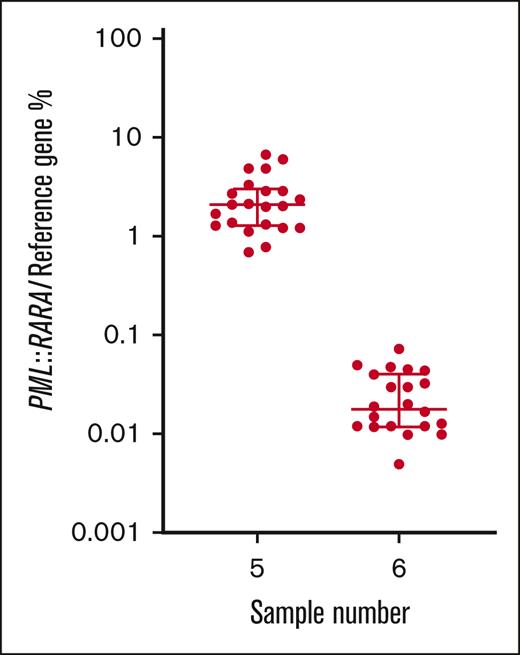
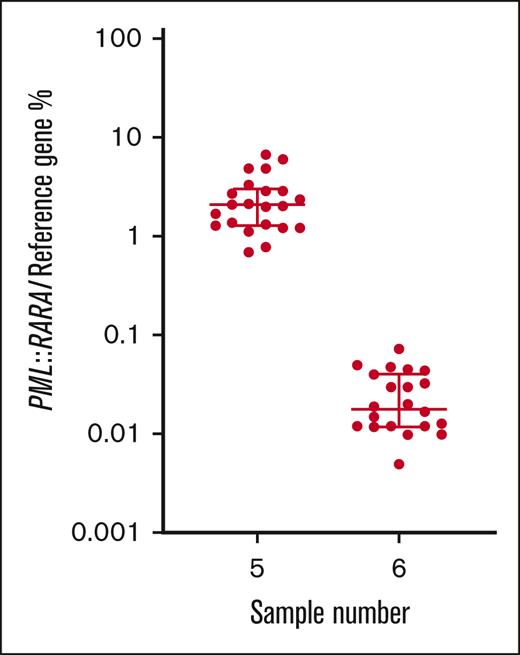
For the t(15;17) PML::RARA rearrangement, all participants that returned results (n = 22) classified the MRD-high sample as positive, 21 of 22 (95.5%) classified the MRD-low sample as positive, and 21 of 22 (95.5%) classified the MRD-negative sample as negative. Calculated from all participants’ results, the mean normalized ratio for the MRD-high sample (5) was 2.5% with a CV of 66.6%. The mean normalized ratio result for the MRD-low sample (6) was 0.026% with a CV of 68.5% (Table 1; Figure 3).
% normalized ratio returned by all participants reporting PML::RARA MRD levels in samples 5 and 6. Long horizontal line represents average. Short horizontal line represents standard deviation.
% normalized ratio returned by all participants reporting PML::RARA MRD levels in samples 5 and 6. Long horizontal line represents average. Short horizontal line represents standard deviation.
NPM1
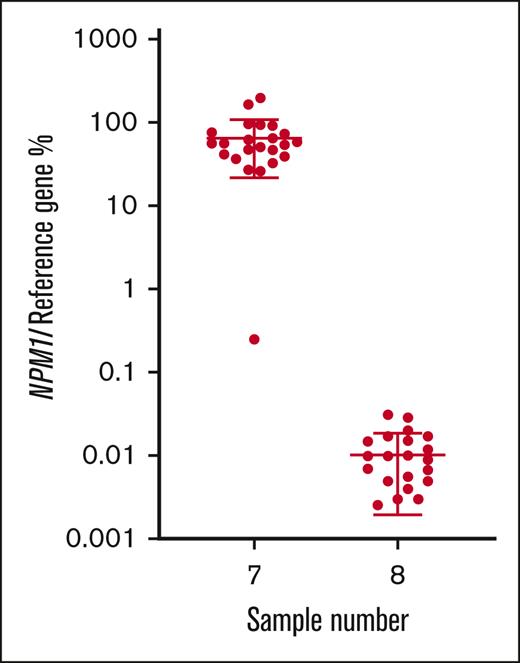
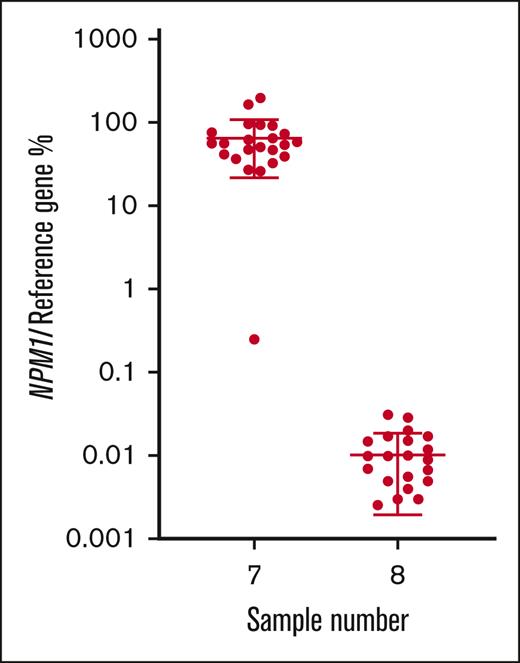
For NPM1, all participants that returned results (n = 23) classified the MRD-high sample as positive, 21 of 23 (91.3%) classified the MRD-low sample as positive, and 17 of 23 (73.9%) classified the MRD-negative sample as negative. Calculated from all participants’ results, the mean normalized ratio for the MRD-high sample (7) was 64.8% with a CV of 66.6%. The mean normalized ratio for the MRD-low sample (8) was 0.011% with a CV of 71.4% (Table 1; Figure 4).
% normalized ratio returned by all participants reporting NPM1 MRD levels in samples 7 and 8. Long horizontal line represents average. Short horizontal line represents standard deviation.
% normalized ratio returned by all participants reporting NPM1 MRD levels in samples 7 and 8. Long horizontal line represents average. Short horizontal line represents standard deviation.
Clinical interpretation of results
Twenty-one laboratories classified their MRD results into different outcome-based criteria defined in the ELN recommendations,3 based on a mock clinical scenario that accompanied each sample (supplemental Data Table 4). Across all 12 samples issued in the study, a total of 243 classifications were returned, with 183 (75.3%) correctly classified in line with the testing consensus and ELN definitions. Sixty (24.7%) interpretations were deemed incorrect, a result of either a testing or interpretation error. Misclassifications were made by 19 of the 21 laboratories that returned classifications. The percentage of participants with the correct definition across the 12 samples ranged from 29.4% to 100%, with an average of 81.3%. When the 60 incorrect definitions across all 12 samples were aggregated and analyzed, 47 (78.3%) errors by 16 different participants were due to misinterpretation of the guidelines, and 13 (21.7%) errors by 9 different participants were the result of an aberrant test result. When the errors that were a result of an aberrant test result were further examined, 7 (53.8%) were due to false-positive results, 3 (23.1%) were due to false-negative results, and 3 (23.1%) were due to quantitative variation (supplemental Data Table 5).
Discussion
Potent new therapies for the treatment of AML have necessitated the development of sensitive, accurate, and precise assays that can be used to measure residual disease present after treatment to assess therapy efficacy and inform posttreatment patient management. EQA/PT is an important tool to establish the performance of different assays and provide ongoing independent oversight.
Here, we have demonstrated the effectiveness of lyophilized cell line-based samples as a viable sample matrix for EQA/PT and other standardization projects. The samples performed well with >95% of participants, using a range of techniques, classifying the samples as suitable for analysis with high median reference gene levels reported.
Most participants in this study were able to detect and accurately quantify MRD when assessing the t(8;21)(q22;q22.1) RUNX1::RUNX1T1, inv(16)(p13.1q22) CBFB::MYH11, t(15;17)(q24.1;q21.2) PML::RARA, and NPM1 markers, with a level of proficiency that would be expected in a clinical trial or standard-of-care setting. However, several testing errors were identified in the study, including false-positive results, false-negative results, and critical quantitative variation. The testing errors identified were widely distributed among participants and were not the result of a small subset of participants producing aberrant results. Of note, a high proportion of participants reported false-positive results in the NPM1 marker MRD-negative sample. The clinical impact of these false-positive results has to some extent been mitigated in a recent update to guidelines,22 which have newly defined patients with complete morphologic remission and NPM1-mutated AML (at levels of) <2% but above the detection limit of the assay as “complete remission with molecular MRD detection at low level” (CR-MRD-LL). The guidelines state that NPM1-mutated patients with stable CR-MRD-LL are associated with a very low relapse risk when measured at the end of consolidation chemotherapy and do not necessarily require a change in treatment. Despite this, some protocols (eg, the UK National Cancer Research Institute AML studies) use MRD positivity in the peripheral blood after 2 cycles of chemotherapy as an indicator of high-risk disease regardless of the level, and patients are selected for complete remission 1 (CR1) transplant based on these results.11 In addition, if false positivity is intermittent, this could lead to the misdiagnosis of molecular relapse. Therefore, in some circumstances, false-positive results could have potentially very serious clinical consequences. Furthermore, the generation of technical false-positive results impedes the ability of clinical studies to assess the risk associated with genuine low-level detection of mutated NPM1. As such, it is essential that laboratories use methods that limit the possibility of false-negative results and comprehensively validate their testing to understand the specificity of testing across all markers, but this is particularly important for NPM1.
The NPM1 type A variant is the only marker in this study that is not a fusion transcript; instead, it is a duplication of a TCTG tetranucleotide in exon 11 of the NPM1 gene (Human Genome Variation Society nomenclature NM_002520.7(NPM1):c.860_863dup, systematic exon numbering of the NPM1 transcript applied) and is vulnerable to false positivity. This has been observed with the widely used Gorello et al method,16,23 but was shown to be reduced with the use of locked nucleic acid probes that improved the specificity of the reaction.9 Compounding this issue, sequence errors can also be induced during the reverse transcription process. As NPM1 is highly expressed in AML,9 cDNA reverse transcribed from RNA has been deemed the template of choice for analysis. However, the process of reverse transcription has a known error rate,24,25 particularly when using highly active transcriptases, such as SuperScript IV (Thermo Fisher Scientific),26 and NPM1 exon 11 errors may be artificially introduced during cDNA synthesis.
Several laboratories reporting false-positive results in this study had used SuperScript IV reverse transcriptase (Thermo Fisher Scientific), and 2 participants commented they had stopped using this polymerase because of observing amplification of NPM1 type A and D transcripts in known negative controls. Further investigation of the role of reverse transcriptase in false-positive results should be the focus of future studies. It is extremely important that laboratories thoroughly validate their NPM1 assays with an understanding of the vulnerability of the assay to false positivity. MRD detection by next-generation sequencing using DNA as a template rather than cDNA could potentially subvert some of these issues and should be the focus of future studies.
Of the 4 false-negative results reported across the 4 markers, 3 participants classified the respective samples as suboptimal, with 1 participant reporting the sample as satisfactory. The participant that reported the samples as satisfactory did not provide information about the reference gene that they used or the respective copy number. Of the 3 remaining false-negative results, all were submitted by a single participant. One result used ABL1 as a reference gene and reported a copy number of 2540, correctly interpreted as suboptimal. Of the remaining 2 samples, both were tested using the GUSB reference gene and were reported as having reference gene copy numbers of 22 400 and 64 700. The ELN guidelines recommend aiming for a minimum of 10 000 copies of the housekeeping gene ABL1. No information is given on what constitutes a satisfactory reference gene copy number level for laboratories that use non-ABL1 reference genes, for example, GUSB. If we can extrapolate from BCR::ABL1 measurement in CML, where 10 000 copies of ABL1 are also used as a minimum reporting threshold, then the equivalent minimum reporting threshold recommended for GUSB users is 24 000.27 As such, for the 2 samples reported as suboptimal using GUSB as a reference gene, 1 would have been reported as suboptimal and 1 as satisfactory. Future iterations of the ELN guidelines should clarify the minimum reporting thresholds for GUSB users.
The potential clinical impact of the interlaboratory variation and error identified in the study was assessed when participants were asked to classify their MRD results for each sample into different outcome-based criteria in line with the clinical scenarios provided for each sample. Most of the errors identified were the result of laboratories misinterpreting the guidelines. The interpretation errors identified were widely distributed among participants and were not the result of a small subset of participants providing aberrant interpretations. In sample 1, 7 participants (33.3%) erroneously classified the sample as “molecular relapse,” even though the clinical scenario did not mention any previous negative results, a requirement for this classification. Subsequent to this study, MRD response criteria have been revised to provide a broader definition of what constitutes an MRD relapse, which should provide a better consensus moving forward.22 However, simple misinterpretations such as this point to the urgent need for education around how to interpret the guidelines with examples covering multiple clinical and technical scenarios, as have been published in CML.28
A lack of understanding of how to interpret the guidelines is compounded by several ambiguities within them, which this study has demonstrated can lead to a wide variety of interpretations being reported for a single sample. A particular lack of clarity was noted around the definition of log changes in MRD level between samples. The guidelines refer interchangeably to changes in copy number, MRD level, and relative increase; however, these terms are not all specifically defined, and it is not clear what is preferred if the results of these subtly different measurands are conflicting. The problematic nature of this ambiguity was exemplified by a participant when interpreting their test results for sample 4. Their % normalized ratio showed a >1 log increase from the previous result, indicating a classification of “molecular relapse”; however, the target rearrangement copy number showed a <1 log increase from the previous samples, indicating a classification of “molecular persistence at low copy number.” This led to the participant erroneously reporting a result of “molecular persistence at low copy number.”
The % normalized ratio would seem unquestionably to be the preferred measurand to classify samples, as the results are standardized to account for inevitable variations in the quality of the extracted template. Large copy number variation between different extractions of the same sample is a common finding in most laboratories, making a normalized ratio preferred for BCR::ABL1 analysis in CML. Copy number thresholds are still used to define relapse, for example, in core-binding factor AMLs,4 by some laboratories; however, it is unknown how widespread this practice is. Copy numbers can be greatly affected by the standard curve used for RT-qPCR studies. The type of standard curves used by participants was not analyzed in this study but will be the focus of future work.
Sample 11 demonstrated further ambiguity in the guidelines. The scenario for this sample was a patient who had tested MRD-negative 8 weeks ago (0 PML::RARA copies/70 000 ABL1 copies) and MRD-positive 4 weeks ago (20 PML::RARA copies/11 000 ABL1 copies [0.18%]). The sample was manufactured to be an MRD-negative sample. Eight participants (47.1%) classified the sample as “complete molecular remission”; however, a classification of “complete molecular remission” requires “2 successive MRD-negative samples obtained within an interval of ≥4 weeks at a sensitivity level of at least 1 in 1000,”3 making a classification of “complete molecular remission” incorrect because of the previous positive sample described in this clinical scenario. A further 4 participants classified the sample as “molecular persistence as low copy number”; 1 of these 4 participants had a false-positive result, making their interpretation of the erroneous results correct. However, the remaining 3 participants all had negative MRD results, so they erroneously based their classification on the 20 PML::RARA copies detected in the previous sample. This sample did, in fact, not fit into any of the outcome-based criteria definitions and should have been reported as MRD-negative only. Five participants (29.4%) correctly did not provide an interpretation.
The guidelines recommend that all reports feature a molecular interpretation for each result but do not provide guidance on what to do when samples do not fit any of the classifications. Classifications should be created for all scenarios, or additional guidance should be provided on how to report results that do not fit into current classifications.
The levels of quantitative interlaboratory variation identified in this study were greater than those that we have reported in similar studies of BCR::ABL1, where testing has been standardized over a number of decades.19,29-33 Although standardization is in its infancy for molecular MRD testing in AML, these findings support the development of similar projects to further reduce interlaboratory variation. This is particularly important when defining cutoffs, such as the distinction between complete remission with positive MRD (CRMRD+) and CR-MRD-LL, which uses 2% NPM1/ABL1 as a quantitative value.22
There are certain caveats to this study. The samples used were lyophilized cell lines, which may not fully reflect peripheral blood and bone marrow samples normally assessed in patients and require minor deviations from laboratories’ standard protocols to process them. It is worth noting, however, that lyophilized cell line-based samples have been used to successfully standardize BCR::ABL1 testing in CML.31,32 It should also be noted that for the inv(16)(p13q22) CBFB::MYH11, t(15;17)(q24.1;q21.2) PML::RARA, and NPM1 markers, multiple transcript types and variants exist and thus performance differences may exist for variant subtypes not featured in this study. dPCR offers the possibility of lower interlaboratory variation; however, with only 1 participant in this study using this technique, we were not able to assess its impact. Although participants in this study were asked to perform testing in line with ELN requirements,3 the validated performance characteristics of each participant’s assay (eg, specificity, limit of detection, limit of quantification) was not requested. As such, the interlaboratory variation, false-positive, and false-negative results detected in this study could not be evaluated with this context.
The findings from this study will provide the basis for an ongoing EQA program for the 4 genetic markers that have been standardized in the ELN recommendations.3 It has established the performance of RT-qPCR-based approaches in this context and set a benchmark from which future standardization projects can look to improve. The study has highlighted several testing and interpretation errors and their associated impact on the clinical management of patients; these should be the focus of laboratory improvement projects at both the local, national, and international levels.
Acknowledgments
The authors thank participants for use of their data in this study and Jacqueline Ryan at Novartis for support in setting up this research collaboration.
This work was supported by an unrestricted research collaboration grant from Novartis Pharmaceuticals.
The supporters did not have any role in the study design or analysis of data or writing in the present work.
Authorship
Contributions: S. Scott designed the research study, analyzed the data, and wrote the manuscript; R.D. and C.T. assisted in the design of the study and reviewed the manuscript; A. Cartwright., H.J.C., S. Sadiq, and D.T. manufactured the samples and reviewed the manuscript; K.M. and N.P. tested the samples before issue and reviewed the manuscript; and A. Chantry and L.W reviewed the manuscript.
Conflict-of-interest disclosure: S. Scott has served on the advisory boards of Novartis and Amgen and has provided educational lectures for Novartis. C.T. is CEO and co-owner of AgenDix GmbH, has served on advisory boards of Novartis, JAZZ, and Astellas, and has received lecture fees from Novartis, JAZZ, Janssen, Astellas, and TEVA. The remaining authors declare no competing financial interests.
Correspondence: Stuart Scott, UK NEQAS for Leucocyte Immunophenotyping, 4th Floor, Pegasus House, 463a Glossop Rd, Sheffield S10 2QD, United Kingdom; e-mail: stuart.scott@ukneqasli.co.uk.
References
Author notes
Presented in abstract form at the 63rd American Society of Hematology Annual Meeting in December 2021.
Data are available on request from the corresponding author, Stuart Scott (stuart.scott@ukneqasli.co.uk).
The full-text version of this article contains a data supplement.